Unglycosylated Soluble SARS-CoV-2 Receptor Binding Domain (RBD) Produced in E. coli Combined with the Army Liposomal Formulation Containing QS21 (ALFQ) Elicits Neutralizing Antibodies against Mismatched Variants
Abstract
1. Introduction
2. Materials and Methods
2.1. E. coli Expression of RBD
2.2. E. coli RBD Purification and Refolding
2.3. Mammalian Expression and Purification
2.4. Endotoxin Content Quantitation
2.5. CR3022 Dot Blot
2.6. Analytical Size Exclusion Chromatography
2.7. Mouse Immunizations
2.8. Human ACE2-Fc and Monoclonal Antibody CR3022 Binding Assay
2.9. Novel Anti-RBD Monoclonal Antibody Binding Assay
2.10. ACE2 Binding Inhibition Assay
2.11. ELISA
2.12. Pseudovirus Neutralization Assay
2.13. Statistical Analysis
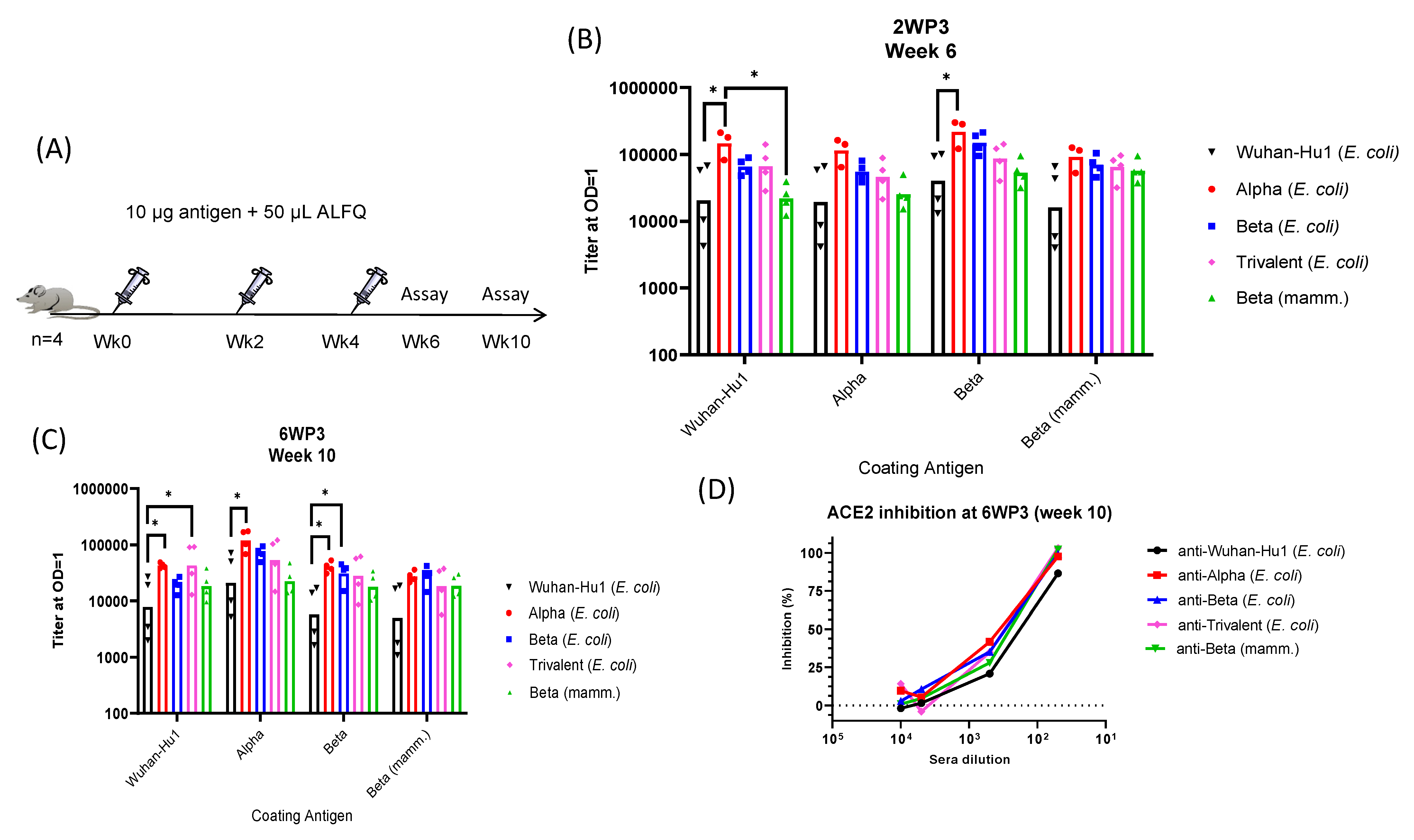
3. Results
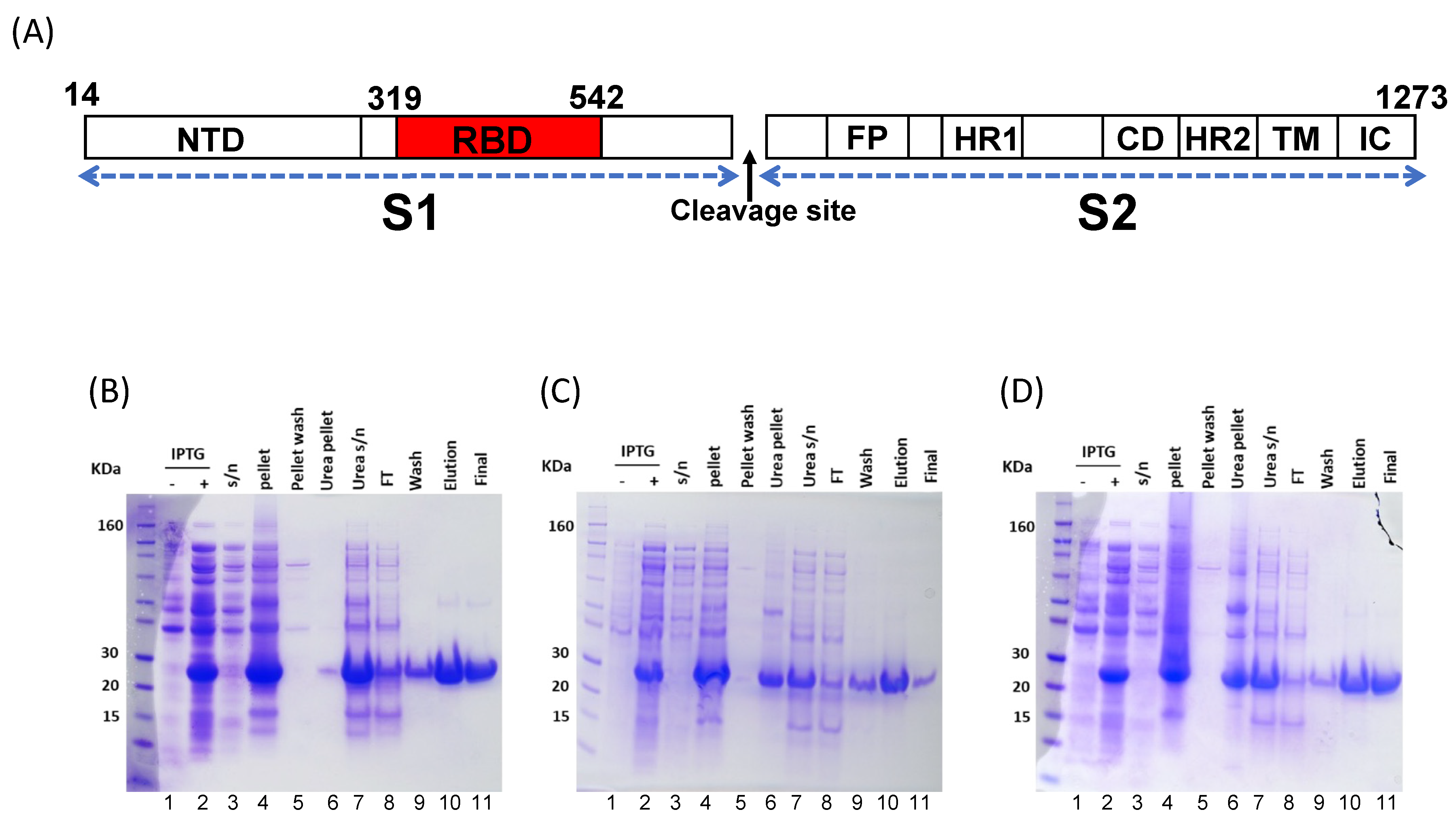
3.1. Wuhan-Hu1 and VoC RBD Expression in E. coli and Mammalian Cells
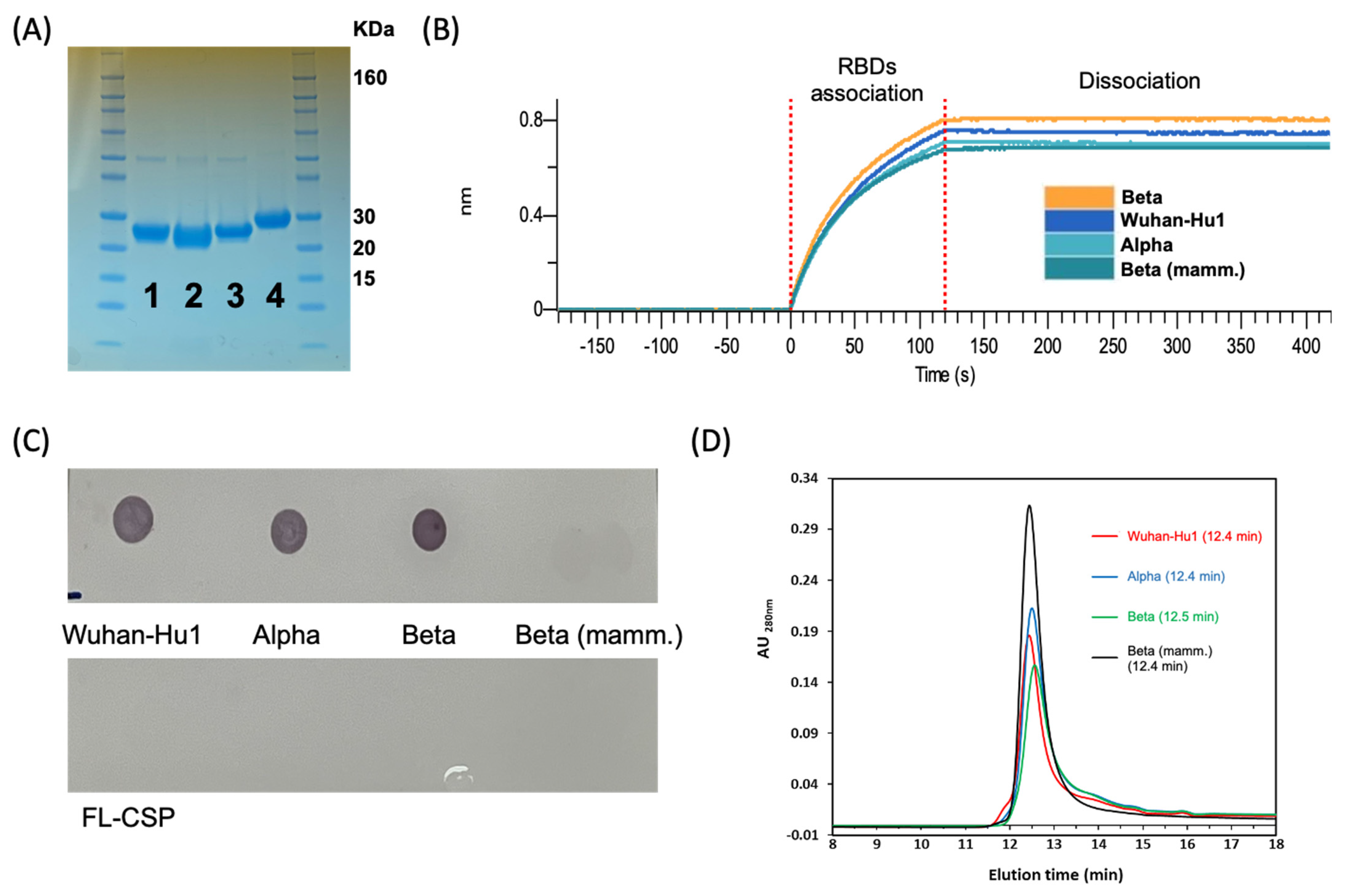
3.2. Biochemical Characterization of RBD Variants Expressed in E. coli
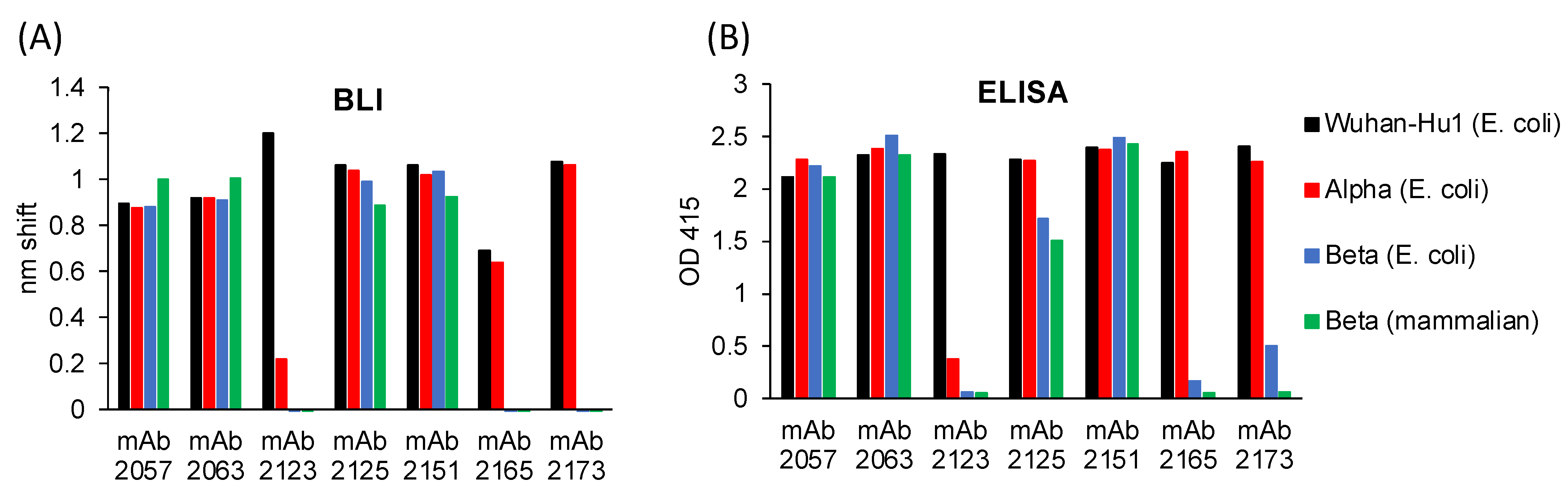
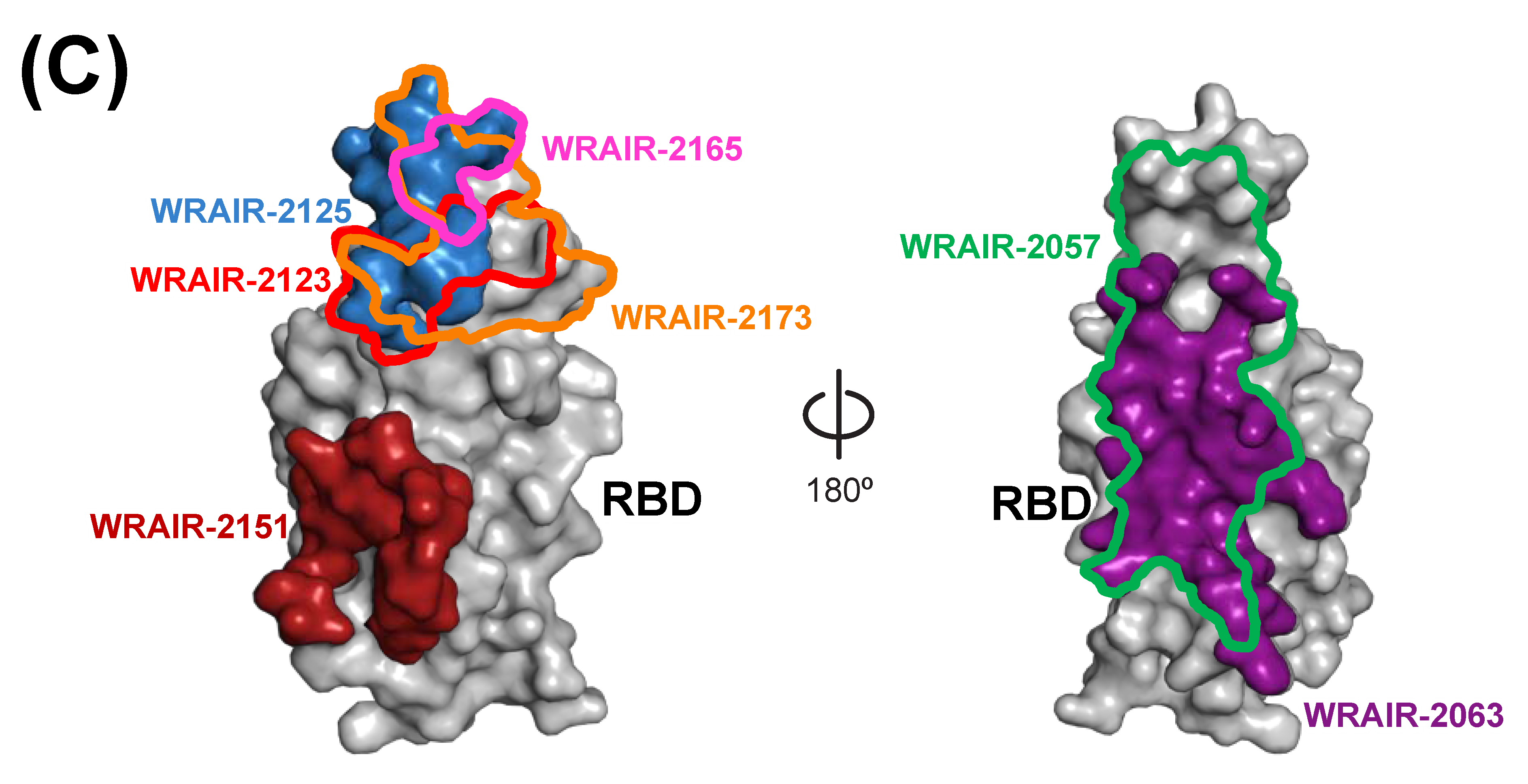
3.3. Antigenicity of the RBD Variants
3.4. Immunogenicity of RBD Variants
3.5. Pseudovirus Neutralization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Skowronski, D.M.; Astell, C.; Brunham, R.C.; Low, D.E.; Petric, M.; Roper, R.L.; Talbot, P.J.; Tam, T.; Babiuk, L. Severe Acute Respiratory Syndrome (SARS): A Year in Review. Annu. Rev. Med. 2005, 56, 357–381. [Google Scholar] [CrossRef] [PubMed]
- Hemida, M.G.; A Perera, R.; Wang, P.; A Alhammadi, M.; Siu, L.Y.; Li, M.; Poon, L.L.; Saif, L.; Alnaeem, A.; Peiris, M. Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Eurosurveillance 2013, 18, 20659. [Google Scholar] [CrossRef] [PubMed]
- Dhama, K.; Patel, S.K.; Sharun, K.; Pathak, M.; Tiwari, R.; Yatoo, M.I.; Malik, Y.S.; Sah, R.; Rabaan, A.A.; Panwar, P.K.; et al. SARS-CoV-2 jumping the species barrier: Zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Travel Med. Infect. Dis. 2020, 37, 101830. [Google Scholar] [CrossRef] [PubMed]
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534. [Google Scholar] [CrossRef]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2 Virus. Cell 2020, 183, 730–738.e13. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, C.; Xu, X.-F.; Xu, W.; Liu, S.-W. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef]
