Membrane Chromatography-Based Downstream Processing for Cell-Culture Produced Influenza Vaccines
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture Conditions and Influenza Strains
2.2. Downstream Processes
2.2.1. Clarification
2.2.2. Ion EXCHANGE chromatography
2.3. Analysis and Quantification for Impurities and Influenza Viruses
2.3.1. Determination of dsDNA
2.3.2. Determination of Total Proteins
2.3.3. Dot Blot Assay
2.3.4. Hemagglutination Assay
3. Results
3.1. Demonstration of a Scalable Depth Filtration as an Alternative to Centrifugation for H1N1 Clarification
3.2. Evaluation of Different Membranes for Ion Exchange Chromatography
3.2.1. NatriFlo® HD Q vs. Sartobind® Q
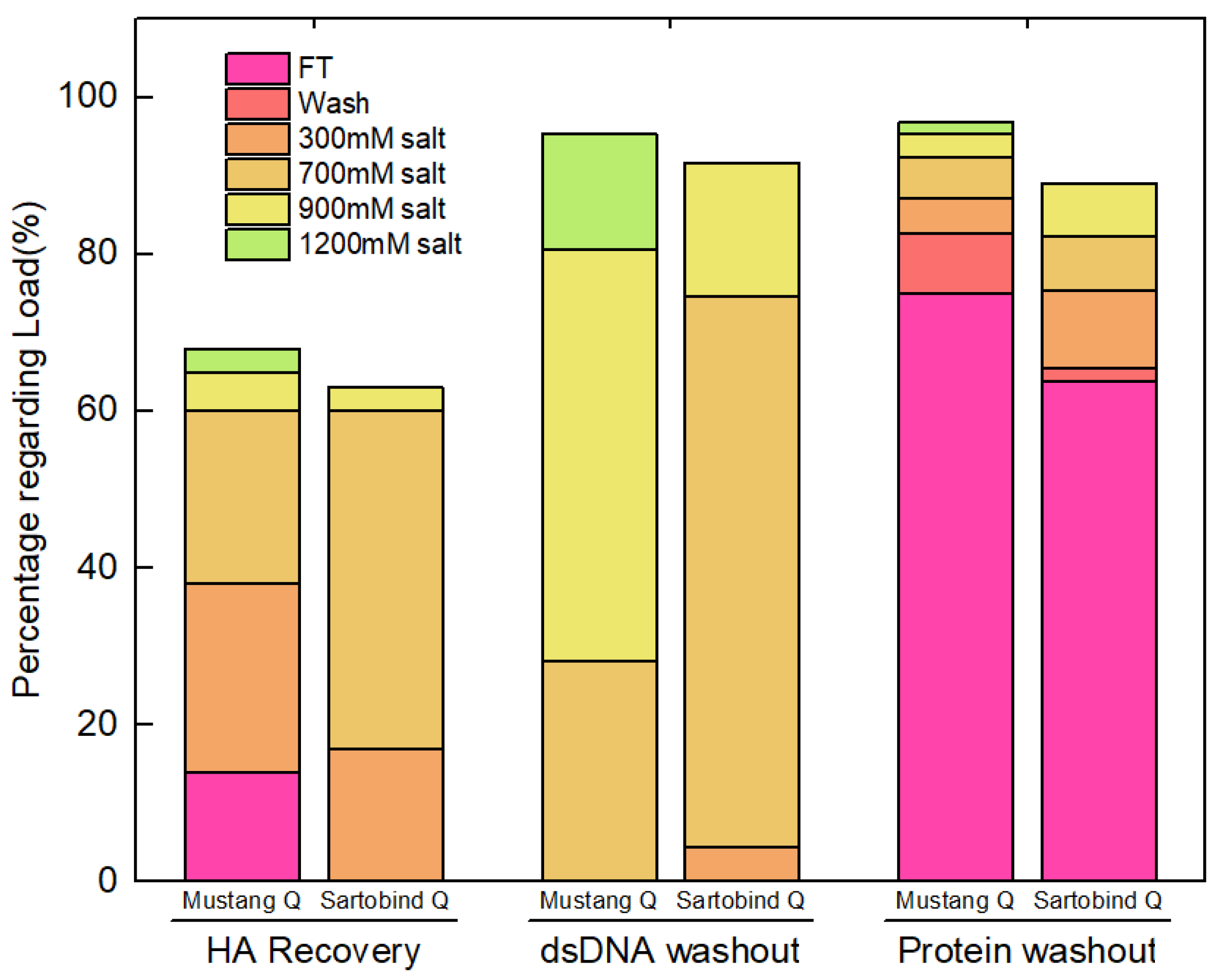
3.2.2. Sartobind® Q vs. Mustang® Q
3.3. Evaluation of Clarification and Membrane Chromatography Steps for Purification of Pandemic Influenza Strains H7N9 and H3N2
3.3.1. Clarification for Influenza H7N9 and H3N2
3.3.2. Implement of Ion Exchange Chromatography for Influenza H7N9 and H3N2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pereyaslov, D.; Zemtsova, G.; Gruessner, C.; Daniels, R.S.; McCauley, J.W.; Brown, C.S. Improving the representativeness of influenza viruses shared within the WHO Global Influenza Surveillance and Response System. Influenza Other Respir. Viruses 2016, 10, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Hong, K.J.; Kim, H.; Nam, J.H. Influenza vaccines: Past, present, and future. Rev. Med. Virol. 2022, 32, e2243. [Google Scholar] [CrossRef]
- Manceur, A.P.; Zou, W.; Marcil, A.; Paquet, E.; Gadoury, C.; Jaentschke, B.; Li, X.; Petiot, E.; Durocher, Y.; Baardsnes, J.; et al. Generation of monoclonal pan-hemagglutinin antibodies for the quantification of multiple strains of influenza. PLoS ONE 2017, 12, e0180314. [Google Scholar] [CrossRef] [PubMed]
- Sharabi, S.; Drori, Y.; Micheli, M.; Friedman, N.; Orzitzer, S.; Bassal, R.; Glatman-Freedman, A.; Shohat, T.; Mendelson, E.; Hindiyeh, M.; et al. Epidemiological and Virological Characterization of Influenza B Virus Infections. PLoS ONE 2016, 11, e0161195. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tong, S.; Zhu, X.; Li, Y.; Shi, M.; Zhang, J.; Bourgeois, M.; Yang, H.; Chen, X.; Recuenco, S.; Gomez, J.; et al. New World Bats Harbor Diverse Influenza A Viruses. PLoS Pathog. 2013, 9, e1003657. [Google Scholar] [CrossRef]
