Gastric Cancer and Viruses: A Fine Line between Friend or Foe
Abstract
1. Introduction
1.1. Gastric Cancers and Onco-Viruses
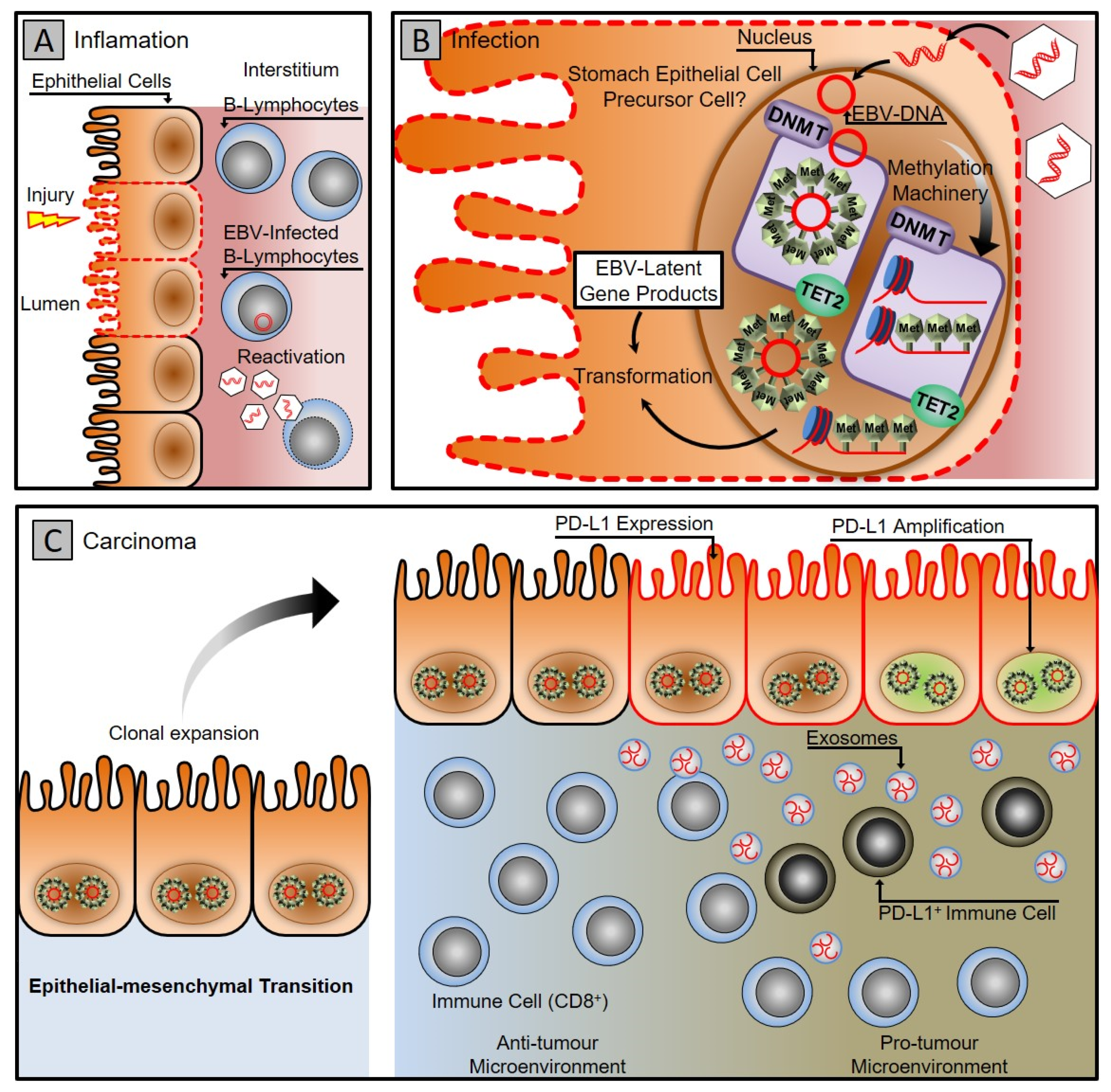
1.2. Epstein–Barr Virus Associated Gastric Cancer
1.3. Hepatitis B Virus and Gastric Cancers
1.4. Oncolytic Viruses
2. General Mechanism
3. Clinical Implication of Oncolytic Viruses in Gastric Cancer
3.1. Herpes Simplex Virus
3.2. Virus of Vesicular Stomatitis
3.3. Virus Vaccinia
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| GC | Gastric cancer |
| OVs | Oncolytic viruses |
| WHO | World Health Organization |
| GERD | Gastroesophageal reflux disease |
| EAC | Esophageal adenocarcinoma |
| HP | Helicobacter pylori |
| EBV | Epstein–Barr virus |
| HCV | Hepatitis C virus |
| HCMV | Human Cytomegalovirus |
| HPV | Human papillomavirus |
| HCC | Hepatocellular carcinoma |
| PAMPs | Pathogen-associated molecular patterns |
| TRAF3 | TNF associated factor 3 |
| RIG-1 | Retinoic acid-inducible gene 1 |
| JAK-SAT | Janus kinase–signal transducer and activator of transcription |
| PKR | Protein kinase R |
| LMP1 | Latent membrane protein 1 |
| EBVaGC | EBV-associated gastric carcinoma |
| TCGA | The Cancer Genome Atlas |
| CD | Cytosine deaminase |
| ADP | Adenovirus death protein |
| PAMPs | Pathogen-associated molecular patterns |
| HMGB1 | High mobility group box 1 |
| DAMPs | Danger-associated molecular pattern signals |
| APCs | Antigen-presenting cells |
| TNFα | Tumor necrosis factor-α |
| APCs | Antigen-presenting cells |
| MHC | Major histocompatibility complex |
| HSV-1 | Herpes simplex virus-1 |
| FDA | Food and Drug Administration |
| T-Vec | Talimogene Laherparepvec |
| MMC | Mitomycin C |
| VSV | Vesicular stomatitis virus |
| VV | Vaccinia virus |
| CAEV | Cell-associated enveloped viruses |
| EEV | Extracellular enveloped viruses |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Mukaisho, K.-I.; Nakayama, T.; Hagiwara, T.; Hattori, T.; Sugihara, H. Two distinct etiologies of gastric cardia adenocarcinoma: Interactions among pH, Helicobacter pylori, and bile acids. Front. Microbiol. 2015, 6, 412. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Chow, W.-H.; Lagergren, J.; Yin, L.; Nyrén, O. Risk of adenocarcinomas of the esophagus and gastric cardia in patients with gastroesophageal reflux diseases and after antireflux surgery. Gastroenterology 2001, 121, 1286–1293. [Google Scholar] [CrossRef] [PubMed]
- Plummer, M.; Franceschi, S.; Vignat, J.; Forman, D.; de Martel, C. Global Burden of Gastric Cancer Attributable to Helicobacter Pylori. Int. J. Cancer 2015, 136, 487–490. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, X.-L.; Liu, K.; Bai, D.; Zhang, W.-H.; Chen, X.-Z.; Hu, J.-K.; on behalf of the SIGES research group. Associations Between Gastric Cancer Risk and Virus Infection Other Than Epstein-Barr Virus: A Systematic Review and Meta-analysis Based on Epidemiological Studies. Clin. Transl. Gastroenterol. 2020, 11, e00201. [Google Scholar] [CrossRef] [PubMed]
- The World Cancer Research Fund (WCRF World Cancer Research Fund/American Institute for Cancer Research). Diet, Nutri-tion, Physical Activity and Cancer: A Global Perspective. Continuous Update Project Expert Report 2018. Available online: https://www.wcrf.org/wp-content/uploads/2021/02/Summary-of-Third-Expert-Report-2018.pdf (accessed on 20 February 2022).
- Lichty, B.D.; Breitbach, C.J.; Stojdl, D.F.; Bell, J.C. Going viral with cancer immunotherapy. Nat. Cancer 2014, 14, 559–567. [Google Scholar] [CrossRef]