- Hwang, Y.C.; Lu, R.M.; Su, S.C.; Chiang, P.Y.; Ko, S.H.; Ke, F.Y.; Liang, K.H.; Hsieh, T.Y.; Wu, H.C. Monoclonal antibodies for COVID-19 therapy and SARS-CoV-2 detection. J. Biomed. Sci. 2022, 29, 1. [Google Scholar] [CrossRef]
- Dussupt, V.; Sankhala, R.S.; Mendez-Rivera, L.; Townsley, S.M.; Schmidt, F.; Wieczorek, L.; Lal, K.G.; Donofrio, G.C.; Tran, U.; Jackson, N.D.; et al. Low-dose in vivo protection and neutralization across SARS-CoV-2 variants by monoclonal antibody combinations. Nat. Immunol. 2021, 22, 1503–1514. [Google Scholar] [CrossRef]
- Westendorf, K.; Žentelis, S.; Wang, L.; Foster, D.; Vaillancourt, P.; Wiggin, M.; Lovett, E.; van der Lee, R.; Hendle, J.; Pustilnik, A.; et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022, 39, 110812. [Google Scholar] [CrossRef]
- Kyriakidis, N.C.; López-Cortés, A.; González, E.V.; Grimaldos, A.B.; Prado, E.O. SARS-CoV-2 vaccines strategies: A comprehensive review of phase 3 candidates. npj Vaccines 2021, 6, 28. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhang, J.; Xiao, T.; Peng, H.; Sterling, S.M.; Walsh, R.M.; Rawson, S.; Rits-Volloch, S.; Chen, B. Distinct conformational states of SARS-CoV-2 spike protein. Science 2020, 369, 1586–1592. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Corey, L.; Beyrer, C.; Cohen, M.S.; Michael, N.L.; Bedford, T.; Rolland, M. SARS-CoV-2 Variants in Patients with Immunosuppression. New Engl. J. Med. 2021, 385, 562–566. [Google Scholar] [CrossRef]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Consortium, C.-G.U.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Galloway, S.E.; Paul, P.; MacCannell, D.R.; Johansson, M.A.; Brooks, J.T.; MacNeil, A.; Slayton, R.B.; Tong, S.; Silk, B.J.; Armstrong, G.L.; et al. Emergence of SARS-CoV-2 B.1.1.7 Lineage—United States, December 29, 2020–January 12, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 95–99. [Google Scholar] [CrossRef]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.T.M.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S.; et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef]
- Cao, Y.; Yisimayi, A.; Jian, F.; Song, W.; Xiao, T.; Wang, L.; Du, S.; Wang, J.; Li, Q.; Chen, X.; et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 2022, 608, 593–602. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R.; et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data—From vision to reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef] [PubMed]
