mRNA Vaccine Designing Using Chikungunya Virus E Glycoprotein through Immunoinformatics-Guided Approaches
Abstract
:1. Introduction
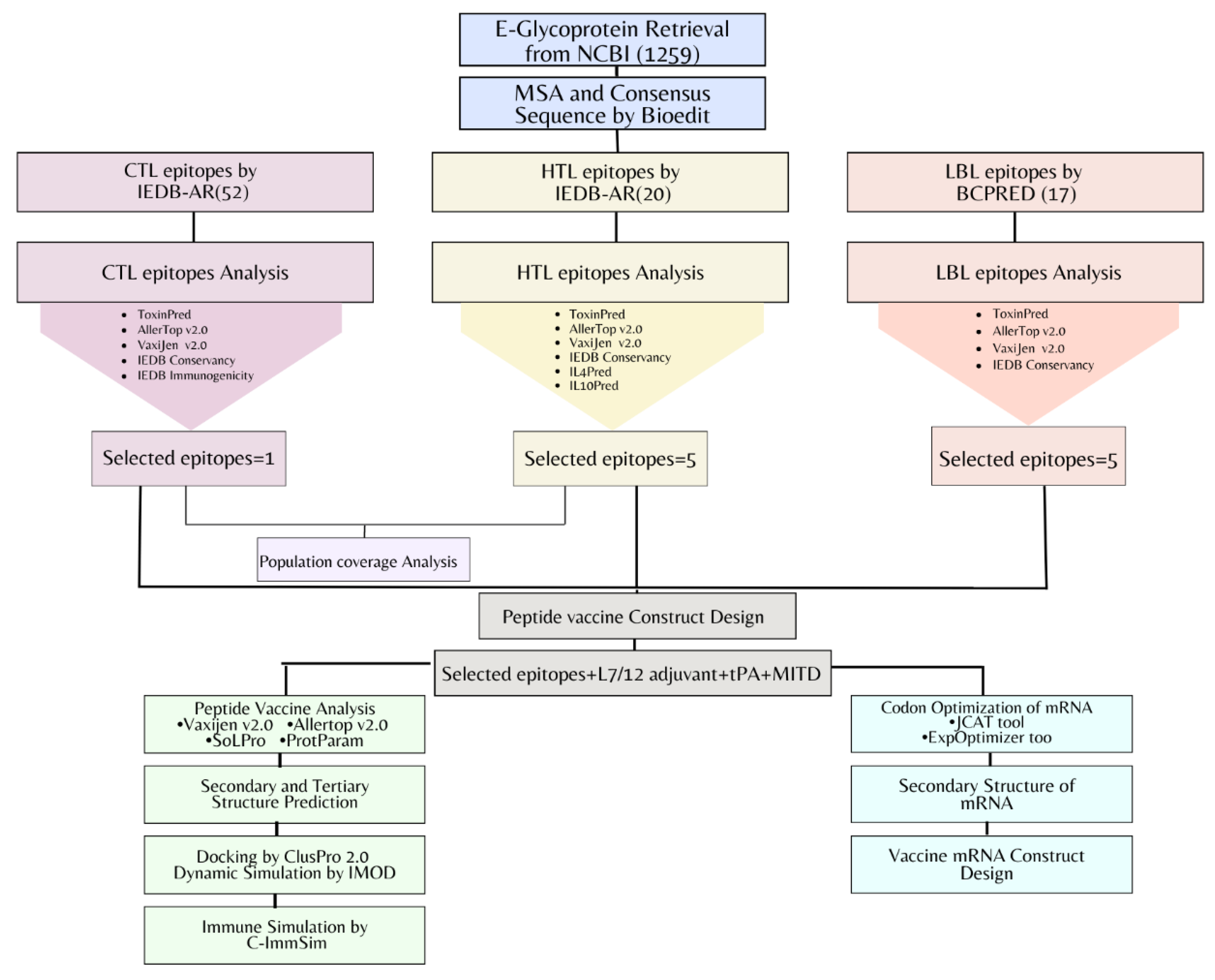
2. Materials and Methods
2.1. Retrieval of Envelope Glycoprotein Sequence
2.2. MSA and Determination of Consensus Sequence
2.3. T-Lymphocytes Epitopes Prediction
2.4. T-Lymphocytes Predicted Epitopes Analysis
2.5. Population Coverage Analysis (PCA) of T-Lymphocytes
2.6. B-Lymphocytes Epitopes Prediction and Analysis
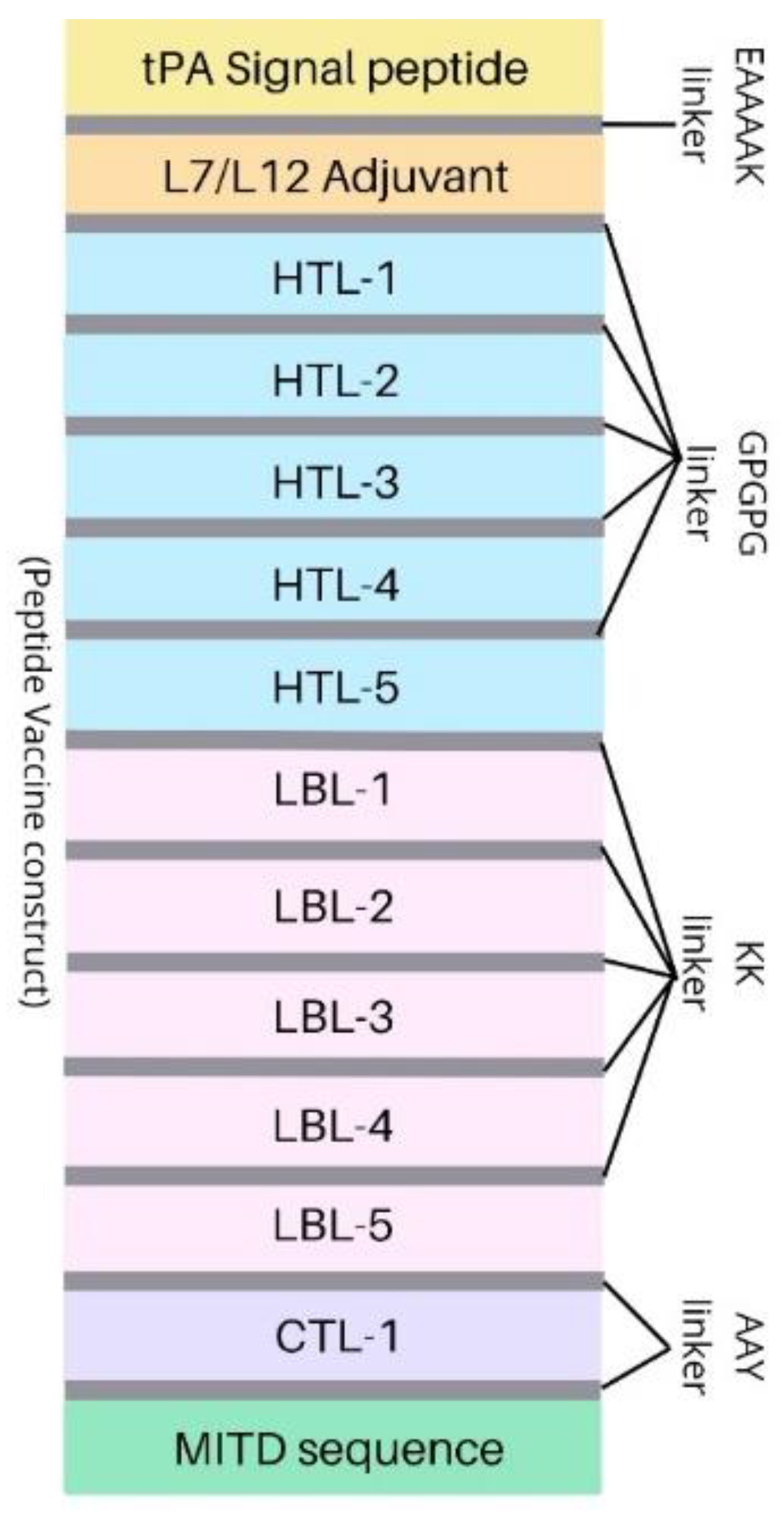
2.7. Multi-Epitope Vaccine Construct Design
2.8. Peptide Vaccine Construct Analysis
2.9. Prediction of Secondary and Tertiary Structures of the Vaccine Construct
2.10. Conformational B-Cell Epitopes Prediction
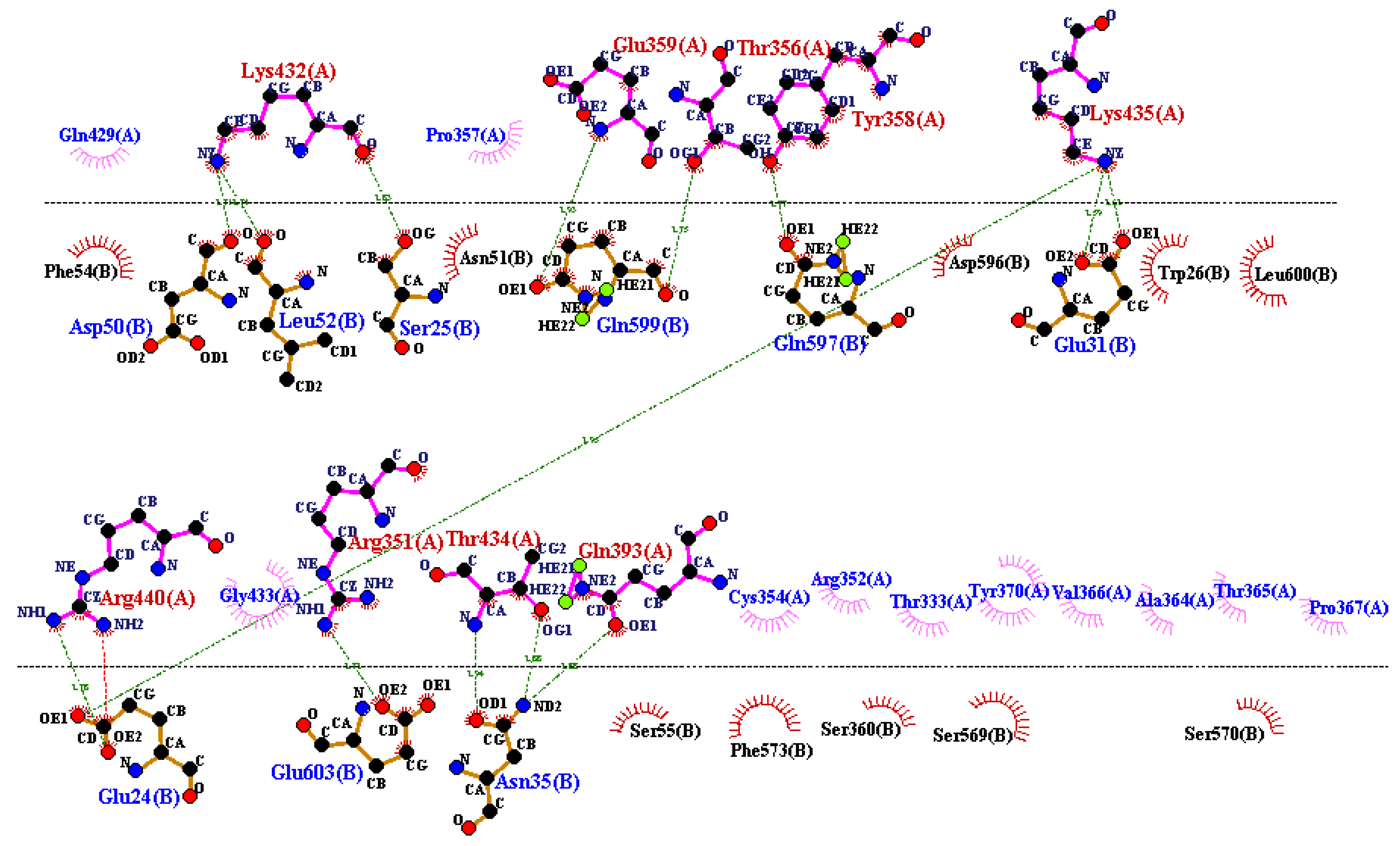
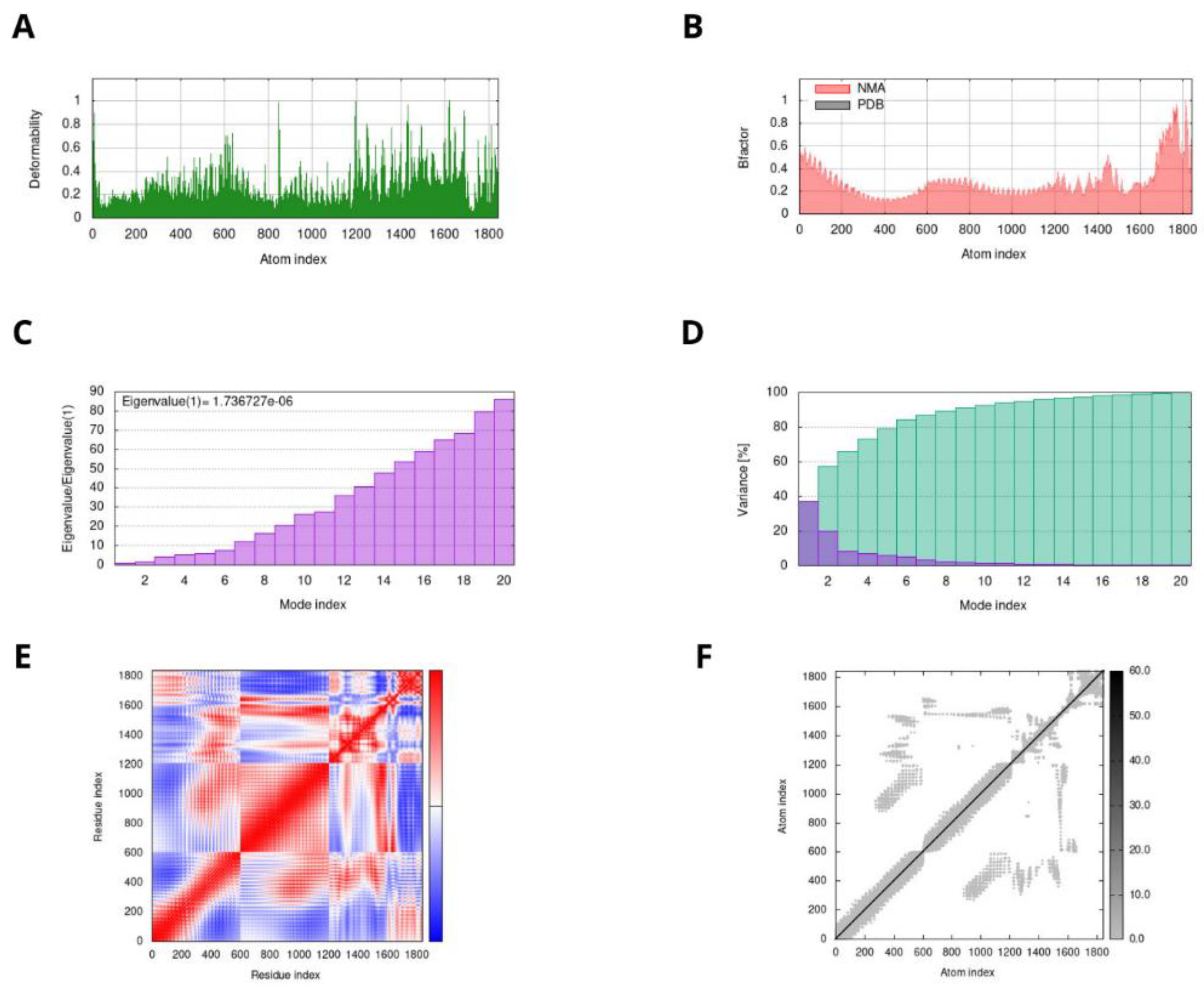
2.11. Molecular Docking and Dynamic Simulation Studies
2.12. Immune Simulation Studies
2.13. Back-Translation and Codon Optimization of Vaccine Construct
2.14. Secondary Structure Prediction of mRNA Sequence
2.15. mRNA Vaccine Construct Design
2.16. In Silico Plasmid Design for Cloning
3. Results
3.1. Determination of Consensus Sequence
3.2. T-Lymphocytes Epitopes Determination
3.3. Population Coverage Analysis (PCA) of Selected T-Lymphocyte Epitopes
3.4. B-Lymphocytes Epitopes Determination
3.5. Peptide Vaccine Construction and Analysis
3.6. Peptide Vaccine Structure Prediction and Validation
3.7. Conformational B-Cell Epitopes Prediction
3.8. Docking and MD Simulation
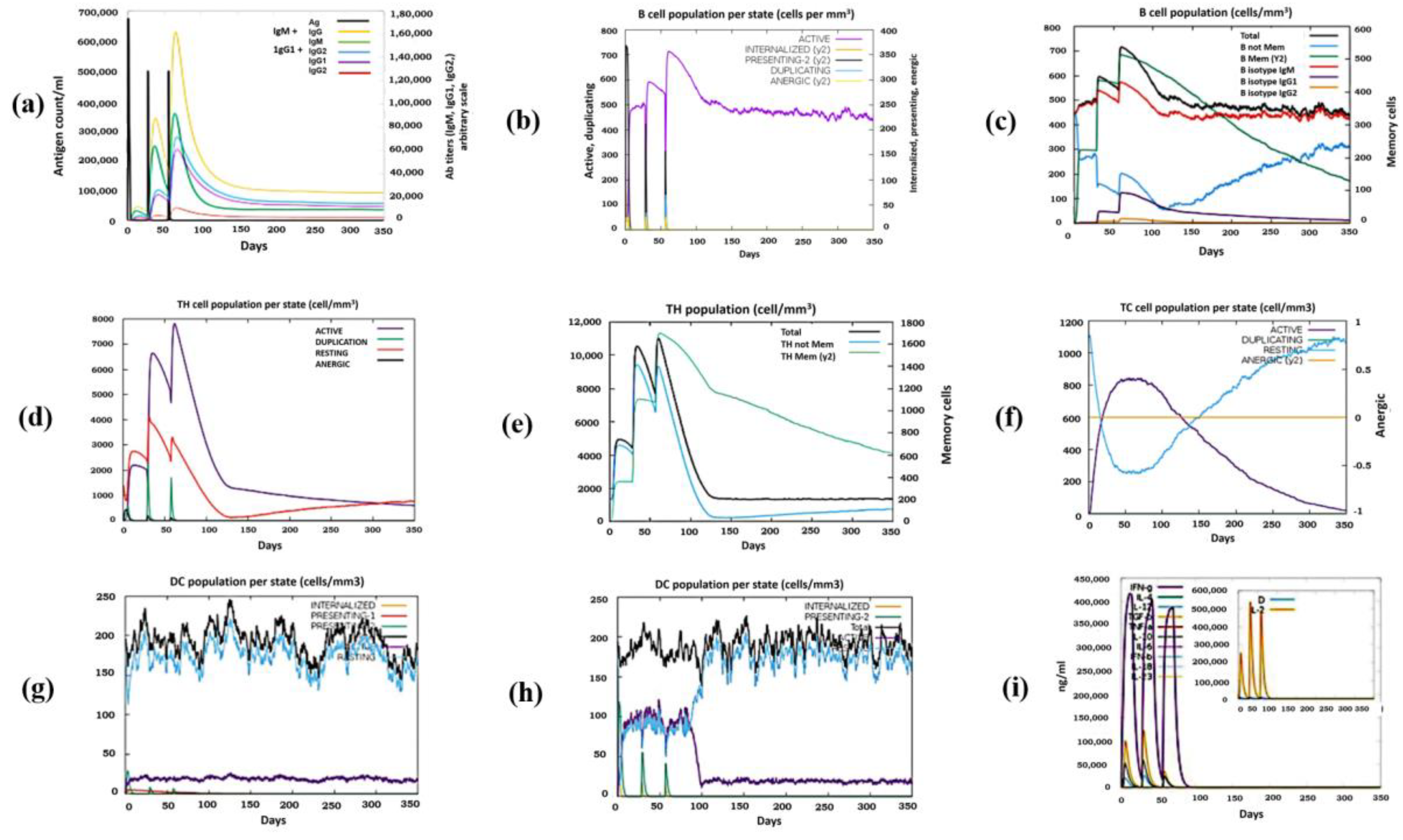
3.9. Immune Simulation
3.10. Optimized mRNA Determination
3.11. Secondary Structure Prediction of Optimized mRNA
3.12. mRNA Vaccine Construct Design
3.13. rPlasmid Design for In Vitro Cloning
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cleton, N.; Koopmans, M.; Reimerink, J.; Godeke, G.-J.; Reusken, C.J.J.O.C.V. Come fly with me: Review of clinically important arboviruses for global travelers. J. Clin. Virol. 2012, 55, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, O.; Albert, M.L.J.N.R.M. Biology and pathogenesis of chikungunya virus. Nat. Rev. Genet. 2010, 8, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Konishi, E.; Hotta, S.J.M. Studies on Structural Proteins of Chikungunya Virus: I. Separation of Three Species of Proteins and Their Preliminary Characterization. Microbiol. Immunol. 1980, 24, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Tsetsarkin, K.A.; Chen, R.; Leal, G.; Forrester, N.; Higgs, S.; Huang, J.; Weaver, S.C. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc. Natl. Acad. Sci. USA 2011, 108, 7872–7877. [Google Scholar] [CrossRef]