- Lamb, R.A.; Takeda, M. Death by influenza virus protein. Nat. Med. 2001, 7, 1286–1288. [Google Scholar] [CrossRef]
- Yuan, W.M.; Krug, R.M. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. Embo J. 2001, 20, 362–371. [Google Scholar] [CrossRef]
- Couch, R.B.; Keitel, W.A.; Cate, T.R.; Quarles, J.A.; Taber, L.A.; Glezen, W.P. Prevention of influenza virus infections by current inactivated influenza virus vaccines. In Proceedings of the 3rd International Conference on Options for the Control of Influenza, Cairns, Australia, 4–9 May 1996; pp. 97–106. [Google Scholar]
- Gubareva, L.V.; Kaiser, L.; Hayden, F.G. Influenza virus neuraminidase inhibitors. Lancet 2000, 355, 827–835. [Google Scholar] [CrossRef]
- Mifsud, E.J.; Hayden, F.G.; Hurt, A.C. Antivirals targeting the polymerase complex of influenza viruses. Antivir. Res. 2019, 169, 104545. [Google Scholar] [CrossRef]
- Krammer, F.; Smith, G.J.D.; Fouchier, R.A.M.; Peiris, M.; Kedzierska, K.; Doherty, P.C.; Palese, P.; Shaw, M.L.; Treanor, J.; Webster, R.G.; et al. Influenza. Nat. Rev. Dis. Primers 2018, 4, 3. [Google Scholar] [CrossRef]
- Kiso, M.; Mitamura, K.; Sakai-Tagawa, Y.; Shiraishi, K.; Kawakami, C.; Kimura, K.; Hayden, F.G.; Sugaya, N.; Kawaoka, Y. Resistant influenza A viruses in children treated with oseltamivir: Descriptive study. Lancet 2004, 364, 759–765. [Google Scholar] [CrossRef]
- Jang, Y.H.; Seong, B.L. The Quest for a Truly Universal Influenza Vaccine. Front. Cell. Infect. Microbiol. 2019, 9, 344. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, C.M.; Marchi, S.; Manini, I.; Lazzeri, G.; Montomoli, E. Challenges in the development of egg-independent vaccines for influenza. Expert Rev. Vaccines 2019, 18, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.A.T.; Kamen, A.A.; Henry, O. Recent advances and current challenges in process intensification of cell culture-based influenza virus vaccine manufacturing. Can. J. Chem. Eng. 2021, 99, 2525–2535. [Google Scholar] [CrossRef]
