Comparison of Bacterial Expression Systems Based on Potato Virus Y-like Particles for Vaccine Generation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cloning of the Fel d 1 Gene with a G4S Linker at the 5′ End of the PVY CP Gene
2.2. Cloning of the PVY CP–NG4S Direct Fusion and Mosaic System with Fel d 1
2.3. Cloning of Covalently Binding Protein Partners SpyCatcher and SpyTag
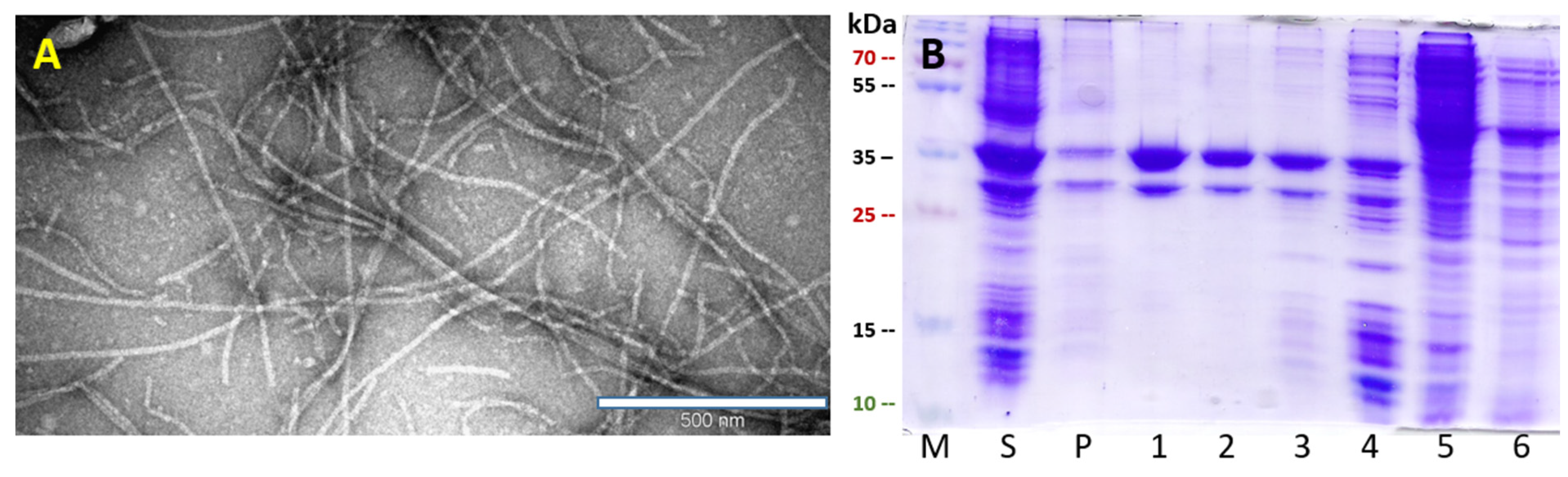
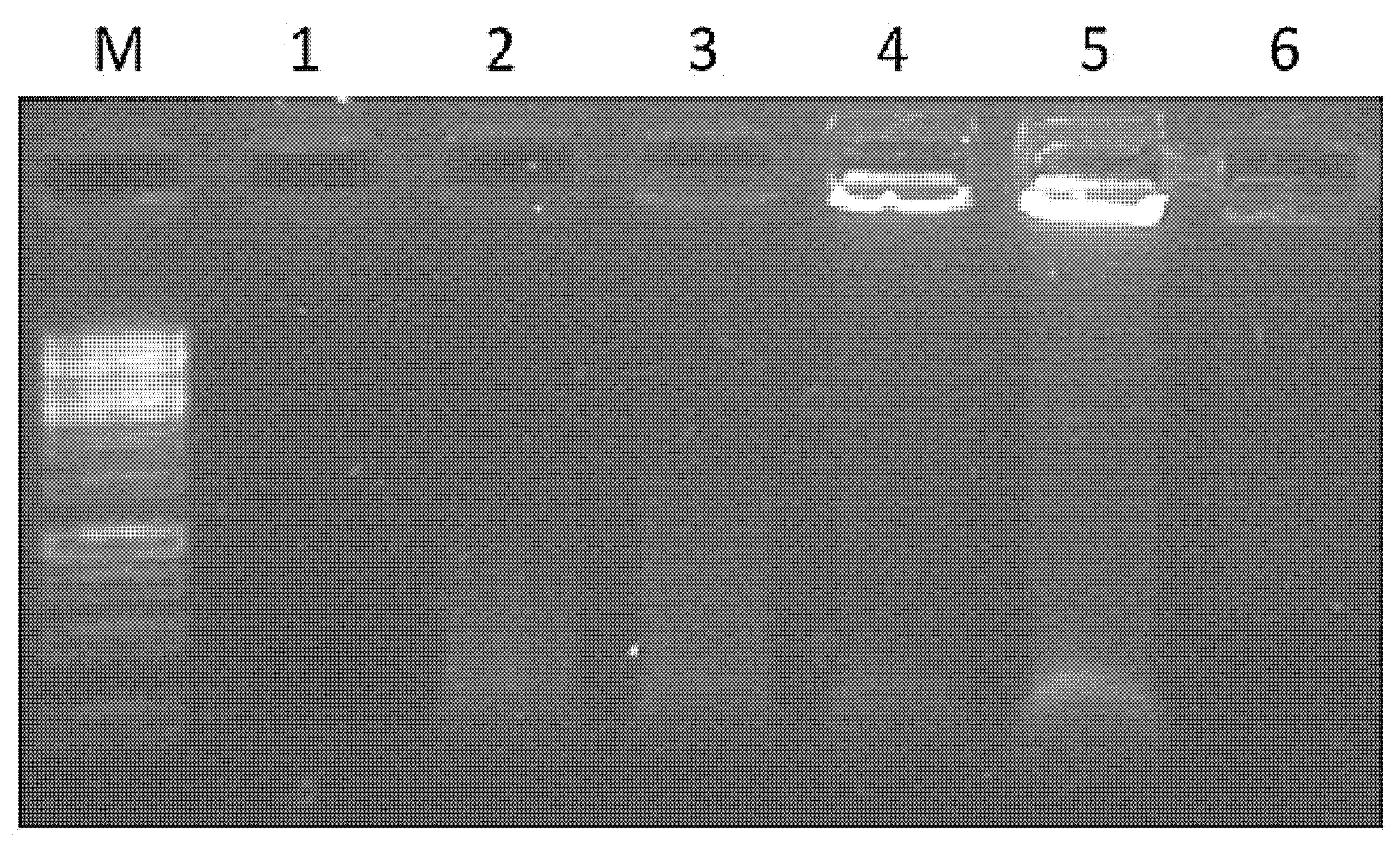
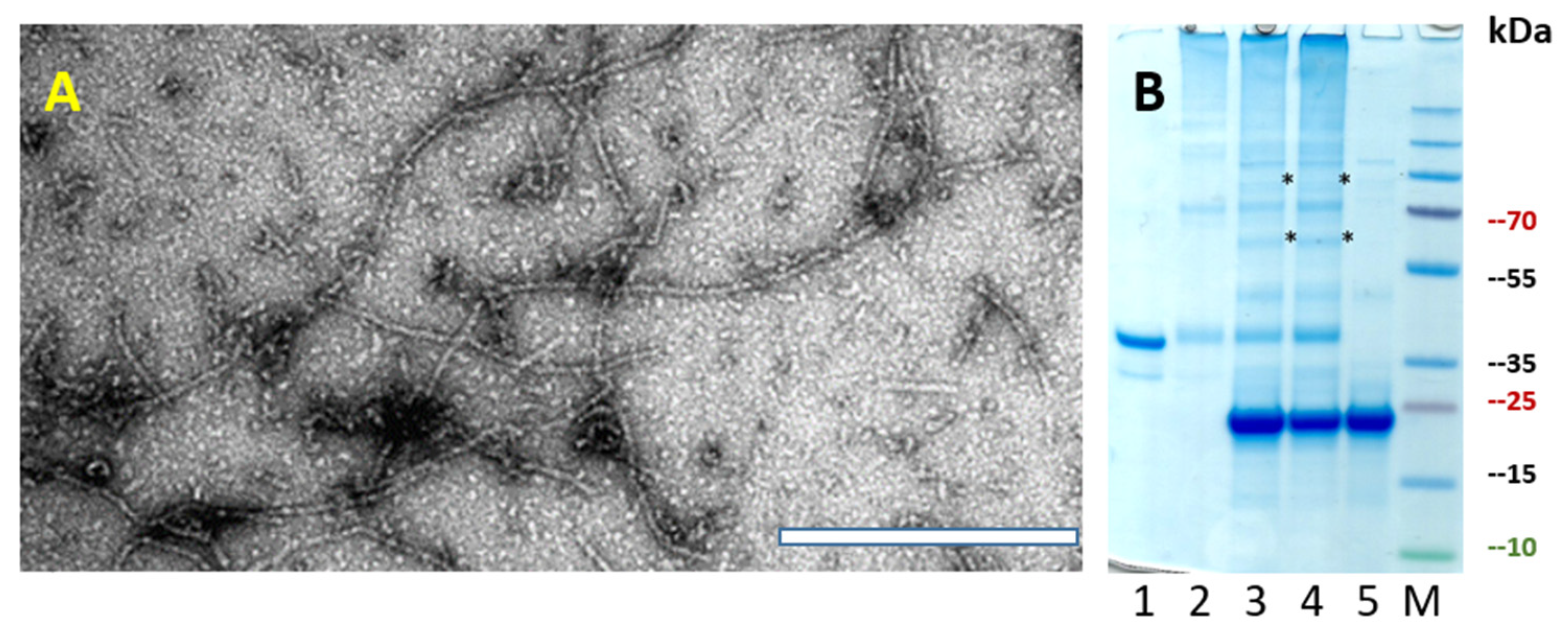
