A New Live Auxotrophic Vaccine Induces Cross-Protection against Klebsiella pneumoniae Infections in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Growth Conditions and Plasmids
2.2. Construction of the murI Deletion Mutant of K. pneumoniae MGH 78578
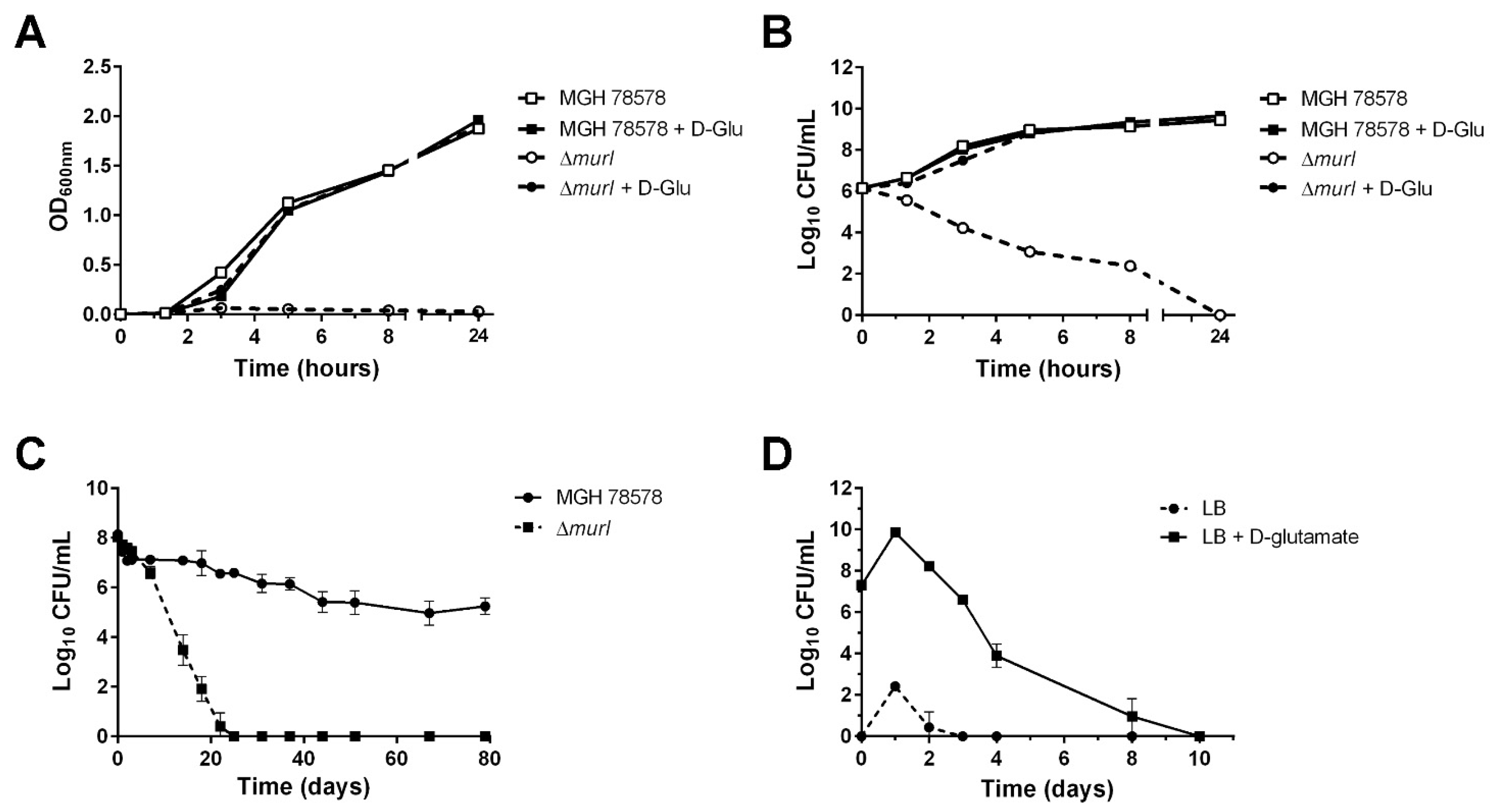
2.3. Growth and Viability Curves
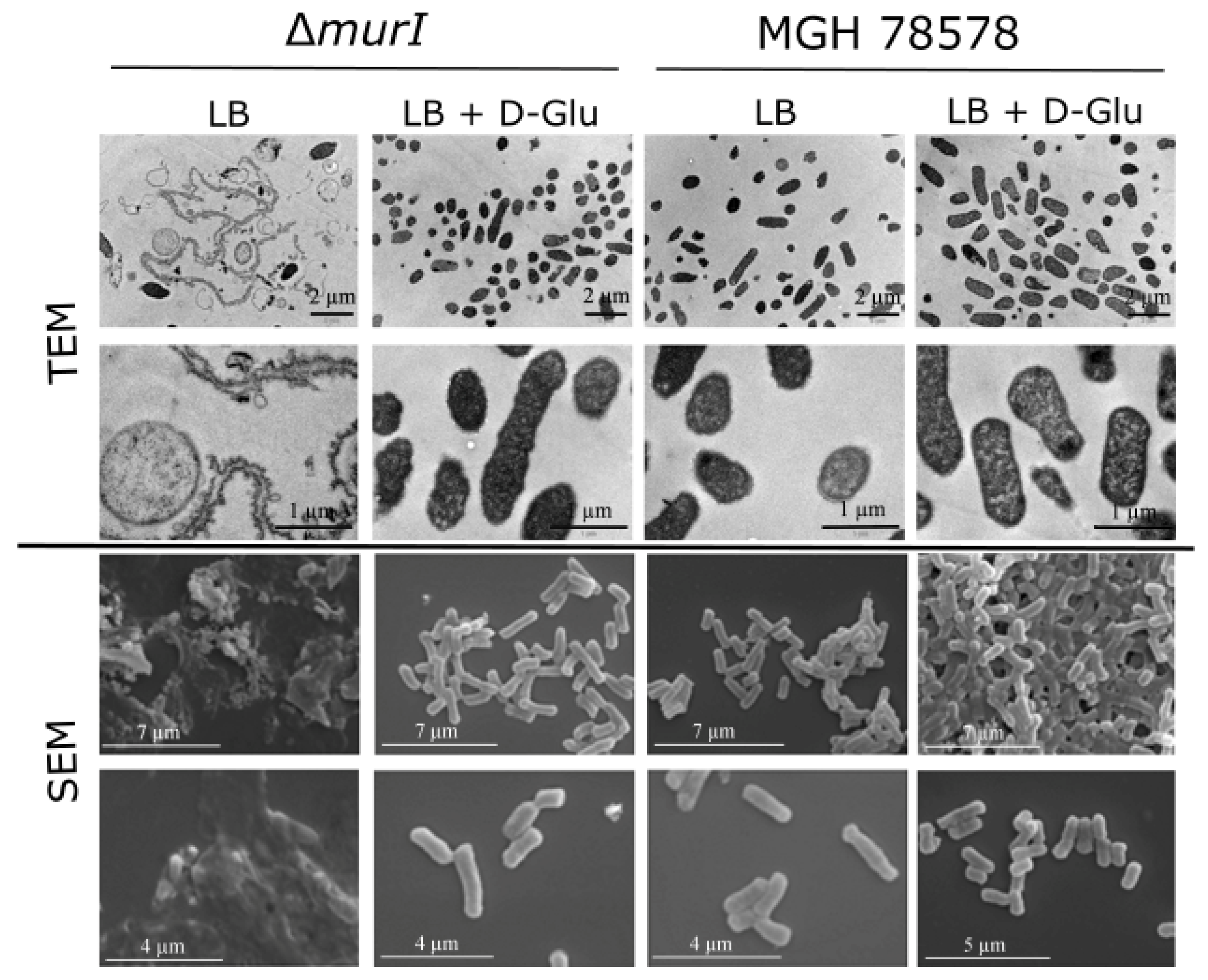
2.4. Electronic Microscopy Analysis
2.5. Control of Phenotypic Stability
2.6. Water Survival Assay
2.7. In Silico Molecular Typing, Capsular Type and Resistance Profile Predictions
2.8. Ethics Statement
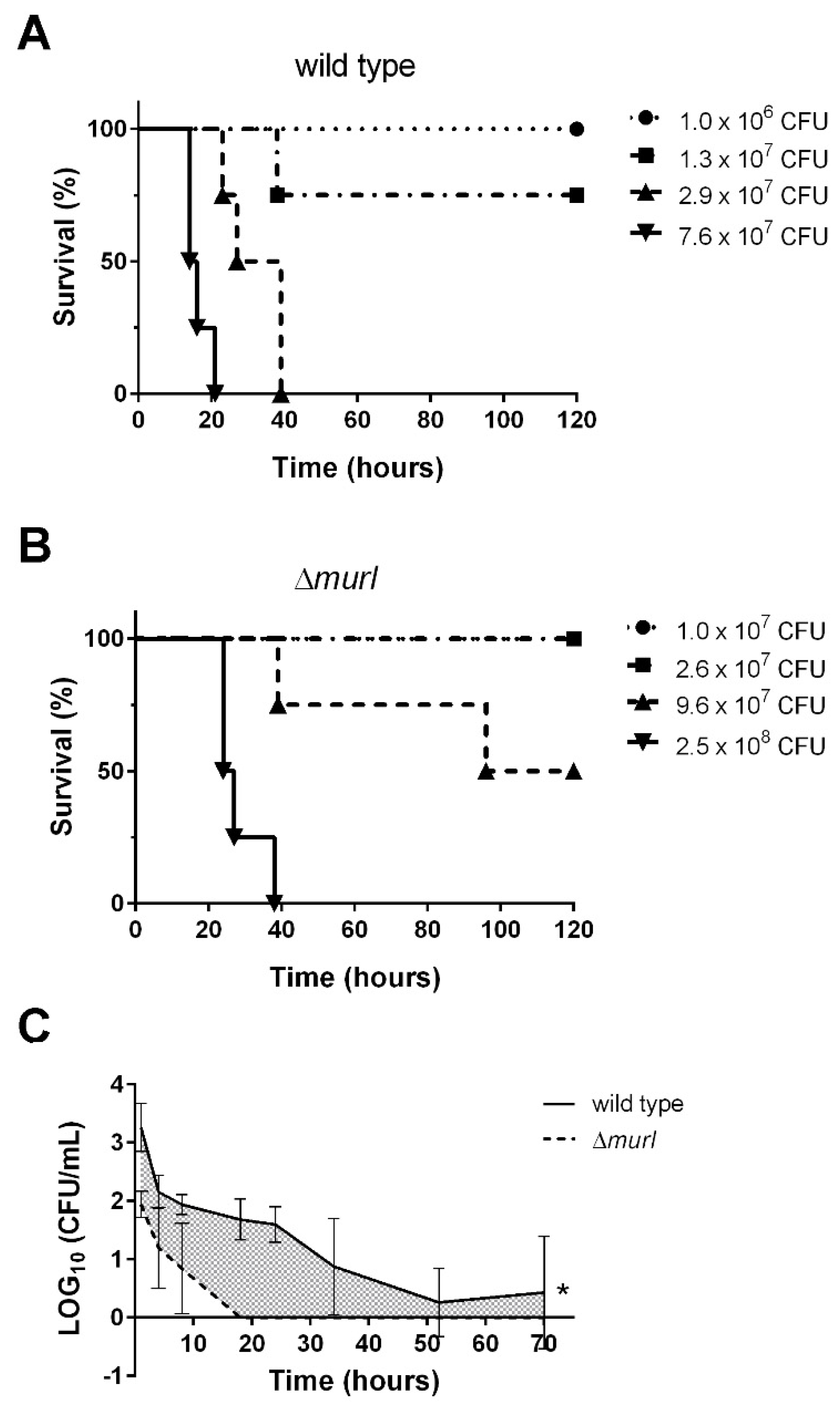
2.9. Mouse Experiments
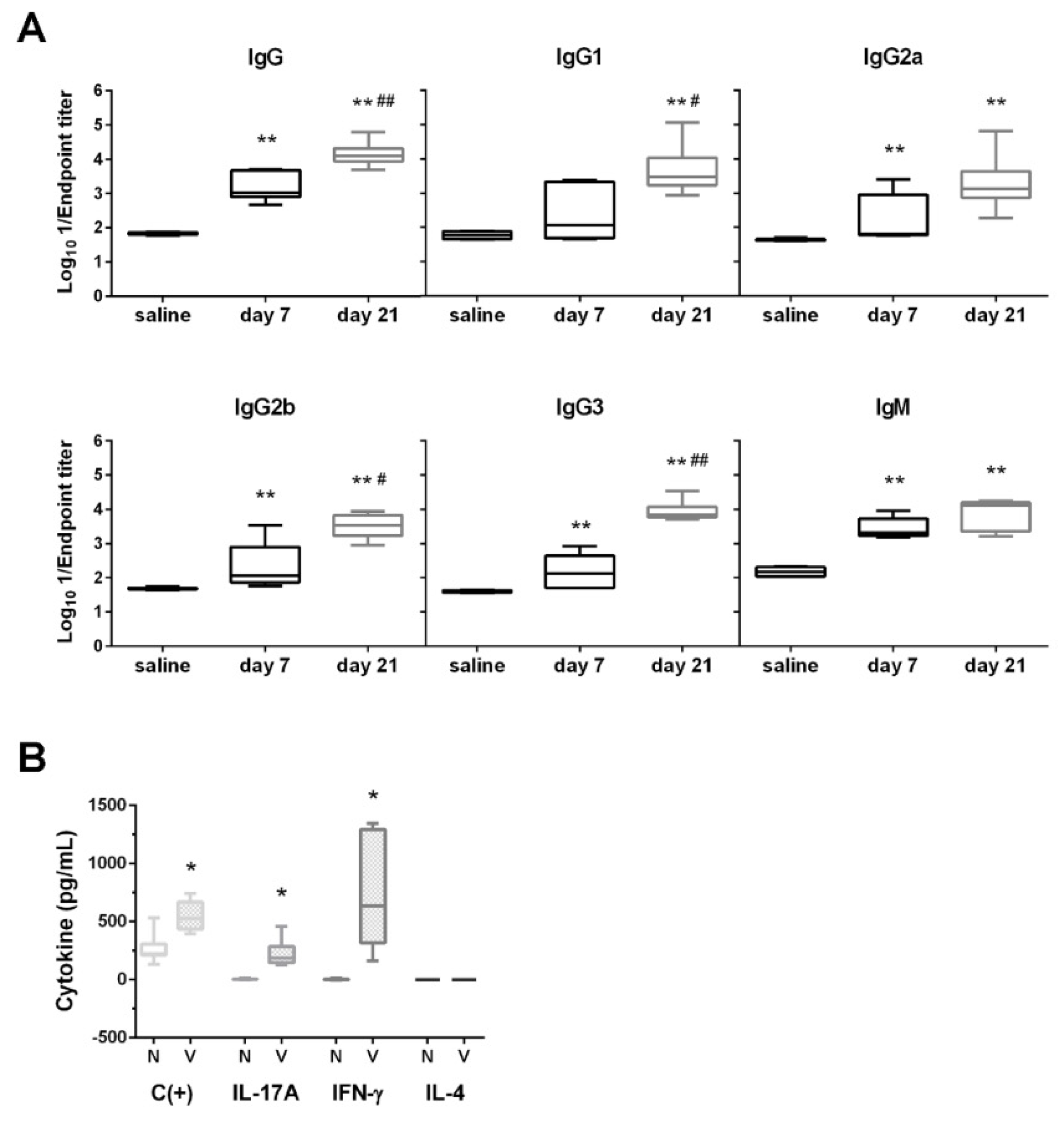
2.10. ELISA
2.11. Statistical Analysis
3. Results
3.1. Characterization of the K. pneumoniae MGH 78578 Glutamate Racemase-Deficient Mutant
3.2. The D-glutamate Auxotroph of K. pneumoniae Is Attenuated in BALB/c Mice
3.3. D-glutamate Auxotrophic Strain Generates Robust Humoral and Cellular Immune Responses against K. pneumoniae
3.4. The D-glutamate Auxotroph Vaccine Generates Cross-Protective Antibodies against K. pneumoniae Heterologous Strains
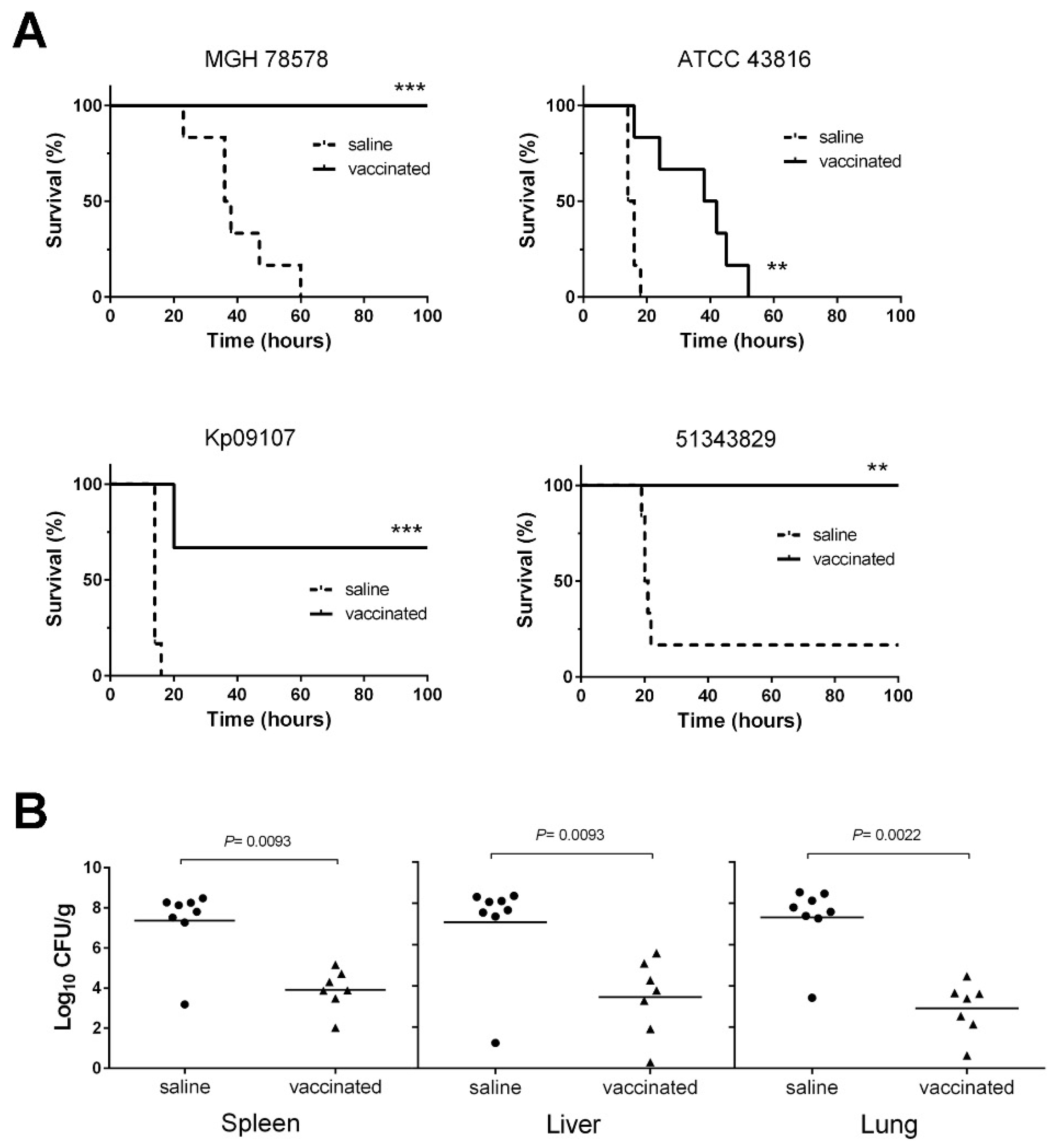
3.5. D-glutamate Auxotrophic Strain of K. pneumoniae MGH 78578 Elicits Protection against Infection with Different K. pneumoniae Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Opoku-Temeng, C.; Malachowa, N.; Kobayashi, S.D.; DeLeo, F.R. Innate Host Defense against Klebsiella pneumoniae and the Outlook for Development of Immunotherapies. J. Innate Immun. 2021, 14, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Lam, M.M.C.; Holt, K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020, 18, 344–359. [Google Scholar] [CrossRef] [PubMed]
- Russo, T.A.; Marr, C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019, 32, e00001-19. [Google Scholar] [CrossRef] [Green Version]
- Russo, T.A.; Olson, R.; Fang, C.T.; Stoesser, N.; Miller, M.; MacDonald, U.; Hutson, A.; Barker, J.H.; La Hoz, R.M.; Johnson, J.R. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J. Clin. Microbiol. 2018, 56, e00776-18. [Google Scholar] [CrossRef] [Green Version]
