FLU-v, a Broad-Spectrum Influenza Vaccine, Induces Cross-Reactive Cellular Immune Responses in Humans Measured by Dual IFN-γ and Granzyme B ELISpot Assay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Vaccination
2.2. Dual ELISpot Assay
2.3. Statistical Analyses
3. Results
3.1. ELISpot Responses to FLU-v
3.2. ELISpot Responses to a Panel of Influenza Strains
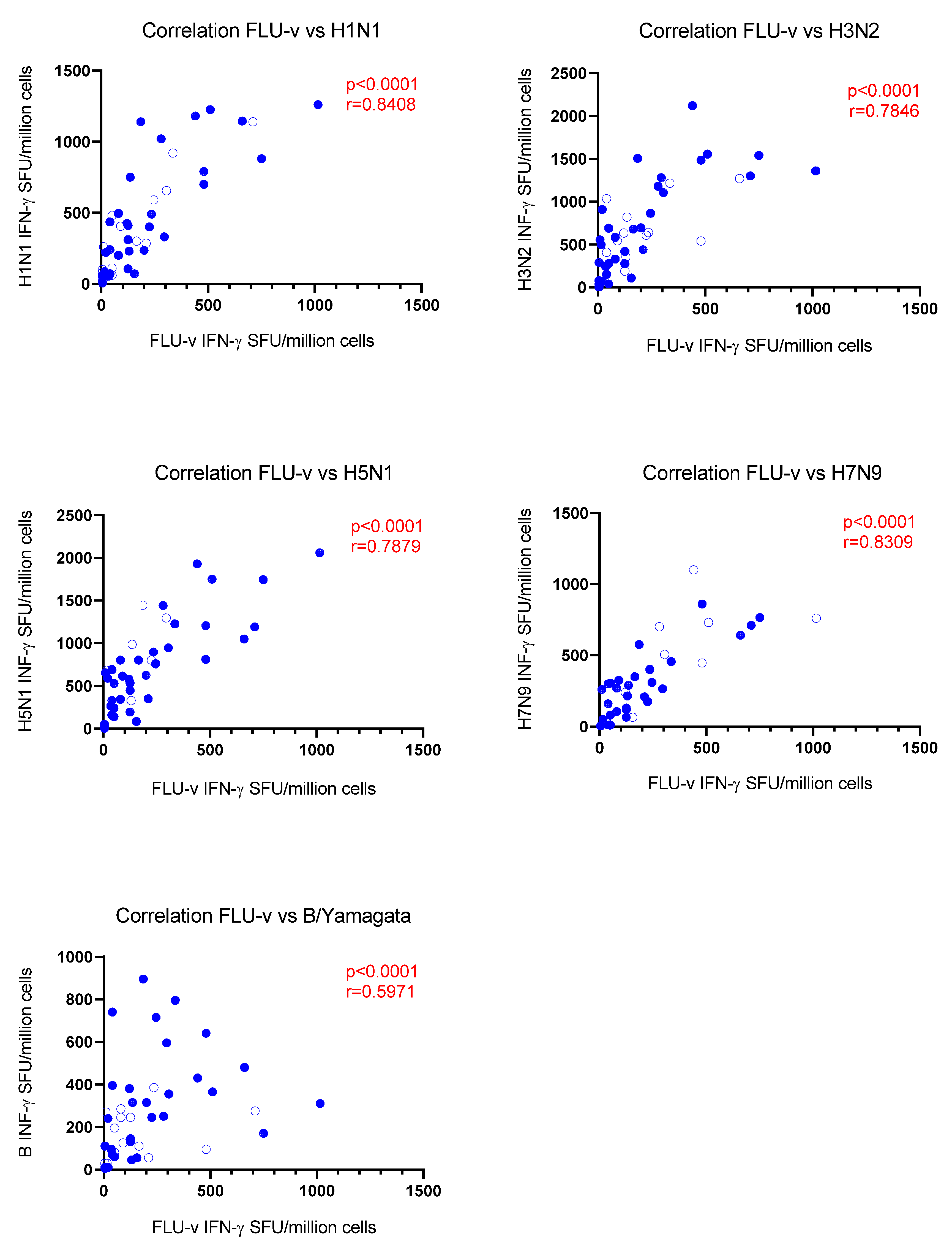
3.3. Correlation Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paules, C.I.; Sullivan, S.G.; Subbarao, K.; Fauci, A.S. Chasing Seasonal Influenza–The Need for a Universal Influenza Vaccine. N. Engl. J. Med. 2018, 378, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Elbahesh, H.; Saletti, G.; Gerlach, T.; Rimmelzwaan, G.F. Broadly protective influenza vaccines: Design and production platforms. Curr. Opin. Virol. 2019, 34, 1–9. [Google Scholar] [CrossRef] [PubMed]
- van Els, C.; Mjaaland, S.; Næss, L.; Sarkadi, J.; Gonczol, E.; Korsholm, K.S.; Hansen, J.; de Jonge, J.; Kersten, G.; Warner, J.; et al. Fast vaccine design and development based on correlates of protection (COPs). Hum. Vaccin. Immunother 2014, 10, 1935–1948. [Google Scholar] [CrossRef] [PubMed]
- Stoloff, G.A.; Caparros-Wanderley, W. Synthetic multi-epitope peptides identified in silico induce protective immunity against multiple influenza serotypes. Eur. J. Immunol. 2007, 37, 2441–2449. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos, O.; Dille, J.; de Groen, S.; Oftung, F.; Niesters, H.G.M.; Islam, M.A.; Næss, L.M.; Hungnes, O.; Aldarij, N.; Idema, D.L.; et al. Immunogenicity, Safety, and Efficacy of a Standalone Universal Influenza Vaccine, FLU-v, in Healthy Adults: A Randomized Clinical Trial. Ann. Intern. Med. 2020, 172, 453–462. [Google Scholar] [CrossRef] [PubMed]
- van Doorn, E.; Pleguezuelos, O.; Liu, H.; Fernandez, A.; Bannister, R.; Stoloff, G.; Oftung, F.; Norley, S.; Huckriede, A.; Frijlink, H.W.; et al. Evaluation of the immunogenicity and safety of different doses and formulations of a broad spectrum influenza vaccine (FLU-v) developed by SEEK: Study protocol for a single-center, randomized, double-blind and placebo-controlled clinical phase IIb trial. BMC Infect. Dis. 2017, 17, 241. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos, O.; Robinson, S.; Stoloff, G.A.; Caparrós-Wanderley, W. Synthetic Influenza vaccine (FLU-v) stimulates cell mediated immunity in a double-blind, randomised, placebo-controlled Phase I trial. Vaccine 2012, 30, 4655–4660. [Google Scholar] [CrossRef]