- Bialy, D.; Shelton, H. Functional neuraminidase inhibitor resistance motifs in avian influenza A(H5Nx) viruses. Antivir. Res. 2020, 182, 104886. [Google Scholar] [CrossRef]
- Nuñez, I.A.; Ross, T.M. A review of H5Nx avian influenza viruses. Ther. Adv. Vaccines Immunother. 2019, 7, 2515135518821625. [Google Scholar] [CrossRef]
- Sparrow, E.; Adetifa, I.; Chaiyakunapruk, N.; Cherian, T.; Fell, D.B.; Graham, B.S.; Innis, B.; Kaslow, D.C.; Karron, R.A.; Nair, H.; et al. WHO preferred product characteristics for monoclonal antibodies for passive immunization against respiratory syncytial virus (RSV) disease in infants—Key considerations for global use. Vaccine 2022, 40, 3506–3510. [Google Scholar] [CrossRef]
- Hegde, N.R. Cell culture-based influenza vaccines: A necessary and indispensable investment for the future. Hum. Vaccines Immunother. 2015, 11, 1223–1234. [Google Scholar] [CrossRef]
- Tzeng, T.T.; Lai, C.C.; Weng, T.C.; Cyue, M.H.; Tsai, S.Y.; Tseng, Y.F.; Sung, W.C.; Lee, M.S.; Hu, A.Y. The stability and immunogenicity of inactivated MDCK cell-derived influenza H7N9 viruses. Vaccine 2019, 37, 7117–7122. [Google Scholar] [CrossRef]
- Chia, M.Y.; Hu, A.Y.; Tseng, Y.F.; Weng, T.C.; Lai, C.C.; Lin, J.Y.; Chen, P.L.; Wang, Y.F.; Chao, S.R.; Chang, J.Y.; et al. Evaluation of MDCK cell-derived influenza H7N9 vaccine candidates in ferrets. PLoS ONE 2015, 10, e0120793. [Google Scholar] [CrossRef]
- Chen, P.L.; Tzeng, T.T.; Hu, A.Y.; Wang, L.H.; Lee, M.S. Development and Evaluation of Vero Cell-Derived Master Donor Viruses for Influenza Pandemic Preparedness. Vaccines 2020, 8, 626. [Google Scholar] [CrossRef] [PubMed]
- Nestola, P.; Peixoto, C.; Silva, R.R.; Alves, P.M.; Mota, J.P.; Carrondo, M.J. Improved virus purification processes for vaccines and gene therapy. Biotechnol. Bioeng. 2015, 112, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Venereo-Sanchez, A.; Gilbert, R.; Simoneau, M.; Caron, A.; Chahal, P.; Chen, W.; Ansorge, S.; Li, X.; Henry, O.; Kamen, A. Hemagglutinin and neuraminidase containing virus-like particles produced in HEK-293 suspension culture: An effective influenza vaccine candidate. Vaccine 2016, 34, 3371–3380. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.M.; Petiot, E.; Mullick, A.; Aucoin, M.G.; Henry, O.; Kamen, A.A. Critical assessment of influenza VLP production in Sf9 and HEK293 expression systems. BMC Biotechnol. 2015, 15, 31. [Google Scholar] [CrossRef]
- Tseng, Y.F.; Weng, T.C.; Lai, C.C.; Chen, P.L.; Lee, M.S.; Hu, A.Y. A fast and efficient purification platform for cell-based influenza viruses by flow-through chromatography. Vaccine 2018, 36, 3146–3152. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.S.; Agarwal, H.; Sharma, A.K.; Pathak, M.; Muthukumar, S. Continuous Processing for Production of Biopharmaceuticals. Prep. Biochem. Biotechnol. 2015, 45, 836–849. [Google Scholar] [CrossRef]
- González-Domínguez, I.; Lorenzo, E.; Bernier, A.; Cervera, L.; Gòdia, F.; Kamen, A. A Four-Step Purification Process for Gag VLPs: From Culture Supernatant to High-Purity Lyophilized Particles. Vaccines 2021, 9, 1154. [Google Scholar] [CrossRef]
- Carvalho, S.B.; Silva, R.J.S.; Moreira, A.S.; Cunha, B.; Clemente, J.J.; Alves, P.M.; Carrondo, M.J.T.; Xenopoulos, A.; Peixoto, C. Efficient filtration strategies for the clarification of influenza virus-like particles derived from insect cells. Sep. Purif. Technol. 2019, 218, 81–88. [Google Scholar] [CrossRef]
- Grein, T.A.; Michalsky, R.; Vega López, M.; Czermak, P. Purification of a recombinant baculovirus of Autographa californica M nucleopolyhedrovirus by ion exchange membrane chromatography. J. Virol. Methods 2012, 183, 117–124. [Google Scholar] [CrossRef]
- Kutner, R.H.; Puthli, S.; Marino, M.P.; Reiser, J. Simplified production and concentration of HIV-1-based lentiviral vectors using HYPERFlask vessels and anion exchange membrane chromatography. BMC Biotechnol. 2009, 9, 10. [Google Scholar] [CrossRef]