- Fodah, R.A.; Scott, J.B.; Tam, H.H.; Yan, P.; Pfeffer, T.L.; Bundschuh, R.; Warawa, J.M. Correlation of Klebsiella pneumoniae comparative genetic analyses with virulence profiles in a murine respiratory disease model. PLoS ONE 2014, 9, e107394. [Google Scholar] [CrossRef] [Green Version]
- Pitout, J.D.; Nordmann, P.; Poirel, L. Carbapenemase-Producing Klebsiella pneumoniae, a Key Pathogen Set for Global Nosocomial Dominance. Antimicrob. Agents Chemother. 2015, 59, 5873–5884. [Google Scholar] [CrossRef] [Green Version]
- Martin, R.M.; Bachman, M.A. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Ferrer, S.; Penaloza, H.F.; Budnick, J.A.; Bain, W.G.; Nordstrom, H.R.; Lee, J.S.; Van Tyne, D. Finding Order in the Chaos: Outstanding Questions in Klebsiella pneumoniae Pathogenesis. Infect. Immun. 2021, 89, e00693-20. [Google Scholar] [CrossRef] [PubMed]
- van Duin, D.; Lok, J.J.; Earley, M.; Cober, E.; Richter, S.S.; Perez, F.; Salata, R.A.; Kalayjian, R.C.; Watkins, R.R.; Doi, Y.; et al. Colistin Versus Ceftazidime-Avibactam in the Treatment of Infections Due to Carbapenem-Resistant Enterobacteriaceae. Clin. Infect. Dis. 2018, 66, 163–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, H.; Chen, L.; Tang, Y.W.; Kreiswirth, B.N. Emergence of the mcr-1 colistin resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet Infect. Dis. 2016, 16, 287–288. [Google Scholar] [CrossRef] [Green Version]
- Shields, R.K.; Chen, L.; Cheng, S.; Chavda, K.D.; Press, E.G.; Snyder, A.; Pandey, R.; Doi, Y.; Kreiswirth, B.N.; Nguyen, M.H.; et al. Emergence of Ceftazidime-Avibactam Resistance Due to Plasmid-Borne blaKPC-3 Mutations during Treatment of Carbapenem-Resistant Klebsiella pneumoniae Infections. Antimicrob. Agents Chemother. 2017, 61, e02097-16. [Google Scholar] [PubMed] [Green Version]