- Pleguezuelos, O.; Robinson, S.; Fernández, A.; Stoloff, G.A.; Mann, A.; Gilbert, A.; Balaratnam, G.; Wilkinson, T.; Lambkin-Williams, R.; Oxford, J.; et al. A Synthetic Influenza Virus Vaccine Induces a Cellular Immune Response That Correlates with Reduction in Symptomatology and Virus Shedding in a Randomized Phase Ib Live-Virus Challenge in Humans. Clin. Vaccine Immunol. 2015, 22, 828–835. [Google Scholar] [CrossRef]
- Pleguezuelos, O.; James, E.; Fernandez, A.; Lopes, V.; Rosas, L.A.; Cervantes-Medina, A.; Cleath, J.; Edwards, K.; Neitzey, D.; Gu, W.; et al. Efficacy of FLU-v, a broad-spectrum influenza vaccine, in a randomized phase IIb human influenza challenge study. NPJ Vaccines 2020, 5, 22. [Google Scholar] [CrossRef]
- Liu, H.; Frijlink, H.W.; Huckriede, A.; van Doorn, E.; Schmidt, E.; Leroy, O.; Rimmelzwaan, G.; McCullough, K.; Whelan, M.; Hak, E. Influenza Vaccine Research funded by the European Commission FP7-Health-2013-Innovation-1 project. Vaccine 2016, 34, 5845–5854. [Google Scholar] [CrossRef] [Green Version]
- Aucouturier, J.; Dupuis, L.; Deville, S.; Ascarateil, S.; Ganne, V. Montanide ISA 720 and 51: A new generation of water in oil emulsions as adjuvants for human vaccines. Expert. Rev. Vaccines 2002, 1, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Podda, A. The adjuvanted influenza vaccines with novel adjuvants: Experience with the MF59-adjuvanted vaccine. Vaccine 2001, 19, 2673–2680. [Google Scholar] [CrossRef]
- Sercarz, E.E.; Lehmann, P.V.; Ametani, A.; Benichou, G.; Miller, A.; Moudgil, K. Dominance and crypticity of T cell antigenic determinants. Annu. Rev. Immunol. 1993, 11, 729–766. [Google Scholar] [CrossRef] [PubMed]
- Yewdell, J.W. Confronting complexity: Real-world immunodominance in antiviral CD8+ T cell responses. Immunity 2006, 25, 533–543. [Google Scholar] [CrossRef]
- Cole, G.A.; Hogg, T.L.; Coppola, M.A.; Woodland, D.L. Efficient priming of CD8+ memory T cells specific for a subdominant epitope following Sendai virus infection. J. Immunol. 1997, 158, 4301–4309. [Google Scholar]
- Oukka, M.; Manuguerra, J.C.; Livaditis, N.; Tourdot, S.; Riche, N.; Vergnon, I.; Cordopatis, P.; Kosmatopoulos, K. Protection against lethal viral infection by vaccination with nonimmunodominant peptides. J. Immunol. 1996, 157, 3039–3045. [Google Scholar]