- Davidoff, A.M.; Ng, C.Y.C.; Sleep, S.; Gray, J.; Azam, S.; Zhao, Y.; McIntosh, J.H.; Karimipoor, M.; Nathwani, A.C. Purification of recombinant adeno-associated virus type 8 vectors by ion exchange chromatography generates clinical grade vector stock. J. Virol. Methods 2004, 121, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Sarsenbayeva, G.; Volgin, Y.; Kassenov, M.; Issagulov, T.; Bogdanov, N.; Nurpeisova, A.; Sagymbay, A.; Abitay, R.; Stukova, M.; Sansyzbay, A.; et al. A novel preservative-free seasonal influenza vaccine safety and immune response study in the frame of preclinical research. J. Med. Virol. 2017, 89, 1168–1173. [Google Scholar] [CrossRef] [PubMed]
- Yang, H. Establishing acceptable limits of residual DNA. PDA J. Pharm. Sci. Technol. 2013, 67, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Kapteyn, J.C.; Porre, A.M.; de Rond, E.J.P.; Hessels, W.B.; Tijms, M.A.; Kessen, H.; Slotboom, A.M.E.; Oerlemans, M.A.; Smit, D.; van der Linden, J.; et al. HPLC-based quantification of haemagglutinin in the production of egg- and MDCK cell-derived influenza virus seasonal and pandemic vaccines. Vaccine 2009, 27, 1468–1477. [Google Scholar] [CrossRef] [PubMed]
- Wolf, M.W.; Reichl, U. Downstream processing of cell culture-derived virus particles. Expert Rev. Vaccines 2011, 10, 1451–1475. [Google Scholar] [CrossRef]
- Kalbfuss, B.; Wolff, M.; Geisler, L.; Tappe, A.; Wickramasinghe, R.; Thom, V.; Reichl, U. Direct capture of influenza A virus from cell culture supernatant with Sartobind anion-exchange membrane adsorbers. J. Membr. Sci. 2007, 299, 251–260. [Google Scholar] [CrossRef]
- Reimer, C.B.; Baker, R.S.; vanFrank, R.M.; Newlin, T.E.; Cline, G.B.; Anderson, N.G. Purification of Large Quantities of Influenza Virus by Density Gradient Centrifugation. J. Virol. 1967, 1, 1207–1216. [Google Scholar] [CrossRef]
- Asanzhanova, N.N.; Ryskeldinova, S.Z.; Chervyakova, O.V.; Khairullin, B.M.; Kasenov, M.M.; Tabynov, K.K. Comparison of Different Methods of Purification and Concentration in Production of Influenza Vaccine. Bull. Exp. Biol. Med. 2017, 164, 229–232. [Google Scholar] [CrossRef]
- Weigel, T.; Solomaier, T.; Peuker, A.; Pathapati, T.; Wolff, M.W.; Reichl, U. A flow-through chromatography process for influenza A and B virus purification. J. Virol. Methods 2014, 207, 45–53. [Google Scholar] [CrossRef]