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.; Yeo, W.; et al. GISAID’s Role in Pandemic Response. China CDC Wkly. 2021, 3, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Xue, W.; Zheng, Q.; Song, S.; Yang, C.; Xiong, H.; Zhang, S.; Hong, M.; Zhang, Y.; Yu, H.; et al. Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants. Nat. Commun. 2021, 12, 5652. [Google Scholar] [CrossRef]
- Reincke, S.M.; Yuan, M.; Kornau, H.-C.; Corman, V.M.; van Hoof, S.; Sánchez-Sendin, E.; Ramberger, M.; Yu, W.; Hua, Y.; Tien, H.; et al. SARS-CoV-2 Beta variant infection elicits potent lineage-specific and cross-reactive antibodies. Science 2022, 375, 782–787. [Google Scholar] [CrossRef]
- Schultheis, K.; Reed, C.C.; Andrade, V.M.; Kalia, R.; Tur, J.; Schouest, B.; Elwood, D.; Maricic, I.; Doan, A.; Eblimit, Z.; et al. 579. Design and Immunogenicity of a Pan-SARS-CoV-2 Synthetic DNA Vaccine. Open Forum Infect. Dis. 2021, 8, S391–S392. [Google Scholar] [CrossRef]
- Walters, J.N.; Schouest, B.; Patel, A.; Reuschel, E.L.; Schultheis, K.; Parzych, E.; Maricic, I.; Gary, E.N.; Purwar, M.; Andrade, V.M.; et al. Prime-boost vaccination regimens with INO-4800 and INO-4802 augment and broaden immune responses against SARS-CoV-2 in nonhuman primates. Vaccine 2022, 40, 2960–2969. [Google Scholar] [CrossRef]
- Núñez-Muñoz, L.G.; Marcelino-Pérez, B.; Calderón-Pérez, M.; Pérez-Saldívar, K.; Acosta-Virgen, H.; González-Conchillos, B.; Vargas-Hernández, A.; Olivares-Martínez, R.; Ruiz-Medrano, D.; Roa-Velázquez, D.; et al. Recombinant antigens based on non-glycosylated regions from rbd sars-cov-2 as potential vaccine candidates against COVID-19. Vaccines 2021, 9, 928. Available online: https://www.mdpi.com/2076-393X/9/8/928 (accessed on 23 August 2022). [CrossRef]
- Gong, Y.; Qin, S.; Dai, L.; Tian, Z. The glycosylation in SARS-CoV-2 and its receptor ACE2. Signal Transduct. Target. Ther. 2021, 6, 396. [Google Scholar] [CrossRef]
- Gstöttner, C.; Zhang, T.; Resemann, A.; Ruben, S.; Pengelley, S.; Suckau, D.; Welsink, T.; Wuhrer, M.; Domínguez-Vega, E. Structural and Functional Characterization of SARS-CoV-2 RBD Domains Produced in Mammalian Cells. Anal. Chem. 2021, 93, 6839–6847. [Google Scholar] [CrossRef]