- Holst, P.J.; Jensen, B.A.; Ragonnaud, E.; Thomsen, A.R.; Christensen, J.P. Targeting of non-dominant antigens as a vaccine strategy to broaden T-cell responses during chronic viral infection. PLoS ONE 2015, 10, e0117242. [Google Scholar] [CrossRef]
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 47–60. [Google Scholar] [CrossRef]
- Sutton, T.C. The Pandemic Threat of Emerging H5 and H7 Avian Influenza Viruses. Viruses 2018, 10, 461. [Google Scholar] [CrossRef]
- Uyeki, T.M.; Katz, J.M.; Jernigan, D.B. Novel influenza A viruses and pandemic threats. Lancet 2017, 389, 2172–2174. [Google Scholar] [CrossRef]
- Greenbaum, J.A.; Kotturi, M.F.; Kim, Y.; Oseroff, C.; Vaughan, K.; Salimi, N.; Vita, R.; Ponomarenko, J.; Scheuermann, R.H.; Sette, A.; et al. Pre-existing immunity against swine-origin H1N1 influenza viruses in the general human population. Proc. Natl. Acad. Sci. USA 2009, 106, 20365–20370. [Google Scholar] [CrossRef] [PubMed]
- Roti, M.; Yang, J.; Berger, D.; Huston, L.; James, E.A.; Kwok, W.W. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J. Immunol. 2008, 180, 1758–1768. [Google Scholar] [CrossRef] [Green Version]
- van de Sandt, C.E.; Kreijtz, J.H.; de Mutsert, G.; Geelhoed-Mieras, M.M.; Hillaire, M.L.; Vogelzang-van Trierum, S.E.; Osterhaus, A.D.; Fouchier, R.A.; Rimmelzwaan, G.F. Human cytotoxic T lymphocytes directed to seasonal influenza A viruses cross-react with the newly emerging H7N9 virus. J. Virol. 2014, 88, 1684–1693. [Google Scholar] [CrossRef] [PubMed]
- Tu, W.; Mao, H.; Zheng, J.; Liu, Y.; Chiu, S.S.; Qin, G.; Chan, P.L.; Lam, K.T.; Guan, J.; Zhang, L.; et al. Cytotoxic T lymphocytes established by seasonal human influenza cross-react against 2009 pandemic H1N1 influenza virus. J. Virol. 2010, 84, 6527–6535. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.Y.; Ha do, L.A.; Simmons, C.; de Jong, M.D.; Chau, N.V.; Schumacher, R.; Peng, Y.C.; McMichael, A.J.; Farrar, J.J.; Smith, G.L.; et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Invest. 2008, 118, 3478–3490. [Google Scholar] [CrossRef]
- Reber, A.; Katz, J. Immunological assessment of influenza vaccines and immune correlates of protection. Expert Rev. Vaccines 2013, 12, 519–536. [Google Scholar] [CrossRef]