- Lorenzo-Gomez, M.F.; Padilla-Fernandez, B.; Garcia-Criado, F.J.; Miron-Canelo, J.A.; Gil-Vicente, A.; Nieto-Huertos, A.; Silva-Abuin, J.M. Evaluation of a therapeutic vaccine for the prevention of recurrent urinary tract infections versus prophylactic treatment with antibiotics. Int. Urogynecol. J. 2013, 24, 127–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsieh, P.F.; Lin, T.L.; Lee, C.Z.; Tsai, S.F.; Wang, J.T. Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 2008, 197, 1717–1727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edelman, R.; Taylor, D.N.; Wasserman, S.S.; McClain, J.B.; Cross, A.S.; Sadoff, J.C.; Que, J.U.; Cryz, S.J. Phase 1 trial of a 24-valent Klebsiella capsular polysaccharide vaccine and an eight-valent Pseudomonas O-polysaccharide conjugate vaccine administered simultaneously. Vaccine 1994, 12, 1288–1294. [Google Scholar] [CrossRef]
- Donta, S.T.; Peduzzi, P.; Cross, A.S.; Sadoff, J.; Haakenson, C.; Cryz, S.J., Jr.; Kauffman, C.; Bradley, S.; Gafford, G.; Elliston, D.; et al. Immunoprophylaxis against Klebsiella and Pseudomonas aeruginosa infections. The Federal Hyperimmune Immunoglobulin Trial Study Group. J. Infect. Dis. 1996, 174, 537–543. [Google Scholar] [CrossRef] [Green Version]
- Zigterman, J.W.; van Dam, J.E.; Snippe, H.; Rotteveel, F.T.; Jansze, M.; Willers, J.M.; Kamerling, J.P.; Vliegenthart, J.F. Immunogenic properties of octasaccharide-protein conjugates derived from Klebsiella serotype 11 capsular polysaccharide. Infect. Immun. 1985, 47, 421–428. [Google Scholar] [CrossRef] [Green Version]
- Seeberger, P.H.; Pereira, C.L.; Khan, N.; Xiao, G.; Diago-Navarro, E.; Reppe, K.; Opitz, B.; Fries, B.C.; Witzenrath, M. A Semi-Synthetic Glycoconjugate Vaccine Candidate for Carbapenem-Resistant Klebsiella pneumoniae. Angew. Chem. Int. Ed. Engl. 2017, 56, 13973–13978. [Google Scholar] [CrossRef] [Green Version]