- Pikor, L.A.; Bell, J.C.; Diallo, J.-S. Oncolytic Viruses: Exploiting Cancer’s Deal with the Devil. Trends Cancer 2015, 1, 266–277. [Google Scholar] [CrossRef]
- Zapatka, M.; Pathogens, P.; Borozan, I.; Brewer, D.S.; Iskar, M.; Grundhoff, A.; Alawi, M.; Desai, N.; Sültmann, H.; Moch, H.; et al. The landscape of viral associations in human cancers. Nat. Genet. 2020, 52, 320–330. [Google Scholar] [CrossRef]
- Rascovan, N.; Duraisamy, R.; Desnues, C. Metagenomics and the Human Virome in Asymptomatic Individuals. Annu. Rev. Microbiol. 2016, 70, 125–141. [Google Scholar] [CrossRef]
- Liang, G.; Bushman, F.D. The human virome: Assembly, composition and host interactions. Nat. Rev. Genet. 2021, 19, 514–527. [Google Scholar] [CrossRef]
- Kuss-Duerkop, S.K.; Westrich, J.A.; Pyeon, D. DNA Tumor Virus Regulation of Host DNA Methylation and Its Implications for Immune Evasion and Oncogenesis. Viruses 2018, 10, 82. [Google Scholar] [CrossRef] [PubMed]
- Shahid, A.; Ali, S.; Zahra, T.; Raza, M.; Shahid, A.; Saeed, M.U.; Javaid, F. Influence of Microbes in Progression of Cancer and DNA Damaging Effects. Haya Saudi J. Life Sci. 2020, 5, 246–252. [Google Scholar] [CrossRef]
- Morales-Sánchez, A.; Fuentes-Pananá, E.M. Human Viruses and Cancer. Viruses 2014, 6, 4047–4079. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.; Serrano, M. Senescence in tumours: Evidence from mice and humans. Nat. Cancer 2010, 10, 51–57. [Google Scholar] [CrossRef]
- Burkhart, D.L.; Sage, J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Cancer 2008, 8, 671–682. [Google Scholar] [CrossRef]
- Guven-Maiorov, E.; Tsai, C.-J.; Nussinov, R. Oncoviruses Can Drive Cancer by Rewiring Signaling Pathways through Interface Mimicry. Front. Oncol. 2019, 9, 1236. [Google Scholar] [CrossRef]
- Krump, N.A.; You, J. Molecular mechanisms of viral oncogenesis in humans. Nat. Rev. Genet. 2018, 16, 684–698. [Google Scholar] [CrossRef]
- Vescovo, T.; Pagni, B.; Piacentini, M.; Fimia, G.M.; Antonioli, M. Regulation of Autophagy in Cells Infected With Oncogenic Human Viruses and Its Impact on Cancer Development. Front. Cell Dev. Biol. 2020, 8, 47. [Google Scholar] [CrossRef]
- Meurs, E.; Chong, K.; Galabru, J.; Thomas, N.B.; Kerr, I.M.; Williams, B.R.G.; Hovanessian, A.G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 1990, 62, 379–390. [Google Scholar] [CrossRef]
- Elde, N.C.; Child, S.J.; Geballe, A.P.; Malik, H.S. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature 2008, 457, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Epstein, M.; Achong, B.; Barr, Y. Virus particles in cultured lymphoblasts from burkitt’s lymphoma. Lancet 1964, 1, 702–703. [Google Scholar] [CrossRef]
- Weidner-Glunde, M.; Kruminis-Kaszkiel, E.; Savanagouder, M. Herpesviral Latency—Common Themes. Pathogens 2020, 9, 125. [Google Scholar] [CrossRef] [PubMed]
- Gulley, M.L.; Tang, W. Laboratory Assays for Epstein-Barr Virus-Related Disease. J. Mol. Diagn. 2008, 10, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Farrell, P.J. Epstein–Barr Virus and Cancer. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 29–53. [Google Scholar] [CrossRef] [PubMed]
- Dunmire, S.K.; Verghese, P.S.; Balfour, H.H. Primary Epstein-Barr Virus Infection. J. Clin. Virol. 2018, 102, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Bogolyubova, A.V. Human Oncogenic Viruses: Old Facts and New Hypotheses. Mol. Biol. 2019, 53, 767–775. [Google Scholar] [CrossRef]