- Agbodzi, B.; Yousseu, F.B.S.; Simo, F.B.N.; Kumordjie, S.; Yeboah, C.; Mosore, M.-T.; Bentil, R.E.; Prieto, K.; Colston, S.M.; Attram, N. Chikungunya viruses containing the A226V mutation detected retrospectively in Cameroon form a new geographical subclade. Int. J. Infect. Dis. 2021, 113, 65–73. [Google Scholar] [CrossRef]
- Kuo, S.-C.; Chen, Y.-J.; Wang, Y.-M.; Tsui, P.-Y.; Kuo, M.-D.; Wu, T.-Y.; Lo, S.J. Cell-based analysis of Chikungunya virus E1 protein in membrane fusion. J. Biomed. Sci. 2012, 19, 44. [Google Scholar] [CrossRef]
- Gao, S.; Song, S.; Zhang, L. Recent Progress in Vaccine Development Against Chikungunya Virus. Front. Microbiol. 2019, 10, 2881. [Google Scholar] [CrossRef]
- Wang, F.; Kream, R.M.; Stefano, G.B. An Evidence Based Perspective on mRNA-SARS-CoV-2 Vaccine Development. Med. Sci. Monit. 2020, 26, e924700-1–e924700-8. [Google Scholar] [CrossRef]
- Xu, S.; Yang, K.; Li, R.; Zhang, L. mRNA Vaccine Era—Mechanisms, Drug Platform and Clinical Prospection. Int. J. Mol. Sci. 2020, 21, 6582. [Google Scholar] [CrossRef]
- Tahir ul Qamar, M.; Bari, A.; Adeel, M.M.; Maryam, A.; Ashfaq, U.A.; Du, X.; Muneer, I.; Ahmad, H.I.; Wang, J. Peptide vaccine against chikungunya virus: Immuno-informatics combined with molecular docking approach. J. Transl. Med. 2018, 16, 298. [Google Scholar] [CrossRef] [Green Version]
- Hoque, H.; Islam, R.; Ghosh, S.; Rahaman, M.M.; Jewel, N.A.; Miah, M.A. Implementation of in silico methods to predict common epitopes for vaccine development against Chikungunya and Mayaro viruses. Heliyon 2021, 7, e06396. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.; Biosciences, I.; Carlsbad, C.J.G.B.B. BioEdit: An important software for molecular biology. GERF Bull Biosci. 2011, 2, 60–61. [Google Scholar]