- Brindha, S.; Kuroda, Y. A Multi-Disulfide Receptor-Binding Domain (RBD) of the SARS-CoV-2 Spike Protein Expressed in E. coli Using a SEP-Tag Produces Antisera Interacting with the Mammalian Cell Expressed Spike (S1) Protein. Int. J. Mol. Sci. 2022, 23, 1703. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Dlugosz, L.S.; Drew, D.R.; Ge, X.; Ababacar, D.; Rovira, Y.I.; Moch, J.K.; Shi, M.; Long, C.A.; Foley, M.; et al. Overcoming Antigenic Diversity by Enhancing the Immunogenicity of Conserved Epitopes on the Malaria Vaccine Candidate Apical Membrane Antigen-1. PLoS Pathog. 2013, 9, e1003840. [Google Scholar] [CrossRef] [PubMed]
- Hutter, J.N.; Robben, P.M.; Lee, C.; Hamer, M.; Moon, J.E.; Merino, K.; Zhu, L.; Galli, H.; Quinn, X.; Brown, D.R.; et al. First-in-human assessment of safety and immunogenicity of low and high doses of Plasmodium falciparum malaria protein 013 (FMP013) administered intramuscularly with ALFQ adjuvant in healthy malaria-naïve adults. Vaccine 2022, 40, 5781–5790. [Google Scholar] [CrossRef]
- Thera, M.A.; Doumbo, O.K.; Coulibaly, D.; Laurens, M.B.; Ouattara, A.; Kone, A.K.; Guindo, A.B.; Traore, K.; Traore, I.; Kouriba, B.; et al. A Field Trial to Assess a Blood-Stage Malaria Vaccine. N. Engl. J. Med. 2011, 365, 1004–1013. [Google Scholar] [CrossRef] [PubMed]
- Alving, C.R.; Peachman, K.K.; Matyas, G.R.; Rao, M.; Beck, Z. Army Liposome Formulation (ALF) family of vaccine adjuvants. Expert Rev. Vaccines 2020, 19, 279–292. [Google Scholar] [CrossRef]
- Stael, S.; Miller, L.P.; Fernández-Fernández, D.; Van Breusegem, F. Detection of Damage-Activated Metacaspase Activity by Western Blot in Plants. Plant Proteases Plant Cell Death 2022, 2447, 127–137. [Google Scholar] [CrossRef]
- ter Meulen, J.; Brink, E.N.V.D.; Poon, L.L.M.; E Marissen, W.; Leung, C.S.W.; Cox, F.; Cheung, C.Y.; Bakker, A.Q.; A Bogaards, J.; van Deventer, E.; et al. Human Monoclonal Antibody Combination against SARS Coronavirus: Synergy and Coverage of Escape Mutants. PLoS Med. 2006, 3, e237. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Wu, N.C.; Zhu, X.; Lee, C.-C.D.; So, R.T.Y.; Lv, H.; Mok, C.K.P.; Wilson, I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science 2020, 368, 630–633. [Google Scholar] [CrossRef]
- Beck, Z.; Matyas, G.R.; Jalah, R.; Rao, M.; Polonis, V.R.; Alving, C.R. Differential immune responses to HIV-1 envelope protein induced by liposomal adjuvant formulations containing monophosphoryl lipid A with or without QS21. Vaccine 2015, 33, 5578–5587. [Google Scholar] [CrossRef]