- Wolff, M.W.; Reichl, U. Downstream Processing: From Egg to Cell Culture-Derived Influenza Virus Particles. Chem. Eng. Technol. 2008, 6, 846–857. [Google Scholar] [CrossRef]
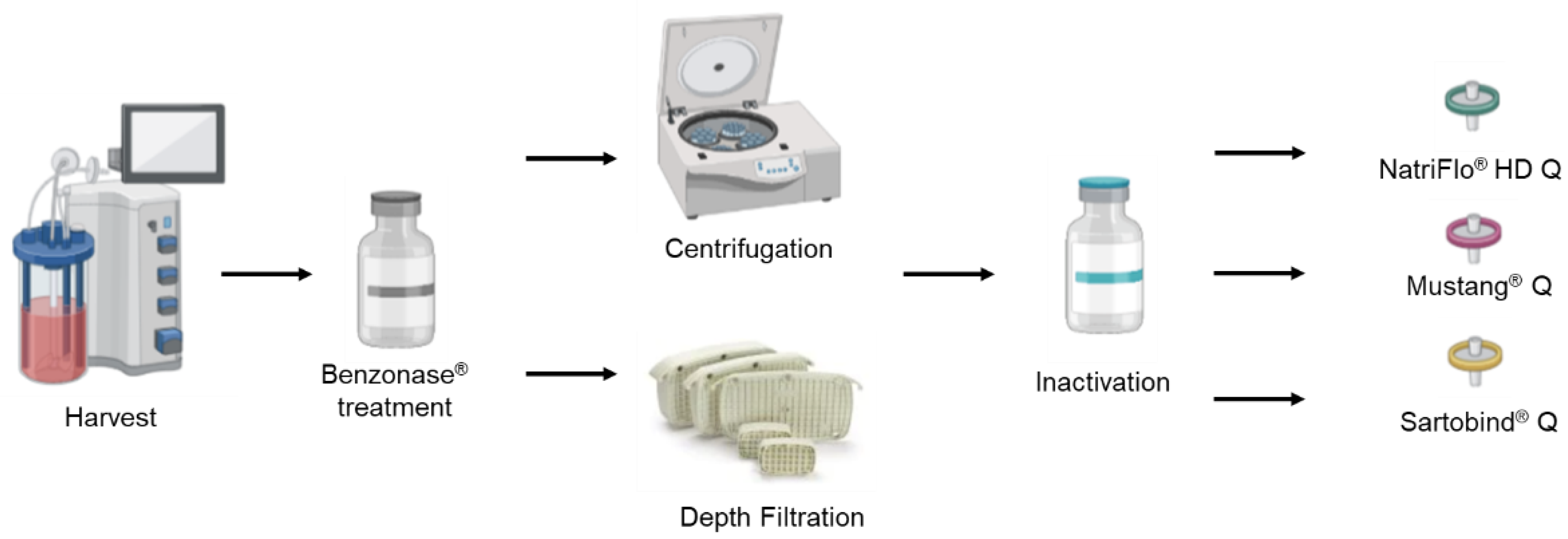
| No. | Strain | Bioreactor Volume | Cell Density at Infection | MOI | TPCK-Trypsin | Temperature at Infection | Time of Harvest | Total Cell Density at Harvest | Cell Viability at Harvest |
|---|---|---|---|---|---|---|---|---|---|
| Run #1 | A/PR/8/34 H1N1 | 2.3 L | 1.3 × 106 cells/mL | 0.001 | 1 μg/mL | 35 °C | 45 hpi | 1.1 × 106 cells/mL | 26% |
| Run #2 | A/PR/8/34 H1N1 | 2.3 L | 1.4 × 106 cells/mL | 0.01 | 1 μg/mL | 35 °C | 40 hpi | 4.7 × 106 cells/mL | 63% |
| Run #3 | A/PR/8/34 H1N1 | 750 mL | 7.5 × 106 cells/mL | 0.001 | 1 μg/mL | 35 °C | 48 hpi | 4.0 × 106 cells/mL | 58% |
| Run #4 | A/Anhui/1/2013 H7N9 | 2.3 L | 6.5 × 106 cells/mL | 0.0001 | 1 μg/mL | 37 °C | 54 hpi | 3.5 × 106 cells/mL | 88% |
| Run #5 | A/Anhui/1/2013 H7N9 | 2.3 L | 1.9 × 106 cells/mL | 0.0001 | 1 μg/mL | 37 °C | 56 hpi | 3.6 × 106 cells/mL | 53% |
| Run #6 | A/Hong-Kong/8/64 H3N2 | 2.3 L | 1.4 × 106 cells/mL | 0.01 | 2 μg/mL | 37 °C | 96 hpi | 1.3 × 106 cells/mL | 72% |
| Run #7 | A/Hong-Kong/8/64 H3N2 | 750 mL | 1.9 × 106 cells/mL | 0.01 | 2 μg/mL | 37 °C | 48 hpi | 3.4 × 106 cells/mL | 66% |
| No. | Sample ID | HA (μg/mL) | DNA (ng/mL) | Protein (μg/mL) | HA Recovery | DNA Removal | Protein Removal |
|---|---|---|---|---|---|---|---|
| Run #1 | Initial | 130.9 ± 17.3 | 7748.0 ± 89.7 | 262.2 ± 14.8 | |||
| Depth filtration | 103.6 ± 15.1 | 268.5 ± 17.0 | 190.3 ± 16.4 | 79.1 ± 11.5% | 96.5 ± 6.1% | 27.4 ± 2.4% | |
| Centrifugation | 92.2 ± 10.4 | 636.3 ± 39.2 | 179.8 ± 9.5 | 70.3 ± 7.9% | 91.8 ± 5.7% | 31.4 ± 1.7% | |
| Run #2 | Initial | 28.5 ± 3.5 | 28,229.1 ± 73.9 | 402.7 ± 28.4 | |||
| Depth filtration | 10.9 ± 1.25 | 1247.2 ± 27.2 | 272.3 ± 16.8 | 35.4 ± 4.1% | 95.6 ± 9.0% | 32.4 ± 2.0% | |
| Centrifugation | 10.4 ± 0.8 | 2012.0 ± 38.1 | 280.7 ± 18.2 | 36.5 ± 2.8% | 92.9 ± 8.7% | 30.3 ± 2.0% |