- Zhang, Q.; Wang, P.; Kim, Y.; Haste-Andersen, P.; Beaver, J.; Bourne, P.E.; Bui, H.-H.; Buus, S.; Frankild, S.; Greenbaum, J.J.N.A.R. Immune epitope database analysis resource (IEDB-AR). Nucleic Acids Res. 2008, 36, W513–W518. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Weissman, D. Recent advances in mRNA vaccine technology. Curr. Opin. Immunol. 2020, 65, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Al Tbeishat, H. Novel In Silico mRNA vaccine design exploiting proteins of M. tuberculosis that modulates host immune responses by inducing epigenetic modifications. Sci. Rep. 2022, 12, 4645. [Google Scholar] [CrossRef]
- Aslam, M.; Shehroz, M.; Hizbullah; Shah, M.; Khan, M.A.; Afridi, S.G.; Khan, A. Potential druggable proteins and chimeric vaccine construct prioritization against Brucella melitensis from species core genome data. Genomics 2020, 112, 1734–1745. [Google Scholar] [CrossRef]
- Dimitrov, I.; Bangov, I.; Flower, D.R.; Doytchinova, I. AllerTOP v. 2—A server for in silico prediction of allergens. J. Mol. Modeling 2014, 20, 2278. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R.J.B.B. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Misra, N.; Panda, P.K.; Shah, K.; Sukla, L.B.; Chaubey, P.J.B. Population coverage analysis of T-Cell epitopes of Neisseria meningitidis serogroup B from Iron acquisition proteins for vaccine design. Bioinformation 2011, 6, 255. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Malick, R.C.; Mondal, N.; Patra, P.; Pal, B.B.; Patra, B.C.; Das, B.K. Computational characterization of epitopic region within the outer membrane protein candidate in Flavobacterium columnare for vaccine development. J. Biomol. Struct. Dyn. 2020, 38, 450–459. [Google Scholar] [CrossRef]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Open Source Drug Discovery Consortium; Raghava, G.P.S. In silico approach for predicting toxicity of peptides and proteins. PLoS ONE 2013, 8, e73957. [Google Scholar]
- Aghajani, Z.; Rasooli, I.; Mousavi Gargari, S.L.J.A. Exploitation of two siderophore receptors, BauA and BfnH, for protection against Acinetobacter baumannii infection. Apmis 2019, 127, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.T.; Zinnia, M.A.; Islam, A. Modeling mRNA-based vaccine YFV.E1988 against yellow fever virus E-protein using immuno-informatics and reverse vaccinology approach. J. Biomol. Struct. Dyn. 2022, 1–22. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A.J.T.P.P.H. Protein identification and analysis tools on the ExPASy server. Proteom. Protoc. Handb. 2005, 571–607. [Google Scholar]