- Vereide, D.; Sugden, B. Insights into the Evolution of Lymphomas Induced by Epstein–Barr Virus. Adv. Cancer Res. 2010, 108, 1–19. [Google Scholar] [CrossRef]
- Vereide, D.T.; Sugden, B. Lymphomas differ in their dependence on Epstein-Barr virus. Blood 2011, 117, 1977–1985. [Google Scholar] [CrossRef]
- Young, L.S.; Rickinson, A.B. Epstein-Barr Virus: 40 Years On. Nat. Rev. Cancer 2004, 4, 757–768. [Google Scholar] [CrossRef]
- Shibata, D.; Tokunaga, M.; Uemura, Y.; Sato, E.; Tanaka, S.; Weiss, L.M. Association of Epstein-Barr virus with undifferentiated gastric carcinomas with intense lymphoid infiltration. Lymphoepithelioma-like carcinoma. Am. J. Pathol. 1991, 139, 469–474. [Google Scholar]
- Nishikawa, J.; Yoshiyama, H.; Iizasa, H.; Kanehiro, Y.; Nakamura, M.; Nishimura, J.; Saito, M.; Okamoto, T.; Sakai, K.; Suehiro, Y.; et al. Epstein-Barr Virus in Gastric Carcinoma. Cancers 2014, 6, 2259–2274. [Google Scholar] [CrossRef]
- Ignatova, E.; Seriak, D.; Fedyanin, M.; Tryakin, A.; Pokataev, I.; Menshikova, S.; Vakhabova, Y.; Smirnova, K.; Tjulandin, S.; Ajani, J.A. Epstein–Barr virus-associated gastric cancer: Disease that requires special approach. Gastric Cancer 2020, 23, 951–960. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Dang, S.; Hou, P. Gene methylation in gastric cancer. Clin. Chim. Acta 2013, 424, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Su, X.; Choi, G.C.G.; Cao, Y.; Ambinder, R.F.; Tao, Q. Methylation profiling of Epstein-Barr virus immediate-early gene promoters, BZLF1 and BRLF1in tumors of epithelial, NK- and B-cell origins. BMC Cancer 2012, 12, 125. [Google Scholar] [CrossRef] [PubMed]
- Geddert, H.; Hausen, A.Z.; Gabbert, H.E.; Sarbia, M. EBV-infection in cardiac and non-cardiac gastric adenocarcinomas is associated with promoter methylation of p16, p14 and APC, but not hMLH1. Cell. Oncol. 2011, 34, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.-S.; Uozaki, H.; Chong, J.-M.; Ushiku, T.; Sakuma, K.; Ishikawa, S.; Hino, R.; Barua, R.R.; Iwasaki, Y.; Arai, K.; et al. CpG Island M’ethylation Status in Gastric Carcinoma with and without Infection of Epstein-Barr Virus. Clin. Cancer Res. 2006, 12, 2995–3002. [Google Scholar] [CrossRef]
- Liang, Q.; Yao, X.; Tang, S.; Zhang, J.; Yau, T.O.; Li, X.; Tang, C.-M.; Kang, W.; Lung, R.W.; Li, J.W.; et al. Integrative Identification of Epstein–Barr Virus–Associated Mutations and Epigenetic Alterations in Gastric Cancer. Gastroenterology 2014, 147, 1350–1362.e4. [Google Scholar] [CrossRef]
- Woellmer, A.; Hammerschmidt, W. Epstein-Barr Virus and Host Cell Methylation: Regulation of Latency, Replication and Virus Reactivation. Current Opin. Virol. 2013, 3, 260–265. [Google Scholar] [CrossRef]
- Niller, H.H.; Tarnai, Z.; Decsi, G.; Zsedényi, Á.; Bánáti, F.; Minarovits, J. Role of epigenetics in EBV regulation and pathogenesis. Futur. Microbiol. 2014, 9, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Nakamura, M.; Nishikawa, J.; Sakai, K.; Zhang, Y.; Saito, M.; Morishige, A.; Oga, A.; Sasaki, K.; Suehiro, Y.; et al. Identification of genes specifically methylated in Epstein-Barr virus-associated gastric carcinomas. Cancer Sci. 2013, 104, 1309–1314. [Google Scholar] [CrossRef] [PubMed]
- Ryan, J.L.; Jones, R.J.; Kenney, S.C.; Rivenbark, A.G.; Tang, W.; Knight, E.R.; Coleman, W.B.; Gulley, M.L. Epstein-Barr virus-specific methylation of human genes in gastric cancer cells. Infect. Agents Cancer 2010, 5, 27. [Google Scholar] [CrossRef] [PubMed]