- Ward, B.J.; Pillet, S.; Charland, N.; Trepanier, S.; Couillard, J.; Landry, N. The establishment of surrogates and correlates of protection: Useful tools for the licensure of effective influenza vaccines? Hum. Vaccin. Immunother. 2018, 14, 647–656. [Google Scholar] [CrossRef]
- Beebe, E.A.; Orr, M.T. Assessment of Antigen-Specific Cellular Immunogenicity Using Intracellular Cytokine Staining, ELISpot, and Culture Supernatants. Methods Mol. Biol. 2017, 1494, 313–320. [Google Scholar] [CrossRef]
- Forrest, B.D.; Pride, M.W.; Dunning, A.J.; Capeding, M.R.; Chotpitayasunondh, T.; Tam, J.S.; Rappaport, R.; Eldridge, J.H.; Gruber, W.C. Correlation of cellular immune responses with protection against culture-confirmed influenza virus in young children. Clin. Vaccine Immunol. 2008, 15, 1042–1053. [Google Scholar] [CrossRef]
- Sridhar, S.; Begom, S.; Bermingham, A.; Hoschler, K.; Adamson, W.; Carman, W.; Bean, T.; Barclay, W.; Deeks, J.J.; Lalvani, A. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 2013, 19, 1305–1312. [Google Scholar] [CrossRef]
- Wilkinson, T.M.; Li, C.K.; Chui, C.S.; Huang, A.K.; Perkins, M.; Liebner, J.C.; Lambkin-Williams, R.; Gilbert, A.; Oxford, J.; Nicholas, B.; et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012, 18, 274–280. [Google Scholar] [CrossRef] [PubMed]
- McMichael, A.J.; Gotch, F.M.; Noble, G.R.; Beare, P.A. Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 1983, 309, 13–17. [Google Scholar] [CrossRef] [PubMed]
- McElhaney, J.E.; Gravenstein, S.; Upshaw, C.M.; Hooton, J.W.; Krause, P.; Drinka, P.; Bleackley, R.C. Granzyme B: A marker of risk for influenza in institutionalized older adults. Vaccine 2001, 19, 3744–3751. [Google Scholar] [CrossRef]
- McElhaney, J.E.; Ewen, C.; Zhou, X.; Kane, K.P.; Xie, D.; Hager, W.D.; Barry, M.B.; Kleppinger, A.; Wang, Y.; Bleackley, R.C. Granzyme B: Correlates with protection and enhanced CTL response to influenza vaccination in older adults. Vaccine 2009, 27, 2418–2425. [Google Scholar] [CrossRef]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.M.; Lee, S.; Garcia-Hernandez Mde, L.; Swain, S.L. Multifunctional CD4 cells expressing gamma interferon and perforin mediate protection against lethal influenza virus infection. J. Virol. 2012, 86, 6792–6803. [Google Scholar] [CrossRef] [Green Version]

| Peptide Name | Protein Origin | Amino Acid Sequence |
|---|---|---|