- Magnan, C.N.; Randall, A.; Baldi, P.J.B. SOLpro: Accurate sequence-based prediction of protein solubility. Bioinformatics 2009, 25, 2200–2207. [Google Scholar] [CrossRef]
- Geourjon, C.; Deleage, G.J.B. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Du, Z.; Su, H.; Wang, W.; Ye, L.; Wei, H.; Peng, Z.; Anishchenko, I.; Baker, D.; Yang, J. The trRosetta server for fast and accurate protein structure prediction. Nat. Protoc. 2021, 16, 5634–5651. [Google Scholar] [CrossRef]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef]
- Shah, M.; Jaan, S.; Fatima, B.; Javed, M.S.; Amjad, A.; Khan, A.; Afridi, S.G.; Nishan, U.; Iqbal, A.; Nawaz, H. Delineating Novel Therapeutic Drug and Vaccine Targets for Staphylococcus cornubiensis NW1T Through Computational Analysis. Int. J. Pept. Res. Ther. 2020, 27, 181–195. [Google Scholar] [CrossRef]
- Vilar, S.; Cozza, G.; Moro, S. Medicinal chemistry and the molecular operating environment (MOE): Application of QSAR and molecular docking to drug discovery. Curr. Top. Med. Chem. 2008, 8, 1555–1572. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S.J.N.P. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Tian, F.; Shang, Z. 2D depiction of nonbonding interactions for protein complexes. J. Comput. Chem. 2009, 30, 940–951. [Google Scholar] [CrossRef] [PubMed]
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef]
- Castiglione, F.; Bernaschi, M. C-immsim: Playing with the immune response. In Proceedings of the Sixteenth International Symposium on Mathematical Theory of Networks and Systems (MTNS2004), Bayreuth, Germany, 12–16 September 2022. [Google Scholar]
- Almofti, Y.A.; Abd-Elrahman, K.A.; Eltilib, E.E.M. Vaccinomic approach for novel multi epitopes vaccine against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). BMC Immunol. 2021, 22, 22. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Denman, R.B.J.B. Using RNAFOLD to predict the activity of small catalytic RNAs. BioTechniques 1993, 15, 1090–1095. [Google Scholar]
- Ahammad, I.; Lira, S.S. Designing a novel mRNA vaccine against SARS-CoV-2: An immunoinformatics approach. Int. J. Biol. Macromol. 2020, 162, 820–837. [Google Scholar] [CrossRef]
- Constant, L.E.C.; Rajsfus, B.F.; Carneiro, P.H.; Sisnande, T.; Mohana-Borges, R.; Allonso, D. Overview on Chikungunya Virus Infection: From Epidemiology to State-of-the-Art Experimental Models. Front. Microbiol. 2021, 12, 2873. [Google Scholar] [CrossRef]