- King, H.A.D.; Joyce, M.G.; Lakhal-Naouar, I.; Ahmed, A.; Cincotta, C.M.; Subra, C.; Peachman, K.K.; Hack, H.R.; Chen, R.E.; Thomas, P.V.; et al. Efficacy and breadth of adjuvanted SARS-CoV-2 receptor-binding domain nanoparticle vaccine in macaques. Proc. Natl. Acad. Sci. USA 2021, 118, e2106433118. [Google Scholar] [CrossRef]
- Joyce, M.G.; Chen, W.-H.; Sankhala, R.S.; Hajduczki, A.; Thomas, P.V.; Choe, M.; Martinez, E.J.; Chang, W.C.; Peterson, C.E.; Morrison, E.B.; et al. SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity. Cell Rep. 2021, 37, 110143. [Google Scholar] [CrossRef] [PubMed]
- Joyce, M.G.; King, H.A.D.; Elakhal-Naouar, I.; Ahmed, A.; Peachman, K.K.; Cincotta, C.M.; Subra, C.; Chen, R.E.; Thomas, P.V.; Chen, W.-H.; et al. A SARS-CoV-2 ferritin nanoparticle vaccine elicits protective immune responses in nonhuman primates. Sci. Transl. Med. 2022, 14, eabi5735. [Google Scholar] [CrossRef] [PubMed]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Rogers, T.F.; Zhao, F.; Huang, D.; Beutler, N.; Burns, A.; He, W.-T.; Limbo, O.; Smith, C.; Song, G.; Woehl, J.; et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science 2020, 369, 956–963. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent Binding of 2019 Novel Coronavirus Spike Protein by a SARS Coronavirus-Specific Human Monoclonal Antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef]
- Du, L.; Zhao, G.; Chan, C.C.; Sun, S.; Chen, M.; Liu, Z.; Guo, H.; He, Y.; Zhou, Y.; Zheng, B.-J.; et al. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology 2009, 393, 144–150. [Google Scholar] [CrossRef]
- Zhao, J.-C.; Zhao, Z.-D.; Wang, W.; Gao, X.-M. Prokaryotic expression, refolding, and purification of fragment 450–650 of the spike protein of SARS-coronavirus. Protein Expr. Purif. 2005, 39, 169–174. [Google Scholar] [CrossRef]
- Fitzgerald, G.A.; Komarov, A.; Kaznadzey, A.; Mazo, I.; Kireeva, M.L. Expression of SARS-CoV-2 surface glycoprotein fragment 319–640 in E. coli, and its refolding and purification. Protein Expr. Purif. 2021, 183, 105861. [Google Scholar] [CrossRef]
- Márquez-Ipiña, A.; González-González, E.; Rodríguez-Sánchez, I.; Lara-Mayorga, I.; Mejía-Manzano, L.; Sánchez-Salazar, M.; González-Valdez, J.; Ortiz-López, R.; Rojas-Martínez, A.; Santiago, G.T.-D.; et al. Serological Test to Determine Exposure to SARS-CoV-2: ELISA Based on the Receptor-Binding Domain of the Spike Protein (S-RBDN318-V510) Expressed in Escherichia coli. Diagnostics 2021, 11, 271. [Google Scholar] [CrossRef]