| FLU-5 acetate | M1 protein | DLEALMEWLKTRPILSPLTKGILGFVFTLTVP |
| FLU-7 acetate | NP protein from A strains | DLIFLARSALILRGSVAHKS |
| FLU-8N acetate | NP protein from B strains | PGIADIEDLTLLARSMVVVR |
| FLU-10 acetate | M2 protein | IIGILHLILWILDRLFFKCIYRLF |
| Antigen | Treatment Group | Median SFU (n) (CI) Day 0 IFN-γ Granzyme B Double Positive | Median SFU (n) (CI) Day 42 IFN-γ Granzyme B Double Positive | Median SFU (n) (CI) Day 180 IFN-γ Granzyme B Double Positive | p-Value (Wilcoxon) Day 0–42 | p-Value (Wilcoxon) Day 0–180 |
|---|---|---|---|---|---|---|
| FLU-v | Adjuvanted FLU-v | 5 (49) (5–5) | 125 (48) (50–200) | 75 (40) (40–130) | <0.0001 | <0.0001 |
| 5 (48) (5–5) | 40 (46) (10–60) | 20 (41) (5–40) | <0.0001 | 0.0047 | ||
| 5 (28) (5–5) | 20 (25) (10–40) | 5 (21) (5–20) | <0.0001 | 0.0059 | ||
| Adjuvanted Placebo | 5 (22) (5–10) | 5 (21) (5–10) | 5 (17) (5–15) | 0.83 | 0.14 | |
| 5 (20) (5–10) | 5 (19) (5–5) | 5 (16) (5–15) | 0.16 | 0.98 | ||
| 5 (12) (5–5) | 5 (12) (5–5) | 5 (7) (5–20) | NA | >0.99 | ||
| H1N1 | Adjuvanted FLU-v | 128 (48) (50–260) | 310 (47) (220–480) | 240 (39) (95–400) | <0.0001 | 0.0163 |
| 50 (47) (20–115) | 240 (46) (95–380) | 90 (40) (30–160) | <0.0001 | 0.16 | ||
| 25 (28) (5–45) | 125 (25) (30–170) | 50 (21) (20–155) | 0.0011 | 0.0025 | ||
| Adjuvanted Placebo | 265 (20) (75–410) | 153 (18) (110–360) | 160 (15) (70–390) | 0.29 | 0.90 | |
| 73 (18) (20–200) | 65 (16) (20–125) | 73 (14) (10–285) | 0.76 | 0.62 | ||
| 23 (12) (10–70) | 28 (12) (10–80) | 30 (7) (5–155) | 0.83 | 0.56 | ||
| H3N2 | Adjuvanted FLU-v | 268 (48) (140–585) | 555 (47) (355–695) | 490 (39) (230–740) | 0.0001 | 0.12 |
| 193 (48) (95–340) | 485 (45) (360–630) | 215 (40) (105–460) | 0.0002 | 0.94 | ||
| 135 (29) (10–200) | 220 (26) (155–385) | 205 (22) (60–350) | 0.0064 | 0.0449 | ||
| Adjuvanted Placebo | 510 (21) (205–800) | 420 (19) (235–575) | 460 (16) (230–865) | 0.25 | >0.99 | |
| 235 (19) (70–530) | 310 (17) (175–435) | 310 (15) (95–480) | 0.49 | 0.61 | ||
| 175 (12) (75–245) | 138 (12) (75–230) | 200 (7) (110–410) | 0.42 | 0.78 | ||
| H5N1 | Adjuvanted FLU-v | 460 (45) (175–550) | 668 (42) (530–810) | 585 (37) (355–845) | 0.0001 | 0.0381 |
| 305 (44) (160–495) | 650 (41) (450–750) | 325 (38) (115–570) | 0.0041 | 0.69 | ||
| 245 (28) (110–335) | 305 (24) (250–455) | 260 (21) (175–465) | 0.0220 | 0.18 | ||