- Schnee, M.; Vogel, A.B.; Voss, D.; Petsch, B.; Baumhof, P.; Kramps, T.; Stitz, L. An mRNA Vaccine Encoding Rabies Virus Glycoprotein Induces Protection against Lethal Infection in Mice and Correlates of Protection in Adult and Newborn Pigs. PLoS Negl. Trop. Dis. 2016, 10, e0004746. [Google Scholar] [CrossRef]
- Le, T.K.; Paris, C.; Khan, K.S.; Robson, F.; Ng, W.L.; Rocchi, P. Nucleic Acid-Based Technologies Targeting Coronaviruses. Trends Biochem. Sci. 2021, 46, 351–365. [Google Scholar] [CrossRef]

| Alleles | Start | End | Epitopes | Score | Rank | Conservancy | Antigenicity | Toxicity | Allergenicity |
|---|---|---|---|---|---|---|---|---|---|
| HLA-C*14:02 | 186 | 194 | YYNWHHGAV | 0.553042 | 0.15 | 100.00% | Antigen | Non-Toxin | Allergen |
| HLA-B*58:02 | 253 | 261 | ITPEGAEEW | 0.191746 | 0.06 | 88.89% | Non-Antigen | Non-Toxin | Allergen |
| HLA-C*15:02 | 1170 | 1178 | ASAEFRVQV | 0.910008 | 0.01 | 77.78% | Antigen | Non-Toxin | Non-Allergen |
| HLA-B*35:03 | 772 | 780 | IPLAALIVL | 0.819695 | 0.05 | 88.89% | Antigen | Non-Toxin | Allergen |
| HLA-C*07:01 | 719 | 727 | RRCITPYEL | 0.309353 | 0.05 | 88.89% | Antigen | Non-Toxin | Non-Allergen |
| HLA-A*24:02 | 195 | 203 | QYSGGRFTI | 0.779379 | 0.06 | 88.89% | Non-Antigen | Non-Toxin | Allergen |
| HLA-A*30:02 | 653 | 661 | VTWGNNEPY | 0.46361 | 0.2 | 77.78% | Antigen | Non-Toxin | Allergen |
| HLA-C*12:03 | 672 | 680 | TAHGHPHEI | 0.919117 | 0.01 | 77.78% | Non-Antigen | Non-Toxin | Non-Allergen |
| HLA-A*32:01 | 888 | 896 | KVFTGVYPF | 0.959162 | 0.01 | 88.89% | Non-Antigen | Non-Toxin | Allergen |
| HLA-B*35:01 | 816 | 824 | IPNTVGVPY | 0.986276 | 0.01 | 88.89% | Antigen | Non-Toxin | Allergen |
| HLA-C*15:02 | 1222 | 1230 | ITGGVGLVV | 0.442712 | 0.17 | 88.89% | Antigen | Non-Toxin | Allergen |
| HLA-B*51:01 | 430 | 438 | CPKGETLTV | 0.706451 | 0.07 | 88.89% | Antigen | Non-Toxin | Non-Allergen |
| HLA-B*08:01 | 219 | 227 | DNKGRVVAI | 0.789156 | 0.04 | 88.89% | Antigen | Non-Toxin | Allergen |
| HLA-C*14:02 | 683 | 691 | YYYELYPTM | 0.960772 | 0.01 | 88.89% | Antigen | Non-Toxin | Allergen |
| HLA-A*33:01 | 385 | 393 | DSHDWTKLR | 0.815075 | 0.03 | 88.89% | Non-Antigen | Non-Toxin | Allergen |