| No. | Sample ID | Volume (mL) | HA (μg/mL) | DNA (ng/mL) | Protein (μg/mL) | HA Recovery | DNA Removal | Protein Removal |
|---|---|---|---|---|---|---|---|---|
| Run #3 | Initial | 124.3 | 18.9 ± 1.8 | 77,237.3 ± 734.6 | 786.0 ± 30.6 | |||
| Load | 112.7 | 12.6 ± 0.7 | 8170.9 ± 162.3 | 698.0 ± 27.3 | 60.4 ± 3.3% | 90.4 ± 7.3% | 19.5 ± 1.8% | |
| NatriFlo®-1 M | 11.4 | 52.3 ± 2.8 | 52,289.7 ± 525.9 | 1116.6 ± 35.4 | 42.0 ± 3.8% | 35.3 ± 2.7% | 83.8 ± 6.3% | |
| Run #3 | Initial | 128.4 | 18.9 ± 1.8 | 77,237.3 ± 734.6 | 786.0 ± 28.5 | |||
| Load | 122.1 | 14.1 ± 1.1 | 7400.6 ± 154.7 | 737.3 ± 26.1 | 70.9 ± 5.3% | 90.9 ± 7.1% | 10.8 ± 0.8% | |
| Sartobind® Q-1 M | 16.4 | 74.2 ± 4.7 | 49,169.3 ± 502.0 | 1377.6 ± 38.9 | 70.7 ± 4.5% | 10.8 ± 0.8% | 74.9 ± 7.0% |
| No. | Sample ID | Volume (mL) | HA (μg/mL) | DNA (ng/mL) | Protein (μg/mL) | HA Recovery | ng DNA/Dose | Protein /HA |
|---|---|---|---|---|---|---|---|---|
| Run #1 | Load | 100.0 | 37.5 ± 2.6 | 80.5 ± 5.0 | 193.5 ± 11.4 | |||
| Mustang® Q | 17.0 | 101.0 ± 7.6 | 132.2 ± 10.7 | 130.9 ± 7.9 | 45.8 ± 3.5% | 19.6 ± 1.6 | 1.3 ± 0.1 | |
| Sartorbind® Q | 37.5 | 62.5 ± 3.2 | 171.1 ± 9.1 | 109.1 ± 8.9 | 62.5 ± 3.2% | 41.1 ± 2.2 | 1.7 ± 0.1 |
| No. | Sample ID | HAU (unit/mL) | DNA (ng/mL) | Protein (μg/mL) | HAU Recovery | DNA Removal | Protein Removal |
|---|---|---|---|---|---|---|---|
| Run #4 | Initial | 1230.5 ± 85.8 | 2793.6 ± 67.2 | 125.1 ± 11.5 | |||
| H7N9 | Depth filtration | 867.4 ± 65.3 | 196.1 ± 10.7 | 104.8 ± 9.9 | 70.5 ± 0.4% | 93.0 ± 5.1% | 16.2 ± 1.5% |
| Run #5 | Initial | 888.4 ± 81.3 | 9276.8 ± 185.0 | 202.7 ± 20.0 | |||
| H7N9 | Centrifugation | 641.1 ± 65.3 | 1867.1 ± 53.7 | 168.8 ± 11.6 | 72.9 ± 13.1% | 79.9 ± 6.5% | 16.7 ± 1.2% |
| Run #6 | Initial | 28.9 ± 1.0 | 2341.6 ± 53.6 | 251.7 ± 22.6 | |||
| H3N2 | Depth filtration | 22.8 ± 4.8 | 275.4 ± 12.3 | 200.6 ± 16.3 | 79.1 ± 18.2% | 88.2 ± 6.3% | 20.3 ± 1.7% |
| Run #7 | Initial | 163.2 ± 5.6 | 4441.4 ± 89.1 | 113.5 ± 10.6 | |||
| H3N2 | Centrifugation | 117.7 ± 4.0 | 248.4 ± 11.2 | 90.0 ± 7.8 | 72.1 ± 2.4% | 94.4 ± 8.4% | 20.7 ± 1.8% |