| Adjuvanted Placebo | 493 (20) (250–890) | 450 (17) (340–700) | 470 (14) (170–1060) | 0.73 | 0.80 | |
| 490 (18) (120–800) | 433 (16) (275–580) | 360 (13) (105–650) | 0.98 | 0.89 | ||
| 135 (11) (50–830) | 230 (11) (80–725) | 263 (6) (130–410) | 0.76 | 0.56 | ||
| H7N9 | Adjuvanted FLU-v | 130 (44) (75–210) | 268 (40) (160–350) | 215 (36) (70–280) | 0.0026 | 0.27 |
| 50 (43) (15–100) | 140 (39) (55–180) | 50 (35) (15–105) | 0.49 | 0.88 | ||
| 20 (28) (10–45) | 60 (23) (20–115) | 23 (20) (5–80) | 0.06 | 0.47 | ||
| Adjuvanted Placebo | 150 (18) (40–430) | 195 (16) (80–455) | 140 (13) (30–265) | 0.75 | 0.79 | |
| 90 (18) (10–135) | 83 (16) (10–260) | 50 (12) (5–100) | 0.99 | 0.58 | ||
| 20 (11) (5–170) | 35 (11) (5–100) | 30 (6) (5–45) | 0.71 | >0.99 | ||
| B | Adjuvanted FLU-v | 130 (47) (75–200) | 243 (46) (110–310) | 135 (39) (60–230) | 0.0085 | 0.63 |
| 25 (48) (10–75) | 75 (45) (45–120) | 40 (39) (15–75) | 0.0601 | 0.27 | ||
| 10 (28) (5–20) | 30 (25) (15–50) | 20 (21) (10–50) | 0.0046 | 0.21 | ||
| Adjuvanted Placebo | 133 (20) (40–270) | 165 (18) (70–375) | 100 (15) (40–265) | 0.96 | 0.79 | |
| 45 (19) (5–80) | 45 (16) (15–110) | 25 (14) (5–45) | 0.99 | 0.49 | ||
| 10 (12) (5–35) | 15 (12) (5–40) | 10 (7) (5–50) | 0.64 | 0.72 |
| Antigen | Median Fold Increase (n) (CI) Day 42 | Median Fold Increase (n) (CI) Day 180 | ||||
|---|---|---|---|---|---|---|
| Adjuvanted FLU-v IFN-γ Granzyme B Double Positive | Adjuvanted Placebo IFN-γ Granzyme B Double Positive | p-Value (MW) | Adjuvanted FLU-v IFN-γ Granzyme B Double Positive | Adjuvanted Placebo IFN-γ Granzyme B Double Positive | p-Value (MW) | |
| FLU-v | 16.3 (48) (9.0–25.0) | 1.0 (21) (1.0–1.0) | <0.0001 | 9.5 (40) (4.0–19.0) | 1.0 (17) (1.0–2.0) | <0.0001 |
| 3.5 (46) (2.0–9.0) | 1.0 (19) (0.5–1.0) | <0.0001 | 2.0 (41) (1.0–5.0) | 1.0 (16) (0.4–3.0) | 0.0461 | |
| 3.0 (25) (1.0–8.0) | 1.0 (12) (1.0–1.0) | 0.0012 | 1.0 (21) (1.0–4.0) | 1.0 (7) (1.0–4.0) | 0.30 | |
| H1N1 | 2.3 (47) (1.8–3.1) | 0.8 (18) (0.5–2.0) | 0.0083 | 1.9 (39) (1.4–3.0) | 0.8 (15) (0.4–3.3) | 0.23 |
| 3.5 (46) (2.1–4.4) | 1.0 (16) (0.5–3.0) | 0.0075 | 1.2 (40) (0.8–2.6) | 0.9 (14) (0.1–5.5) | 0.52 | |
| 2.9 (25) (2.0–5.0) | 1.0 (12) (0.6–1.3) | 0.0219 | 1.8 (21) (1.0–6.6) | 0.5 (7) (0.1–7.8) | 0.11 | |
| H3N2 | 1.7 (47) (1.3–2.0) | 0.8 (19) (0.5–1.5) | 0.0178 | 1.3 ((39) 0.8–2.5) | 1.2 (16) (0.2–2.2) | 0.43 |