- Ke, Q.; Sun, P.; Wang, T.; Mi, T.; Xu, H.; Wu, J.; Liu, B. Non-glycosylated SARS-CoV-2 RBD elicited a robust neutralizing antibody response in mice. J. Immunol. Methods 2022, 506, 113279. [Google Scholar] [CrossRef] [PubMed]
- Bellone, M.L.; Puglisi, A.; Piaz, F.D.; Hochkoeppler, A. Production in Escherichia coli of recombinant COVID-19 spike protein fragments fused to CRM197. Biochem. Biophys. Res. Commun. 2021, 558, 79–85. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Qi, J.; Xiao, L.; Shen, L.; Yu, W.; Hu, T. Purification and characterization of the receptor-binding domain of SARS-CoV-2 spike protein from Escherichia coli. Eng. Life Sci. 2021, 21, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Ghaemi, A.; Asl, P.R.; Zargaran, H.; Ahmadi, D.; Hashimi, A.A.; Abdolalipour, E.; Bathaeian, S.; Miri, S.M. Recombinant COVID-19 vaccine based on recombinant RBD/Nucleoprotein and saponin adjuvant induces long-lasting neutralizing antibodies and cellular immunity. Front. Immunol. 2022, 13, 974364. [Google Scholar] [CrossRef] [PubMed]
- Wang, P. Natural and Synthetic Saponins as Vaccine Adjuvants. Vaccines 2021, 9, 222. [Google Scholar] [CrossRef]
- Huang, B.; Wang, W.; Li, R.; Wang, X.; Jiang, T.; Qi, X.; Gao, Y.; Tan, W.; Ruan, L. Influenza A virus nucleoprotein derived from Escherichia coli or recombinant vaccinia (Tiantan) virus elicits robust cross-protection in mice. Virol. J. 2012, 9, 322. [Google Scholar] [CrossRef]
- Aguilar-Yáñez, J.M.; Portillo-Lara, R.; I Mendoza-Ochoa, G.; García-Echauri, S.A.; López-Pacheco, F.; Bulnes-Abundis, D.; Salgado-Gallegos, J.; Lara-Mayorga, I.M.; Webb-Vargas, Y.; León-Angel, F.O.; et al. An Influenza A/H1N1/2009 Hemagglutinin Vaccine Produced in Escherichia coli. PLoS ONE 2010, 5, e11694. [Google Scholar] [CrossRef]
- Babu, J.P.; Pattnaik, P.; Gupta, N.; Shrivastava, A.; Khan, M.; Rao, P.L. Immunogenicity of a recombinant envelope domain III protein of dengue virus type-4 with various adjuvants in mice. Vaccine 2008, 26, 4655–4663. [Google Scholar] [CrossRef]
- Tan, L.C.M.; Chua, A.J.S.; Goh, L.S.L.; Pua, S.M.; Cheong, Y.K.; Ng, M.L. Rapid purification of recombinant dengue and West Nile virus envelope Domain III proteins by metal affinity membrane chromatography. Protein Expr. Purif. 2010, 74, 129–137. [Google Scholar] [CrossRef]
- Tripathi, N.; Shrivastava, A. Recent Developments in Recombinant Protein–Based Dengue Vaccines. Front. Immunol. 2018, 9, 1919. [Google Scholar] [CrossRef]