- Feldman, M.F.; Mayer Bridwell, A.E.; Scott, N.E.; Vinogradov, E.; McKee, S.R.; Chavez, S.M.; Twentyman, J.; Stallings, C.L.; Rosen, D.A.; Harding, C.M. A promising bioconjugate vaccine against hypervirulent Klebsiella pneumoniae. Proc. Natl. Acad. Sci. USA 2019, 116, 18655–18663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, M.; Tennant, S.M.; Simon, R.; Cross, A.S. Progress towards the development of Klebsiella vaccines. Expert. Rev. Vaccines 2019, 18, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Corral-Lugo, A.; McConnell, M.J. Vaccines for multidrug resistant Gram negative bacteria: Lessons from the past for guiding future success. FEMS Microbiol. Rev. 2021, 45, fuaa054. [Google Scholar] [CrossRef] [PubMed]
- Cubero, M.; Grau, I.; Tubau, F.; Pallares, R.; Dominguez, M.A.; Linares, J.; Ardanuy, C. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007–2013). Clin. Microbiol. Infect. 2016, 22, 154–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gato, E.; Vazquez-Ucha, J.C.; Rumbo-Feal, S.; Alvarez-Fraga, L.; Vallejo, J.A.; Martinez-Guitian, M.; Beceiro, A.; Ramos Vivas, J.; Sola Campoy, P.J.; Perez-Vazquez, M.; et al. Kpi, a chaperone-usher pili system associated with the worldwide-disseminated high-risk clone Klebsiella pneumoniae ST-15. Proc. Natl. Acad. Sci. USA 2020, 117, 17249–17259. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.T.; Chuang, Y.P.; Shun, C.T.; Chang, S.C.; Wang, J.T. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 2004, 199, 697–705. [Google Scholar] [CrossRef] [Green Version]
- Gust, B.; Challis, G.L.; Fowler, K.; Kieser, T.; Chater, K.F. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 2003, 100, 1541–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, T.W.; Lam, I.; Chang, H.Y.; Tsai, S.F.; Palsson, B.O.; Charusanti, P. Capsule deletion via a lambda-Red knockout system perturbs biofilm formation and fimbriae expression in Klebsiella pneumoniae MGH 78578. BMC Res. Notes 2014, 7, 13. [Google Scholar] [CrossRef] [Green Version]
- Cabral, M.P.; Garcia, P.; Beceiro, A.; Rumbo, C.; Perez, A.; Moscoso, M.; Bou, G. Design of live attenuated bacterial vaccines based on D-glutamate auxotrophy. Nat. Commun. 2017, 8, 15480. [Google Scholar] [CrossRef] [Green Version]