| 1.8 (45) (1.1–2.6) | 1.2 (17) (0.5–3.3) | 0.33 | 0.9 (40) (0.7–1.6) | 2.0 (15) (0.4–3.6) | 0.39 | |
| 1.7 (26) (1.0–2.6) | 0.9 (12) (0.6–1.4) | 0.0136 | 1.3 (22) (0.8–4.0) | 1.5 (7) (0.2–4.0) | 0.56 | |
| H5N1 | 1.7 (42) (1.2–2.4) | 1.0 (17) (0.7–1.7) | 0.0441 | 1.5 (37) (1.1–1.8) | 1.2 (14) (0.4–3.0) | 0.57 |
| 1.5 (41) (1.3–2.6) | 0.9 (16) (0.7–2.6) | 0.28 | 0.9 (38) (0.5–1.6) | 1.6 (13) (0.6–3.2) | 0.31 | |
| 1.4 (24) (1.1–2.6) | 0.9 (11) (0.6–2.1) | 0.25 | 1.2 (21) (0.9–2.5) | 2.2 (6) (0.5–12.7) | 0.60 | |
| H7N9 | 2.2 (40) (1.2–3.3) | 0.9 (16) (0.5–4.3) | 0.25 | 1.2 (35) (0.9–3.0) | 1.1 (13) (0.2–3.5) | 0.42 |
| 1.7 (39) (0.6–4.0) | 0.8 (16) (0.2–11.0) | 0.62 | 1.0 (35) (0.3–3.0) | 1.1 (12) (0.1–4.0) | 0.75 | |
| 2.1 (23) (0.5–3.3) | 0.6 (11) (0.3–10.0) | 0.59 | 1.0 (20) (1.0–2.0) | 1.6 (6) (0.0–9.0) | 0.68 | |
| B | 1.5 (46) (1.0–2.5) | 1.2 (18) (0.6–2.3) | 0.51 | 1.3 (39) (0.6–2.3) | 1.1 (15) (0.3–3.5) | 0.97 |
| 2.0 (45) (1.0–3.3) | 0.9 (16) (0.3–5.0) | 0.24 | 0.8 (39) (0.4–1.6) | 1.0 (14) (0.1–6.1) | 0.88 | |
| 2.0 (25) (1.0–6.0) | 0.8 (12) (0.5–2.0) | 0.0227 | 1.2 (21) (1.0–5.0) | 1.5 (7) (0.1–5.0) | 0.61 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oftung, F.; Næss, L.M.; Laake, I.; Stoloff, G.; Pleguezuelos, O. FLU-v, a Broad-Spectrum Influenza Vaccine, Induces Cross-Reactive Cellular Immune Responses in Humans Measured by Dual IFN-γ and Granzyme B ELISpot Assay. Vaccines 2022, 10, 1528. https://doi.org/10.3390/vaccines10091528
Oftung F, Næss LM, Laake I, Stoloff G, Pleguezuelos O. FLU-v, a Broad-Spectrum Influenza Vaccine, Induces Cross-Reactive Cellular Immune Responses in Humans Measured by Dual IFN-γ and Granzyme B ELISpot Assay. Vaccines. 2022; 10(9):1528. https://doi.org/10.3390/vaccines10091528
Chicago/Turabian StyleOftung, Fredrik, Lisbeth M. Næss, Ida Laake, Gregory Stoloff, and Olga Pleguezuelos. 2022. "FLU-v, a Broad-Spectrum Influenza Vaccine, Induces Cross-Reactive Cellular Immune Responses in Humans Measured by Dual IFN-γ and Granzyme B ELISpot Assay" Vaccines 10, no. 9: 1528. https://doi.org/10.3390/vaccines10091528
APA StyleOftung, F., Næss, L. M., Laake, I., Stoloff, G., & Pleguezuelos, O. (2022). FLU-v, a Broad-Spectrum Influenza Vaccine, Induces Cross-Reactive Cellular Immune Responses in Humans Measured by Dual IFN-γ and Granzyme B ELISpot Assay. Vaccines, 10(9), 1528. https://doi.org/10.3390/vaccines10091528