2.4. Expression and Purification of PVY VLPs and PVY Containing Fel d 1 VLPs
2.5. Transmission Electron Microscopy (TEM)
2.6. Chemical Coupling of PVY CP VLPs and Fel d 1
2.7. Immunological Studies
2.7.1. Western Blot (WB) Analysis
2.7.2. Mouse Vaccination
2.7.3. The Enzyme-Linked Immunosorbent Assay (ELISA)
2.7.4. Avidity ELISA
2.7.5. ELISA for Native Fel d 1
3. Results and Discussion
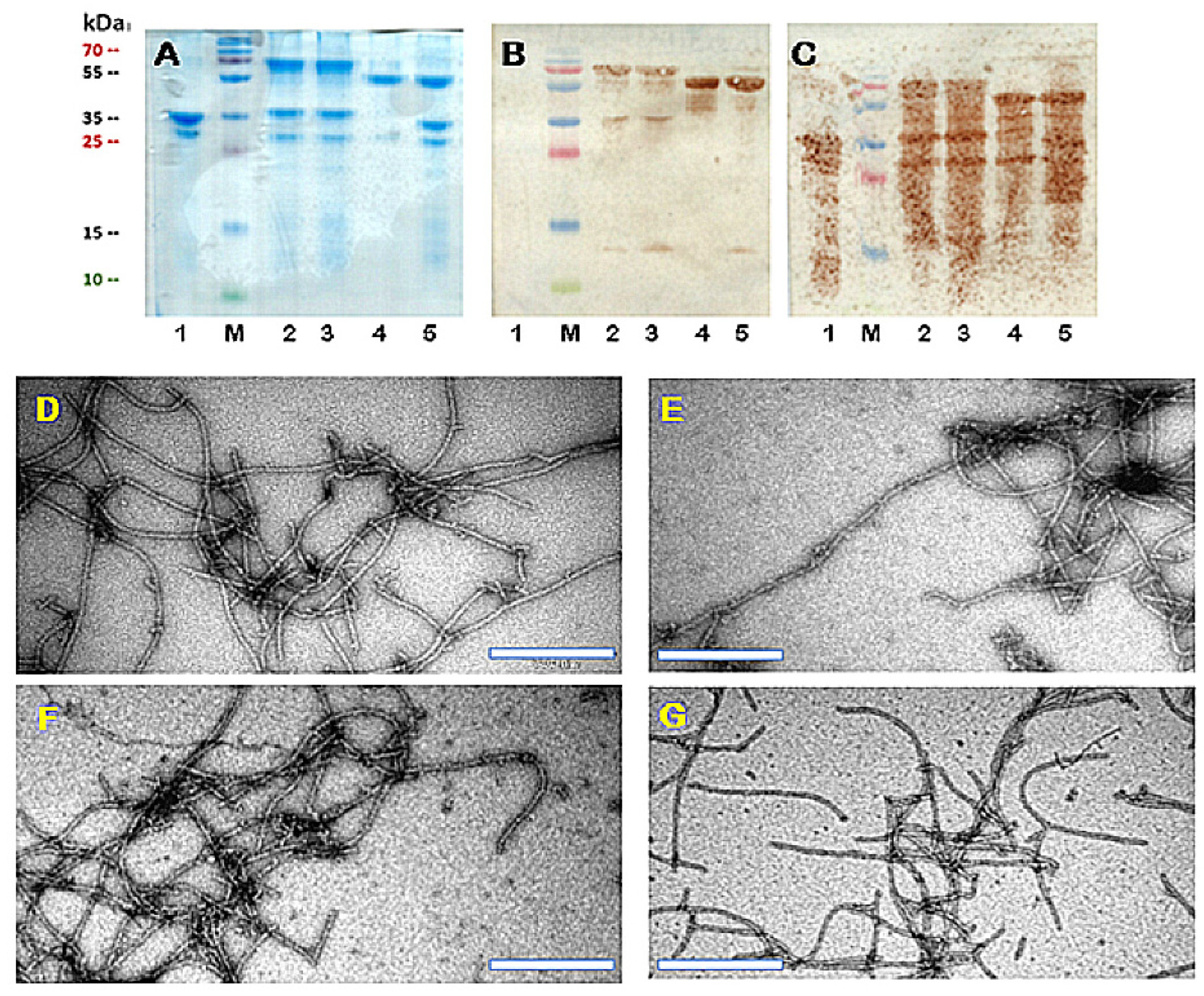
3.1. Construction and Characterization of PVY CP VLPs Containing Fel d 1
3.2. Chemical Coupling of PVY CP with Fel d 1