| HLA-C*15:02 | 1140 | 1148 | HSMTNAVTI | 0.500844 | 0.14 | 88.89% | Non-Antigen | Non-Toxin | Non-Allergen |
| HLA-C*14:02 | 612 | 620 | LYPDHPTLL | 0.969246 | 0.01 | 88.89% | Non-Antigen | Non-Toxin | Non-Allergen |
| HLA-B*08:01 | 1052 | 1060 | WLKERGASL | 0.974106 | 0.01 | 88.89% | Antigen | Non-Toxin | Non-Allergen |
| HLA-A*68:02 | 1062 | 1070 | HTAPFGCQI | 0.79801 | 0.05 | 88.89% | Antigen | Non-Toxin | Allergen |
| HLA-B*08:01 | 593 | 601 | VPKARNPTV | 0.792978 | 0.04 | 88.89% | Non-Antigen | Non-Toxin | Non-Allergen |
| HLA-C*15:02 | 845 | 853 | VTLEPTLSL | 0.887273 | 0.01 | 88.89% | Antigen | Non-Toxin | Non-Allergen |
| *HLA-C*04:01 | 159 | 167 | KYDLECAQI | 0.366935 | 0.11 | 100.00% | Antigen | Non-Toxin | Non-Allergen |
| Alleles | Start | End | Peptide | Rank | Conservancy | Antigenicity | Toxicity | Allergenicity | IL4 Inducer | IL10 Inducer |
|---|---|---|---|---|---|---|---|---|---|---|
| HLA-DPA1*01:03/DPB1*02:01 | 677 | 691 | PHEIILYYYELYPTM | 0.04 | 66.67% | Antigen | Non-Toxin | Non-Allergen | IL4 inducer | IL10 non-inducer |
| HLA-DQA1*05:01/DQB1*03:01 | 1219 | 1233 | VQKITGGVGLVVAVA | 0.85 | 86.67% | Non-Antigen | Non-Toxin | Non-Allergen | Non IL4 inducer | IL10 non-inducer |
| HLA-DRB1*09:01 | 723 | 737 | TPYELTPGATVPFLL | 0.88 | 93.33% | Antigen | Non-Toxin | Non-Allergen | Non IL4 inducer | IL10 non-inducer |
| *HLA-DRB1*13:02 | 213 | 227 | SGRPIFDNKGRVVAI | 2 | 93.33% | Antigen | Non-Toxin | Non-Allergen | IL4 inducer | IL10 non-inducer |
| HLA-DQA1*01:02/DQB1*06:02 | 230 | 244 | GGANEGARTALSVVT | 1.1 | 93.33% | Non-Antigen | Non-Toxin | Allergen | IL4 inducer | IL10 non-inducer |
| HLA-DRB3*02:02 | 405 | 419 | RAGLFVRTSAPCTIT | 1.1 | 86.67% | Antigen | Non-Toxin | Non-Allergen | IL4 inducer | IL10 non-inducer |
| HLA-DQA1*01:02/DQB1*06:02 | 1137 | 1151 | CAVHSMTNAVTIREA | 0.86 | 93.33% | Non-Antigen | Non-Toxin | Non-Allergen | Non IL4 inducer | IL10 non-inducer |
| HLA-DRB1*04:04 | 1234 | 1248 | ALILIVVLCVSFSRH | 1.1 | 93.33% | Antigen | Non-Toxin | Allergen | Non IL4 inducer | IL10 inducer |
| *HLA-DRB3*01:01 | 103 | 117 | ERMCMKIENDCIFEV | 2 | 100.00% | Antigen | Non-Toxin | Non-Allergen | IL4 inducer | IL10 inducer |
| HLA-DRB3*02:02 | 557 | 571 | HKKWQYNSPLVPRNA | 1.6 | 86.67% | Antigen | Non-Toxin | Non-Allergen | IL4 inducer | IL10 non-inducer |