- Akinyemiju, T.; Abera, S.; Ahmed, M.; Alam, N.; Alemayohu, M.A.; Allen, C.; Al-Raddadi, R.; Alvis-Guzman, N.; Amoako, Y.; Artaman, A.; et al. The Burden of Primary Liver Cancer and Underlying Etiologies from 1990 to 2015 at the Global, Regional, and National Level: Results from the Global Burden of Disease Study 2015. JAMA Oncol. 2017, 3, 1683–1691. [Google Scholar]
- Wong, Y.; Meehan, M.T.; Burrows, S.R.; Doolan, D.L.; Miles, J.J. Estimating the global burden of Epstein–Barr virus-related cancers. J. Cancer Res. Clin. Oncol. 2021, 148, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Z.-H.; Jiang, F. Hepatitis B virus infection increases the risk of pancreatic cancer: A meta-analysis. Scand. J. Gastroenterol. 2021, 56, 252–258. [Google Scholar] [CrossRef] [PubMed]
- Su, F.-H.; Bai, C.-H.; Le, T.N.; Muo, C.-H.; Chang, S.-N.; Te, A.; Sung, F.-C.; Yeh, C.-C. Patients with Chronic Hepatitis C Virus Infection Are at an Increased Risk of Colorectal Cancer: A Nationwide Population-Based Case-Control Study in Taiwan. Front. Oncol. 2021, 10, 3002. [Google Scholar] [CrossRef]
- Cui, H.; Jin, Y.; Chen, F.; Ni, H.; Hu, C.; Xu, Y.; Xuan, H.; Hu, D.; Deng, W.; Zhang, Y.; et al. Clinicopathological evidence of hepatitis B virus infection in the development of gastric adenocarcinoma. J. Med Virol. 2019, 92, 71–77. [Google Scholar] [CrossRef]
- An, J.; Kim, J.W.; Shim, J.H.; Han, S.; Yu, C.S.; Choe, J.; Lee, D.; Kim, K.M.; Lim, Y.-S.; Chung, Y.-H.; et al. Chronic hepatitis B infection and non-hepatocellular cancers: A hospital registry-based, case-control study. PLoS ONE 2018, 13, e0193232. [Google Scholar] [CrossRef]
- Baghbanian, M.; Mousa, S.A.H.; Doosti, M.; Moghimi, M. Association between Gastric Pathology and Hepatitis B Virus Infection in Patients with or without Helicobacter Pylori. Asian Pac. J. Cancer Prev. 2019, 20, 2177–2180. [Google Scholar] [CrossRef]
- Ghasemi, M.; Vahedi Larijani, L.; Abediankenari, S. Investigation of Relationship between Hepatitis B Virus and Gastric Ad-enocarcinoma. Iran. Red Crescent Med. J. 2012, 14, 453. [Google Scholar] [PubMed]
- He, Y.; Mao, M.; Shi, W.; He, Z.; Zhang, L.; Wang, X. Development and validation of a prognostic nomogram in gastric cancer with hepatitis B virus infection. J. Transl. Med. 2019, 17, 98. [Google Scholar] [CrossRef] [PubMed]
- Shalapour, S.; Lin, X.J.; Bastian, I.N.; Brain, J.; Burt, A.D.; Aksenov, A.A.; Vrbanac, A.F.; Li, W.; Perkins, A.; Matsutani, T.; et al. Inflammation-induced IgA+ cells dismantle anti-liver cancer immunity. Nature 2017, 551, 340–345. [Google Scholar] [CrossRef] [PubMed]
- Vasmehjani, A.A.; Javeshghani, D.; Baharlou, R.; Shayestehpour, M.; Mousavinasab, S.D.; Joharinia, N.; Enderami, S.E. Hepatitis A infection in patients with chronic viral liver disease: A cross-sectional study in Jahrom, Iran. Epidemiol. Infect. 2014, 143, 534–539. [Google Scholar] [CrossRef]
- Lu, T.; Yang, Q.; Li, M.; Zhang, J.; Zou, J.; Huang, L.; Lin, J.; Jin, H.; He, J. HBV infection and extra-hepatic cancers in adolescents and 20s: A retrospective study in China. Cancer Epidemiol. 2018, 55, 149–155. [Google Scholar] [CrossRef]
- Wei, X.-L.; Qiu, M.-Z.; Jin, Y.; Huang, Y.-X.; Wang, R.-Y.; Chen, W.-W.; Wang, D.-S.; Wang, F.-H.; Luo, H.-Y.; Zhang, D.-S.; et al. Hepatitis B virus infection is associated with gastric cancer in China: An endemic area of both diseases. Br. J. Cancer 2015, 112, 1283–1290. [Google Scholar] [CrossRef]