| No. | Sample ID | Volume (mL) | HAU (unit/mL) | DNA (ng/mL) | Protein (μg/mL) | HAU Recovery | DNA Residual | Protein Residual |
|---|---|---|---|---|---|---|---|---|
| Run #4 | Load | 147.5 | 797.3 ± 4.6 | 185.3 ± 17.9 | 103.2 ± 5.2 | |||
| H7N9 | 0.3 M | 7.0 | 1771.4 ± 171.4 | 79.7 ± 4.9 | 42.5 ± 3.0 | 10.5 ± 1.0% | 2.0 ± 0.1% | 1.9 ± 0.1% |
| 0.7 M | 7.0 | 5223.5 ± 179.3 | 2175.8 ± 195.8 | 49.6 ± 3.1 | 31.1 ± 1.0% | 55.7 ± 5.0% | 2.3 ± 0.1% | |
| Total HAU Recovery: 41.6% | ||||||||
| Run #6 | Load | 100 | 22.8 ± 4.8 | 174.4 ± 17.0 | 202.9 ± 18.1 | |||
| H3N2 | 0.3 M | 8.5 | 29.4 ± 1.0 | 21.2 ± 1.3 | 131.9 ± 12.8 | 11.3 ± 2.5% | 1.0 ± 0.1% | 5.5 ± 0.5% |
| 0.7 M | 7.5 | 89.6 ± 8.2 | 1383.8 ± 102.5 | 82.2 ± 6.0 | 30.0 ± 3.2% | 59.5 ± 4.4% | 3.0 ± 0.2% | |
| Total HAU Recovery: 41.3% | ||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Z.; Xu, X.; Silva, C.A.T.; Farnos, O.; Venereo-Sanchez, A.; Toussaint, C.; Dash, S.; González-Domínguez, I.; Bernier, A.; Henry, O.; et al. Membrane Chromatography-Based Downstream Processing for Cell-Culture Produced Influenza Vaccines. Vaccines 2022, 10, 1310. https://doi.org/10.3390/vaccines10081310
Yang Z, Xu X, Silva CAT, Farnos O, Venereo-Sanchez A, Toussaint C, Dash S, González-Domínguez I, Bernier A, Henry O, et al. Membrane Chromatography-Based Downstream Processing for Cell-Culture Produced Influenza Vaccines. Vaccines. 2022; 10(8):1310. https://doi.org/10.3390/vaccines10081310
Chicago/Turabian StyleYang, Zeyu, Xingge Xu, Cristina A. T. Silva, Omar Farnos, Alina Venereo-Sanchez, Cécile Toussaint, Shantoshini Dash, Irene González-Domínguez, Alice Bernier, Olivier Henry, and et al. 2022. "Membrane Chromatography-Based Downstream Processing for Cell-Culture Produced Influenza Vaccines" Vaccines 10, no. 8: 1310. https://doi.org/10.3390/vaccines10081310
APA StyleYang, Z., Xu, X., Silva, C. A. T., Farnos, O., Venereo-Sanchez, A., Toussaint, C., Dash, S., González-Domínguez, I., Bernier, A., Henry, O., & Kamen, A. (2022). Membrane Chromatography-Based Downstream Processing for Cell-Culture Produced Influenza Vaccines. Vaccines, 10(8), 1310. https://doi.org/10.3390/vaccines10081310