| *HLA-DRB1*04:05 | 856 | 870 | ITCEYKTVIPSPYVK | 1.4 | 93.33% | Antigen | Non-Toxin | Non-Allergen | IL4 inducer | IL10 non-inducer |
| *HLA-DRB3*01:01 | 988 | 1002 | VYKGDVYNMDYPPFG | 1.8 | 93.33% | Antigen | Non-Toxin | Non-Allergen | IL4 inducer | IL10 non-inducer |
| HLA-DRB1*11:01 | 821 | 835 | GVPYKTLVNRPGYSP | 1.4 | 93.33% | Non-Antigen | Non-Toxin | Allergen | Non IL4 inducer | IL10 inducer |
| HLA-DPA1*02:01/DPB1*14:01 | 929 | 943 | SAYRAHTASASAKLR | 0.74 | 86.67% | Antigen | Non-Toxin | Non-Allergen | Non IL4 inducer | IL10 non-inducer |
| HLA-DRB3*02:02 | 599 | 613 | PTVTYGKNQVIMLLY | 1.7 | 86.67% | Antigen | Non-Toxin | Allergen | Non IL4 inducer | IL10 non-inducer |
| HLA-DRB4*01:01 | 365 | 379 | EATDGTLKIQVSLQI | 0.61 | 93.33% | Antigen | Non-Toxin | Non-Allergen | Non IL4 inducer | IL10 non-inducer |
| HLA-DRB5*01:01 | 1039 | 1053 | HVPYSQAPSGFKYWL | 0.37 | 93.33% | Non-Antigen | Non-Toxin | Non-Allergen | IL4 inducer | IL10 non-inducer |
| HLA-DRB1*07:01 | 1155 | 1169 | VEGNSQLQISFSTAL | 0.27 | 86.67% | Antigen | Non-Toxin | Non-Allergen | IL4 inducer | IL10 inducer |
| *HLA-DRB1*03:01 | 126 | 140 | YACLVGDKVMKPAHV | 1.1 | 93.33% | Antigen | Non-Toxin | Non-Allergen | IL4 inducer | IL10 inducer |
| HLA-DRB1*12:01 | 146 | 160 | NADLAKLAFKRSSKY | 1.9 | 86.67% | Antigen | Non-Toxin | Non-Allergen | IL4 inducer | IL10 non-inducer |
| Start | End | Epitopes | Score | Antigenicity | Allergenicity | Toxicity | Conservancy |
|---|---|---|---|---|---|---|---|
| 198 | 217 | GGRFTIPTGAGKPGDSGRPI | 1 | Non-Antigen | Non-Allergen | Non-Toxin | 95.00% |
| 993 | 1012 | VYNMDYPPFGAGRPGQFGDI | 1 | Antigen | Allergen | Non-Toxin | 90.00% |
| 490 | 509 | EEIEVHMPPDTPDRTLMSQQ | 0.996 | Non-Antigen | Non-Allergen | Non-Toxin | 85.00% |
| 447 | 466 | SHSCTHPFHHDPPVIGREKF | 0.983 | Antigen | Non-Allergen | Toxin | 80.00% |
| 242 | 261 | VVTWNKDIVTKITPEGAEEW | 0.98 | Non-Antigen | Allergen | Non-Toxin | 90.00% |
| 1058 | 1077 | *ASLQHTAPFGCQIATNPVRA | 0.976 | Antigen | Non-Allergen | Non-Toxin | 95.00% |
| 972 | 991 | *VGPMSSAWTPFDNKIVVYKG | 0.974 | Antigen | Non-Allergen | Non-Toxin | 95.00% |
| 177 | 196 | *KFTHEKPEGYYNWHHGAVQY | 0.968 | Antigen | Non-Allergen | Non-Toxin | 95.00% |
| 654 | 673 | TWGNNEPYKYWPQLSTNGTA | 0.958 | Non-Antigen | Non-Allergen | Non-Toxin | 90.00% |
| 1149 | 1168 | REAEIEVEGNSQLQISFSTA | 0.925 | Antigen | Non-Allergen | Non-Toxin | 85.00% |