- Russell, S.J.; Peng, K.-W.; Bell, J.C. Oncolytic virotherapy. Nat. Biotechnol. 2012, 30, 658–670. [Google Scholar] [CrossRef]
- Tripodi, L.; Vitale, M.; Cerullo, V.; Pastore, L. Oncolytic Adenoviruses for Cancer Therapy. Int. J. Mol. Sci. 2021, 22, 2517. [Google Scholar] [CrossRef]
- Aldrak, N.; Alsaab, S.; Algethami, A.; Bhere, D.; Wakimoto, H.; Shah, K.; Alomary, M.; Zaidan, N. Oncolytic Herpes Simplex Virus-Based Therapies for Cancer. Cells 2021, 10, 1541. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, F. Advances and Potential Pitfalls of Oncolytic Viruses Expressing Immunomodulatory Transgene Therapy for Malignant Gliomas. Cell Death Dis. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Kaufman, H.L.; Andtbacka, R.H.I.; Collichio, F.A.; Wolf, M.; Zhao, Z.; Shilkrut, M.; Puzanov, I.; Ross, M. Durable response rate as an endpoint in cancer immunotherapy: Insights from oncolytic virus clinical trials. J. Immunother. Cancer 2017, 5, 72. [Google Scholar] [CrossRef] [PubMed]
- Kelly, E.; Russell, S.J. History of Oncolytic Viruses: Genesis to Genetic Engineering. Mol. Ther. 2007, 15, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Jun, K.-H.; Gholami, S.; Song, T.-J.; Au, J.; Haddad, D.; Carson, J.; Chen, C.-H.; Mojica, K.; Zanzonico, P.; Chen, N.G.; et al. A novel oncolytic viral therapy and imaging technique for gastric cancer using a genetically engineered vaccinia virus carrying the human sodium iodide symporter. J. Exp. Clin. Cancer Res. 2014, 33, 2. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, D.C.; Shevde, L.A. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019, 79, 4557–4566. [Google Scholar] [CrossRef]
- Arneth, B. Tumor Microenvironment. Medicina 2019, 56, 15. [Google Scholar] [CrossRef]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-Related Inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, Inflammation, and Cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef]
- Doronin, K.; Toth, K.; Kuppuswamy, M.; Ward, P.; Tollefson, A.E.; Wold, W.S.M. Tumor-Specific, Replication-Competent Adenovirus Vectors Overexpressing the Adenovirus Death Protein. J. Virol. 2000, 74, 6147–6155. [Google Scholar] [CrossRef]
- Kaufman, H.L.; Kohlhapp, F.J.; Zloza, A. Oncolytic viruses: A new class of immunotherapy drugs. Nat. Rev. Drug Discov. 2015, 14, 642–662. [Google Scholar] [CrossRef]
- Wang, G.; Kang, X.; Chen, K.S.; Jehng, T.; Jones, L.; Chen, J.; Huang, X.F.; Chen, S.-Y. An engineered oncolytic virus expressing PD-L1 inhibitors activates tumor neoantigen-specific T cell responses. Nat. Commun. 2020, 11, 1395. [Google Scholar] [CrossRef]
- Andtbacka, R.H.I.; Collichio, F.; Harrington, K.J.; Middleton, M.R.; Downey, G.; Öhrling, K.; Kaufman, H.L. Final analyses of OPTiM: A randomized phase III trial of talimogene laherparepvec versus granulocyte-macrophage colony-stimulating factor in unresectable stage III–IV melanoma. J. Immunother. Cancer 2019, 7, 145. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Kim, J.-H.; Shin, H.-Y.; Lee, H.; Yang, J.M.; Kim, J.; Sohn, J.-H.; Kim, H.; Yun, C.-O. Ad-mTERT-Δ19, a Conditional Replication-Competent Adenovirus Driven by the Human Telomerase Promoter, Selectively Replicates in and Elicits Cytopathic Effect in a Cancer Cell-Specific Manner. Hum. Gene Ther. 2003, 14, 1415–1428. [Google Scholar] [CrossRef] [PubMed]