- Lam, M.M.C.; Wick, R.R.; Watts, S.C.; Cerdeira, L.T.; Wyres, K.L.; Holt, K.E. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 2021, 12, 4188. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Wick, R.R.; Gorrie, C.; Jenney, A.; Follador, R.; Thomson, N.R.; Holt, K.E. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2016, 2, e000102. [Google Scholar] [CrossRef] [PubMed]
- Moscoso, M.; Garcia, P.; Cabral, M.P.; Rumbo, C.; Bou, G. A D-Alanine auxotrophic live vaccine is effective against lethal infection caused by Staphylococcus aureus. Virulence 2018, 9, 604–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.C.; Lee, C.H.; Hong, M.Y.; Hsieh, C.C.; Tang, H.J.; Ko, W.C. Propensity-matched analysis of the impact of extended-spectrum beta-lactamase production on adults with community-onset Escherichia coli, Klebsiella species, and Proteus mirabilis bacteremia. J. Microbiol. Immunol. Infect. 2018, 51, 519–526. [Google Scholar] [CrossRef]
- Tzouvelekis, L.S.; Markogiannakis, A.; Psichogiou, M.; Tassios, P.T.; Daikos, G.L. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: An evolving crisis of global dimensions. Clin. Microbiol. Rev. 2012, 25, 682–707. [Google Scholar] [CrossRef] [Green Version]
- Yao, B.; Xiao, X.; Wang, F.; Zhou, L.; Zhang, X.; Zhang, J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int. J. Infect. Dis. 2015, 37, 107–112. [Google Scholar] [CrossRef] [Green Version]
- Gu, D.; Dong, N.; Zheng, Z.; Lin, D.; Huang, M.; Wang, L.; Chan, E.W.; Shu, L.; Yu, J.; Zhang, R.; et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: A molecular epidemiological study. Lancet Infect. Dis. 2018, 18, 37–46. [Google Scholar] [CrossRef]
- Karlsson, M.; Stanton, R.A.; Ansari, U.; McAllister, G.; Chan, M.Y.; Sula, E.; Grass, J.E.; Duffy, N.; Anacker, M.L.; Witwer, M.L.; et al. Identification of a Carbapenemase-Producing Hypervirulent Klebsiella pneumoniae Isolate in the United States. Antimicrob. Agents Chemother. 2019, 63, e00519-19. [Google Scholar] [CrossRef] [Green Version]