| 856 | 875 | ITCEYKTVIPSPYVKCCGTA | 0.917 | Non-Antigen | Allergen | Toxin | 80.00% |
| 807 | 826 | VSAYEHVTVIPNTVGVPYKT | 0.912 | Antigen | Non-Allergen | Non-Toxin | 90.00% |
| 1037 | 1056 | TVHVPYSQAPSGFKYWLKER | 0.875 | Non-Antigen | Non-Allergen | Non-Toxin | 95.00% |
| 883 | 902 | PDYSCKVFTGVYPFMWGGAY | 0.819 | Antigen | Allergen | Non-Toxin | 95.00% |
| 360 | 379 | *ERIRNEATDGTLKIQVSLQI | 0.812 | Antigen | Non-Allergen | Non-Toxin | 95.00% |
| 715 | 734 | *MCARRRCITPYELTPGATVP | 0.757 | Antigen | Non-Allergen | Non-Toxin | 95.00% |
| 129 | 148 | LVGDKVMKPAHVKGTIDNAD | 0.752 | Non-Antigen | Allergen | Non-Toxin | 95.00% |
| Physiochemical Properties | Measurement | Indication |
|---|---|---|
| Total Number of Amino Acids | 631 | Appropriate |
| Molecular Weight | 70,036.77 | Appropriate |
| Theoretical pI | 8.52 | Basic |
| Total Number of Negatively Charged Residues (Asp + Glu) | 82 | - |
| Total Number of Positively Charged Residues (Arg + Lys) | 90 | - |
| Aliphatic Index (AI) | 80.55 | Thermostable |
| Grand Average of Hydropathicity (GRAVY) | –0.361 | Hydrophilic |
| Antigenicity (using Vaxijen) | 0.4711 | Antigenic |
| Solubility upon overexpression | 0.722294 | Soluble |
| Allergenicity | Non-allergen | Non-Allergenic |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jaan, S.; Zaman, A.; Ahmed, S.; Shah, M.; Ojha, S.C. mRNA Vaccine Designing Using Chikungunya Virus E Glycoprotein through Immunoinformatics-Guided Approaches. Vaccines 2022, 10, 1476. https://doi.org/10.3390/vaccines10091476
Jaan S, Zaman A, Ahmed S, Shah M, Ojha SC. mRNA Vaccine Designing Using Chikungunya Virus E Glycoprotein through Immunoinformatics-Guided Approaches. Vaccines. 2022; 10(9):1476. https://doi.org/10.3390/vaccines10091476
Chicago/Turabian StyleJaan, Samavia, Aqal Zaman, Sarfraz Ahmed, Mohibullah Shah, and Suvash Chandra Ojha. 2022. "mRNA Vaccine Designing Using Chikungunya Virus E Glycoprotein through Immunoinformatics-Guided Approaches" Vaccines 10, no. 9: 1476. https://doi.org/10.3390/vaccines10091476
APA StyleJaan, S., Zaman, A., Ahmed, S., Shah, M., & Ojha, S. C. (2022). mRNA Vaccine Designing Using Chikungunya Virus E Glycoprotein through Immunoinformatics-Guided Approaches. Vaccines, 10(9), 1476. https://doi.org/10.3390/vaccines10091476