- Wirth, T.; Zender, L.; Schulte, B.; Mundt, B.; Plentz, R.; Lenhard Rudolph, K.; Manns, M.; Kubicka, S.; Kühnel, F. A Telomer-ase-Dependent Conditionally Replicating Adenovirus for Selective Treatment of Cancer. Cancer Res. 2003, 63, 3181–3188. [Google Scholar] [PubMed]
- Parato, K.A.; Breitbach, C.J.; Le Boeuf, F.; Wang, J.; Storbeck, C.; Ilkow, C.; Diallo, J.-S.; Falls, T.; Burns, J.; Garcia, V.; et al. The Oncolytic Poxvirus JX-594 Selectively Replicates in and Destroys Cancer Cells Driven by Genetic Pathways Commonly Activated in Cancers. Mol. Ther. 2012, 20, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Martin-Vilchez, S.; Lara-Pezzi, E.; Trapero-Marugán, M.; Moreno-Otero, R.; Sanz-Cameno, P. The Molecular and Patho-physiological Implications of Hepatitis B X Antigen in Chronic Hepatitis B Virus Infection. Rev. Med. Virol. 2011, 21, 315–329. [Google Scholar]
- Petrovic, B.; Leoni, V.; Gatta, V.; Zaghini, A.; Vannini, A.; Campadelli-Fiume, G. Dual Ligand Insertion in gB and gD of Oncolytic Herpes Simplex Viruses for Retargeting to a Producer Vero Cell Line and to Cancer Cells. J. Virol. 2018, 92, e02122-17. [Google Scholar] [CrossRef]
- Bhatia, S.; O’Bryan, S.M.; Rivera, A.; Curiel, D.T.; Mathis, J.M. CXCL12 retargeting of an adenovirus vector to cancer cells using a bispecific adapter. Oncol. Virotherapy 2016, 5, 99–113. [Google Scholar] [CrossRef]
- Puhlmann, M.; Gnant, M.; Brown, C.K.; Alexander, H.R.; Bartlett, D.L. Thymidine Kinase-Deleted Vaccinia Virus Expressing Purine Nucleoside Phosphorylase as a Vector for Tumor-Directed Gene Therapy. Hum. Gene Ther. 1999, 10, 649–657. [Google Scholar] [CrossRef]
- Toth, K.; Dhar, D.; Wold, W.S. Oncolytic (replication-competent) adenoviruses as anticancer agents. Expert Opin. Biol. Ther. 2010, 10, 353–368. [Google Scholar] [CrossRef]
- Freytag, S.O.; Rogulski, K.R.; Paielli, D.L.; Gilbert, J.D.; Kim, J.H. A Novel Three-Pronged Approach to Kill Cancer Cells Selectively: Concomitant Viral, Double Suicide Gene, and Radiotherapy. Hum. Gene Ther. 1998, 9, 1323–1333. [Google Scholar] [CrossRef]
- Freytag, S.O.; Stricker, H.; Pegg, J.; Paielli, D.; Pradhan, D.G.; Peabody, J.; DePeralta-Venturina, M.; Xia, X.; Brown, S.; Lu, M.; et al. Phase I study of replication-competent adenovirus-mediated double-suicide gene therapy in combination with conventional-dose three-dimensional conformal radiation therapy for the treatment of newly diagnosed, intermediate- to high-risk prostate cancer. Cancer Res. 2003, 63, 7497–7506. [Google Scholar] [PubMed]
- Foloppe, J.; Kintz, J.; Futin, N.; Findeli, A.; Cordier, P.; Schlesinger, Y.; Hoffmann, C.; Tosch, C.; Balloul, J.-M.; Erbs, P. Targeted delivery of a suicide gene to human colorectal tumors by a conditionally replicating vaccinia virus. Gene Ther. 2008, 15, 1361–1371. [Google Scholar] [CrossRef] [PubMed]
- Sova, P.; Ren, X.-W.; Ni, S.; Bernt, K.M.; Mi, J.; Kiviat, N.; Lieber, A. A Tumor-Targeted and Conditionally Replicating Oncolytic Adenovirus Vector Expressing TRAIL for Treatment of Liver Metastases. Mol. Ther. 2004, 9, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Hirvinen, M.; Rajecki, M.; Kapanen, M.; Parviainen, S.; Rouvinen-Lagerström, N.; Diaconu, I.; Nokisalmi, P.; Tenhunen, M.; Hemminki, A.; Cerullo, V. Immunological Effects of a Tumor Necrosis Factor Alpha–Armed Oncolytic Adenovirus. Hum. Gene Ther. 2015, 26, 134–144. [Google Scholar] [CrossRef]