- Kurupati, P.; Ramachandran, N.P.; Poh, C.L. Protective efficacy of DNA vaccines encoding outer membrane protein A and OmpK36 of Klebsiella pneumoniae in mice. Clin. Vaccine Immunol. 2011, 18, 82–88. [Google Scholar] [CrossRef] [Green Version]
- Vidarsson, G.; Dekkers, G.; Rispens, T. IgG subclasses and allotypes: From structure to effector functions. Front. Immunol. 2014, 5, 520. [Google Scholar] [CrossRef] [Green Version]
- Damelang, T.; Rogerson, S.J.; Kent, S.J.; Chung, A.W. Role of IgG3 in Infectious Diseases. Trends Immunol. 2019, 40, 197–211. [Google Scholar] [CrossRef]
- Moore, T.A.; Perry, M.L.; Getsoian, A.G.; Newstead, M.W.; Standiford, T.J. Divergent role of gamma interferon in a murine model of pulmonary versus systemic Klebsiella pneumoniae infection. Infect. Immun. 2002, 70, 6310–6318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivin, M.; Dumigan, A.; de Vasconcelos, F.N.; Ebner, F.; Borroni, M.; Kavirayani, A.; Przybyszewska, K.N.; Ingram, R.J.; Lienenklaus, S.; Kalinke, U.; et al. Natural killer cell-intrinsic type I IFN signaling controls Klebsiella pneumoniae growth during lung infection. PLoS Pathog. 2017, 13, e1006696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengoechea, J.A.; Sa Pessoa, J. Klebsiella pneumoniae infection biology: Living to counteract host defences. FEMS Microbiol. Rev. 2019, 43, 123–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, W.H.; Choi, H.I.; Hong, S.W.; Kim, K.S.; Gho, Y.S.; Jeon, S.G. Vaccination with Klebsiella pneumoniae-derived extracellular vesicles protects against bacteria-induced lethality via both humoral and cellular immunity. Exp. Mol. Med. 2015, 47, e183. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.Z.; Hu, D.; Dou, Y.; Xiong, L.; Wang, X.; Hu, J.; Xing, S.Z.; Li, W.; Cai, J.P.; Jin, M.; et al. Identification and Evaluation of Recombinant Outer Membrane Proteins as Vaccine Candidates Against Klebsiella pneumoniae. Front. Immunol. 2021, 12, 730116. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, T.A.; El-Sayed, L.H.; Haroun, M.; Hussein, A.A.; El Ashry el, S.H. Development of immunization trials against Klebsiella pneumoniae. Vaccine 2012, 30, 2411–2420. [Google Scholar] [CrossRef] [PubMed]