- Khoury, D.S.; Cromer, D.; Reynaldi, A.; Schlub, T.E.; Wheatley, A.K.; Juno, J.A.; Subbarao, K.; Kent, S.J.; Triccas, J.A.; Davenport, M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021, 27, 1205–1211. [Google Scholar] [CrossRef] [PubMed]
- Cromer, D.; Steain, M.; Reynaldi, A.; E Schlub, T.; Wheatley, A.K.; A Juno, J.; Kent, S.J.; A Triccas, J.; Khoury, D.S.; Davenport, M.P. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: A meta-analysis. Lancet Microbe 2021, 3, e52–e61. [Google Scholar] [CrossRef] [PubMed]
- DiPiazza, A.T.; Leist, S.R.; Abiona, O.M.; Moliva, J.I.; Werner, A.; Minai, M.; Nagata, B.M.; Bock, K.W.; Phung, E.; Schäfer, A.; et al. COVID-19 vaccine mRNA-1273 elicits a protective immune profile in mice that is not associated with vaccine-enhanced disease upon SARS-CoV-2 challenge. Immunity 2021, 54, 1869–1882.e6. [Google Scholar] [CrossRef]
- Spencer, A.J.; McKay, P.F.; Belij-Rammerstorfer, S.; Ulaszewska, M.; Bissett, C.D.; Hu, K.; Samnuan, K.; Blakney, A.K.; Wright, D.; Sharpe, H.R.; et al. Heterologous vaccination regimens with self-amplifying RNA and adenoviral COVID vaccines induce robust immune responses in mice. Nat. Commun. 2021, 12, 2893. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Z.; Cui, T.; Huang, M.; Liu, S.; Su, X.; Li, G.; Song, T.; Li, W.; Zhong, N.; et al. Heterologous boosting with third dose of coronavirus disease recombinant subunit vaccine increases neutralizing antibodies and T cell immunity against different severe acute respiratory syndrome coronavirus 2 variants. Emerg. Microbes Infect. 2022, 11, 829–840. [Google Scholar] [CrossRef] [PubMed]
- Groß, R.; Zanoni, M.; Seidel, A.; Conzelmann, C.; Gilg, A.; Krnavek, D.; Erdemci-Evin, S.; Mayer, B.; Hoffmann, M.; Pöhlmann, S.; et al. Heterologous ChAdOx1 nCoV-19 and BNT162b2 prime-boost vaccination elicits potent neutralizing antibody responses and T cell reactivity against prevalent SARS-CoV-2 variants. eBioMedicine 2021, 75, 103761. [Google Scholar] [CrossRef]
- Sridhar, S.; Chicz, R.M.; Warren, W.; Tartaglia, J.; Savarino, S.; Gurunathan, S.; Toussaint, J.-F. The potential of Beta variant containing COVID booster vaccines for chasing Omicron in 2022. Nat. Commun. 2022, 13, 5794. [Google Scholar] [CrossRef]
- Javadi, M.M.; Hosseinzadeh, M.T.; Soleimani, N.; Rommasi, F. Evaluating the immunogenicity of gold nanoparticles conjugated RBD with Freund’s adjuvant as a potential vaccine against SARS-CoV-2. Microb. Pathog. 2022, 170, 105687. [Google Scholar] [CrossRef]