- Freytag, S.O.; Khil, M.; Stricker, H.; Peabody, J.; Menon, M.; DePeralta-Venturina, M.; Nafziger, D.; Pegg, J.; Paielli, D.; Brown, S.; et al. Phase I study of replication-competent adenovirus-mediated double suicide gene therapy for the treatment of locally recurrent prostate cancer. Cancer Res. 2002, 62, 4968–4976. [Google Scholar] [PubMed]
- Kubo, H.; Gardner, T.A.; Wada, Y.; Koeneman, K.S.; Gotoh, A.; Yang, L.; Kao, C.; Lim, S.D.; Amin, M.B.; Yang, H.; et al. Phase I Dose Escalation Clinical Trial of Adenovirus Vector Carrying Osteocalcin Promoter-Driven Herpes Simplex Virus Thymidine Kinase in Localized and Metastatic Hormone-Refractory Prostate Cancer. Hum. Gene Ther. 2003, 14, 227–241. [Google Scholar] [CrossRef]
- Chiocca, E.A.; Rabkin, S.D. Oncolytic Viruses and Their Application to Cancer Immunotherapy. Cancer Immunol. Res. 2014, 2, 295–300. [Google Scholar] [CrossRef]
- Khare, R.; May, S.M.; Vetrini, F.; Weaver, E.A.; Palmer, D.; Rosewell, A.; Grove, N.; Ng, P.; Barry, M.A. Generation of a Kupffer Cell-evading Adenovirus for Systemic and Liver-directed Gene Transfer. Mol. Ther. 2011, 19, 1254–1262. [Google Scholar] [CrossRef]
- Marchini, A.; Daeffler, L.; Pozdeev, V.I.; Angelova, A.; Rommelaere, J. Immune Conversion of Tumor Microenvironment by Oncolytic Viruses: The Protoparvovirus H-1PV Case Study. Front. Immunol. 2019, 10, 1848. [Google Scholar] [CrossRef]
- Conlon, K.C.; Miljkovic, M.D.; Waldmann, T.A. Cytokines in the Treatment of Cancer. J. Interferon Cytokine Res. 2019, 39, 6–21. [Google Scholar] [CrossRef]
- Kim, J.-H.; Lee, K.-J.; Lee, S.-W. Cancer immunotherapy with T-cell targeting cytokines: IL-2 and IL-7. BMB Rep. 2021, 54, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Xiao, Z.; Zhao, Q.; Li, M.; Wu, X.; Zhang, L.; Hu, W.; Cho, C.H. Anti-Cancer Therapy with TNFα and IFNγ: A Comprehensive Review. Cell Prolif. 2018, 51, 7497–7506. [Google Scholar] [CrossRef] [PubMed]
- Zamarin, D.; Holmgaard, R.B.; Subudhi, S.K.; Park, J.S.; Mansour, M.; Palese, P.; Merghoub, T.; Wolchok, J.D.; Allison, J.P. Localized Oncolytic Virotherapy Overcomes Systemic Tumor Resistance to Immune Checkpoint Blockade Immunotherapy. Sci. Transl. Med. 2014, 6, 226ra32. [Google Scholar] [CrossRef] [PubMed]
- Kärre, K.; Ljunggren, H.G.; Piontek, G.; Kiessling, R. Selective rejection of H–2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 1986, 319, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Ljunggren, H.G.; Kärre, K. Host resistance directed selectively against H-2-deficient lymphoma variants. Analysis of the mechanism. J. Exp. Med. 1985, 162, 1745–1759. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; He, H.; Wang, H. Oncolytic herpes simplex virus and immunotherapy. BMC Immunol. 2018, 19, 40. [Google Scholar] [CrossRef]
- Watson, G.; Xu, W.; Reed, A.; Babra, B.; Putman, T.; Wick, E.; Wechsler, S.; Rohrmann, G.; Jin, L. Sequence and comparative analysis of the genome of HSV-1 strain McKrae. Virology 2012, 433, 528–537. [Google Scholar] [CrossRef][Green Version]
- Menotti, L.; Avitabile, E. Herpes Simplex Virus Oncolytic Immunovirotherapy: The Blossoming Branch of Multimodal Therapy. Int. J. Mol. Sci. 2020, 21, 8310. [Google Scholar] [CrossRef]
- Mondal, M.; Guo, J.; He, P.; Zhou, D. Recent advances of oncolytic virus in cancer therapy. Hum. Vaccines Immunother. 2020, 16, 2389–2402. [Google Scholar] [CrossRef]
- Liu, B.L.; Robinson, M.; Han, Z.-Q.; Branston, R.H.; English, C.; Reay, P.; McGrath, Y.; Thomas, S.K.; Thornton, M.; Bullock, P.; et al. ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther. 2003, 10, 292–303. [Google Scholar] [CrossRef]