| Strain, Plasmid or Primer | Relevant Features or Sequence (5′ → 3′) | Reference |
|---|---|---|
| K. pneumoniaestrains | ||
| MGH 78578 | ATCC 700721, isolate from the sputum of a patient with pneumonia (ST38, K52) | American Type Culture Collection (ATCC) |
| MGH 78578 ΔmurI | MGH 78578 derivative, ΔKPN_04256 | This study |
| ATCC 43816 | A K2 clinical pneumonia isolate (ST493, K2) | ATCC |
| ATCC 43816 KPPR1 | A rifampin-resistant mutant of ATCC 43816 | ATCC |
| ATCC 700603 | Klebsiella quasipneumoniae (formerly known as K. pneumoniae K6), a clinical isolate from a hospitalized patient with urinary tract infection; ESBL reference strain | ATCC |
| H9548 | A hypervirulent strain isolated from a patient with bacteraemia in Barcelona (ST493, K2) | [24] |
| H14721 | A hypermucoviscous strain causing bacteraemia in adults in Barcelona (ST23, K1) | [24] |
| Kp09107 | Isolate from rectal swabs of patients hospitalized in Spain (ST101, K17) | [25] |
| Kp727 | Clinical isolate recovered from blood cultures in Spain (ST405) | [25] |
| Kp924 | Clinical isolate from bronchoalveolar lavage fluid samples of patients in Spain (ST11, K24) | [25] |
| Kp1278 | Isolate from multiple urine cultures in a hospital outbreak in Spain (ST15, K24) | [25] |
| NTUH-K2044 | Isolate from a patient with liver abscess and meningitis in Taiwan (ST23, K1) | [26] |
| 51343829 | Hypermucoviscous strain from rectal swabs of patients in the A Coruña University Hospital (ST15) | Laboratory collection |
| E. colistrains | ||
| E. coli | DH5α competent cells (F− φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK−, mK+) phoA supE44 λ−thi-1 gyrA96 relA1) | Thermo Fisher Scientific |
| Plasmids | ||
| pIJ773 | pBluescript II SK(+) derivative containing an apramycin-resistance cassette (aac(3)IV) and the oriT from RK2 flanked by FLP recognition target (FRT) sites | [27] |