- Thuluva, S.; Paradkar, V.; Gunneri, S.R.; Yerroju, V.; Mogulla, R.; Turaga, K.; Kyasani, M.; Manoharan, S.K.; Medigeshi, G.; Singh, J.; et al. Evaluation of safety and immunogenicity of receptor-binding domain-based COVID-19 vaccine (Corbevax) to select the optimum formulation in open-label, multicentre, and randomised phase-1/2 and phase-2 clinical trials. eBioMedicine 2022, 83, 104217. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balasubramaniyam, A.; Ryan, E.; Brown, D.; Hamza, T.; Harrison, W.; Gan, M.; Sankhala, R.S.; Chen, W.-H.; Martinez, E.J.; Jensen, J.L.; et al. Unglycosylated Soluble SARS-CoV-2 Receptor Binding Domain (RBD) Produced in E. coli Combined with the Army Liposomal Formulation Containing QS21 (ALFQ) Elicits Neutralizing Antibodies against Mismatched Variants. Vaccines 2023, 11, 42. https://doi.org/10.3390/vaccines11010042
Balasubramaniyam A, Ryan E, Brown D, Hamza T, Harrison W, Gan M, Sankhala RS, Chen W-H, Martinez EJ, Jensen JL, et al. Unglycosylated Soluble SARS-CoV-2 Receptor Binding Domain (RBD) Produced in E. coli Combined with the Army Liposomal Formulation Containing QS21 (ALFQ) Elicits Neutralizing Antibodies against Mismatched Variants. Vaccines. 2023; 11(1):42. https://doi.org/10.3390/vaccines11010042
Chicago/Turabian StyleBalasubramaniyam, Arasu, Emma Ryan, Dallas Brown, Therwa Hamza, William Harrison, Michael Gan, Rajeshwer S. Sankhala, Wei-Hung Chen, Elizabeth J. Martinez, Jaime L. Jensen, and et al. 2023. "Unglycosylated Soluble SARS-CoV-2 Receptor Binding Domain (RBD) Produced in E. coli Combined with the Army Liposomal Formulation Containing QS21 (ALFQ) Elicits Neutralizing Antibodies against Mismatched Variants" Vaccines 11, no. 1: 42. https://doi.org/10.3390/vaccines11010042
APA StyleBalasubramaniyam, A., Ryan, E., Brown, D., Hamza, T., Harrison, W., Gan, M., Sankhala, R. S., Chen, W.-H., Martinez, E. J., Jensen, J. L., Dussupt, V., Mendez-Rivera, L., Mayer, S., King, J., Michael, N. L., Regules, J., Krebs, S., Rao, M., Matyas, G. R., ... Dutta, S. (2023). Unglycosylated Soluble SARS-CoV-2 Receptor Binding Domain (RBD) Produced in E. coli Combined with the Army Liposomal Formulation Containing QS21 (ALFQ) Elicits Neutralizing Antibodies against Mismatched Variants. Vaccines, 11(1), 42. https://doi.org/10.3390/vaccines11010042