- Koch, M.S.; Lawler, S.E.; Chiocca, E.A. HSV-1 Oncolytic Viruses from Bench to Bedside: An Overview of Current Clinical Trials. Cancers 2020, 12, 3514. [Google Scholar] [CrossRef] [PubMed]
- Bommareddy, P.K.; Patel, A.; Hossain, S.; Kaufman, H.L. Talimogene Laherparepvec (T-VEC) and Other Oncolytic Viruses for the Treatment of Melanoma. Am. J. Clin. Dermatol. 2016, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Penheiter, A.R.; Wegman, T.R.; Classic, K.L.; Dingli, D.; Bender, C.E.; Russell, S.J.; Carlson, S.K. Sodium Iodide Symporter (NIS)-Mediated Radiovirotherapy for Pancreatic Cancer. Am. J. Roentgenol. 2010, 195, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, K.; Iwai, M.; Yajima, S.; Tanaka, M.; Yanagihara, K.; Seto, Y.; Todo, T. Efficacy of a Third-Generation Oncolytic Herpes Virus G47Δ in Advanced Stage Models of Human Gastric Cancer. Mol. Ther. Oncolytics 2020, 17, 205–215. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, T.; Nakamori, M.; Iwahashi, M.; Nakamura, M.; Ojima, T.; Iida, T.; Katsuda, M.; Hayata, K.; Ino, Y.; Todo, T.; et al. An armed oncolytic herpes simplex virus expressing thrombospondin-1 has an enhancedin vivoantitumor effect against human gastric cancer. Int. J. Cancer 2012, 132, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Nakamori, M.; Matsumura, S.; Nakamura, M.; Ojima, T.; Fukuhara, H.; Ino, Y.; Todo, T.; Yamaue, H. Oncolytic virotherapy with human telomerase reverse transcriptase promoter regulation enhances cytotoxic effects against gastric cancer. Oncol. Lett. 2021, 21, 490. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, S.; Nakamori, M.; Tsuji, T.; Kato, T.; Nakamura, M.; Ojima, T.; Fukuhara, H.; Ino, Y.; Todo, T.; Yamaue, H. Oncolytic virotherapy with SOCS3 enhances viral replicative potency and oncolysis for gastric cancer. Oncotarget 2021, 12, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Zhang, T.; Zhou, S.; Hu, H.; Li, J.; Huang, K.; Lei, Y.; Wang, K.; Zhao, Y.; Liu, R.; et al. Proteomic Analyses of Gastric Cancer Cells Treated with Vesicular Stomatitis Virus Matrix Protein. J. Protein Chem. 2011, 30, 308–317. [Google Scholar] [CrossRef]
- Broyles, S.S. Vaccinia Virus Transcription. J. Gen. Virol. 2003, 84, 2293–2303. [Google Scholar] [CrossRef]
- Huang, H.; Hu, X.-F.; Zhao, F.-H.; Garland, S.M.; Bhatla, N.; Qiao, Y.-L. Estimation of Cancer Burden Attributable to Infection in Asia. J. Epidemiology 2015, 25, 626–638. [Google Scholar] [CrossRef]
- Niedźwiedzka-Rystwej, P.; Grywalska, E.; Hrynkiewicz, R.; Wołącewicz, M.; Becht, R.; Roliński, J. The Double-Edged Sword Role of Viruses in Gastric Cancer. Cancers 2020, 12, 1680. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Firoz, A.; Ali, H.M.; Rehman, S.; Rather, I.A. Gastric Cancer and Viruses: A Fine Line between Friend or Foe. Vaccines 2022, 10, 600. https://doi.org/10.3390/vaccines10040600
Firoz A, Ali HM, Rehman S, Rather IA. Gastric Cancer and Viruses: A Fine Line between Friend or Foe. Vaccines. 2022; 10(4):600. https://doi.org/10.3390/vaccines10040600
Chicago/Turabian StyleFiroz, Ahmad, Hani Mohammed Ali, Suriya Rehman, and Irfan A. Rather. 2022. "Gastric Cancer and Viruses: A Fine Line between Friend or Foe" Vaccines 10, no. 4: 600. https://doi.org/10.3390/vaccines10040600
APA StyleFiroz, A., Ali, H. M., Rehman, S., & Rather, I. A. (2022). Gastric Cancer and Viruses: A Fine Line between Friend or Foe. Vaccines, 10(4), 600. https://doi.org/10.3390/vaccines10040600