| pACBSR-Hyg | A p15A replicon plasmid containing an arabinose-inducible λ-Red recombinase and a hygromycin-resistance marker (hph) | [28] |
| pFLP-Hyg | A p15A replicon plasmid bearing a heat shock-inducible FLP recombinase and a hygromycin-resistance marker (hph) | [28] |
| Primer | ||
| murIKOFW | CTGCAGGACGGGAATACACCTTGTCTGGCAGCTACACCTTCTGATCCACGTCCCACCATGATTCCGGGGATCCGTCGACC | |
| murIKORV | TGACAAGCCCTGTTTTCAAAAAATTATTCAACCGTCTCTTTTTTGTGCAAAACGCCCTTATGTAGGCTGGAGCTGCTTC | |
| EXTMURIFW | ATGATGATACTGGCAAGG | |
| EXTMURIRV | ATAGGGAGTTCTGAGACGT | |
| RT(KbrpoB) Fw | GGTGAAACTGCCTCCTTCG | |
| RT(KbrpoB) Rv | ACGGCCTTTCTCAACGTACA | |
| RT(KbMurI) Fw | GTGGGTTGTCGGTTTATAATGAG | |
| RT(KbMurI) Rv | GGCAACGTTATCGAAAGCATA | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moscoso, M.; Vallejo, J.A.; Cabral, M.P.; García, P.; Fuentes-Valverde, V.; Gato, E.; Arca-Suárez, J.; Aja-Macaya, P.; Bou, G. A New Live Auxotrophic Vaccine Induces Cross-Protection against Klebsiella pneumoniae Infections in Mice. Vaccines 2022, 10, 953. https://doi.org/10.3390/vaccines10060953
Moscoso M, Vallejo JA, Cabral MP, García P, Fuentes-Valverde V, Gato E, Arca-Suárez J, Aja-Macaya P, Bou G. A New Live Auxotrophic Vaccine Induces Cross-Protection against Klebsiella pneumoniae Infections in Mice. Vaccines. 2022; 10(6):953. https://doi.org/10.3390/vaccines10060953
Chicago/Turabian StyleMoscoso, Miriam, Juan A. Vallejo, Maria P. Cabral, Patricia García, Víctor Fuentes-Valverde, Eva Gato, Jorge Arca-Suárez, Pablo Aja-Macaya, and Germán Bou. 2022. "A New Live Auxotrophic Vaccine Induces Cross-Protection against Klebsiella pneumoniae Infections in Mice" Vaccines 10, no. 6: 953. https://doi.org/10.3390/vaccines10060953
APA StyleMoscoso, M., Vallejo, J. A., Cabral, M. P., García, P., Fuentes-Valverde, V., Gato, E., Arca-Suárez, J., Aja-Macaya, P., & Bou, G. (2022). A New Live Auxotrophic Vaccine Induces Cross-Protection against Klebsiella pneumoniae Infections in Mice. Vaccines, 10(6), 953. https://doi.org/10.3390/vaccines10060953