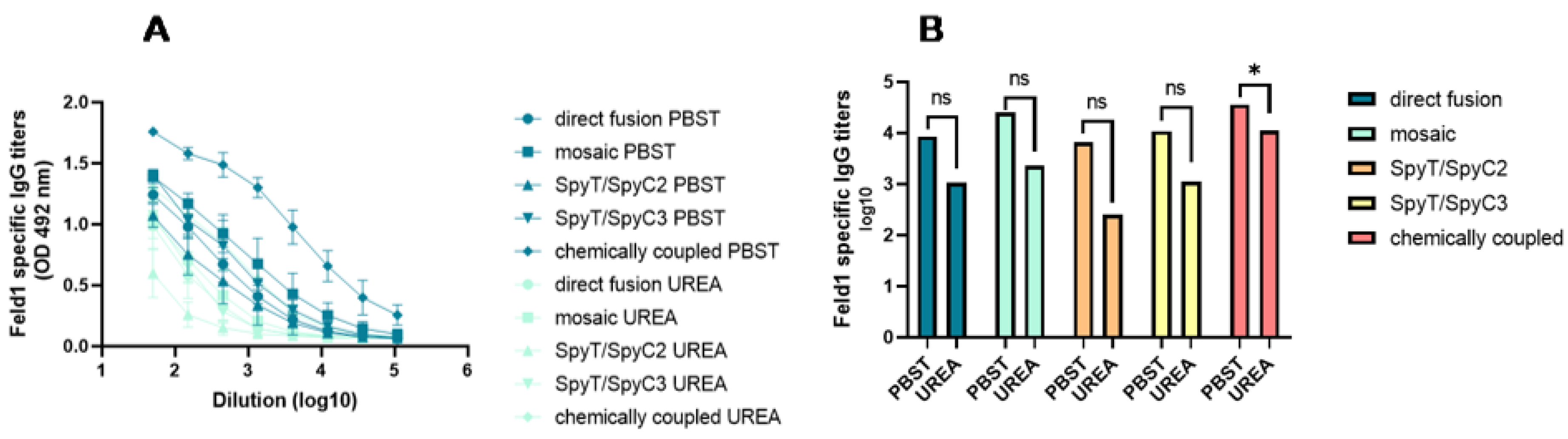
3.3. Immunological Characterization of the PVY–Fel d 1 Vaccine Candidates
3.3.1. Monoclonal Antibody ELISA
3.3.2. Total Levels of IgG
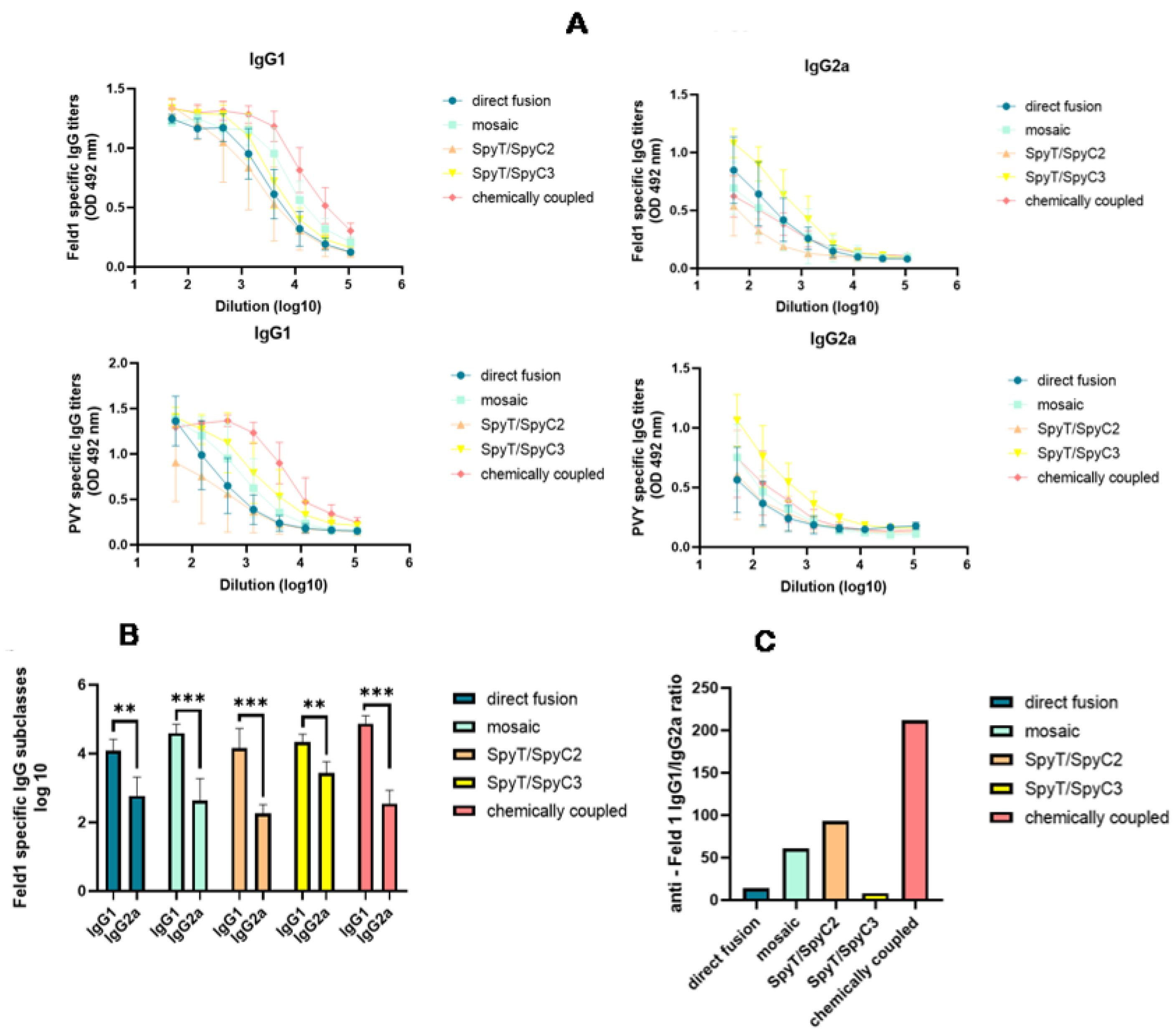
3.3.3. Subclass Specific Antibody Production
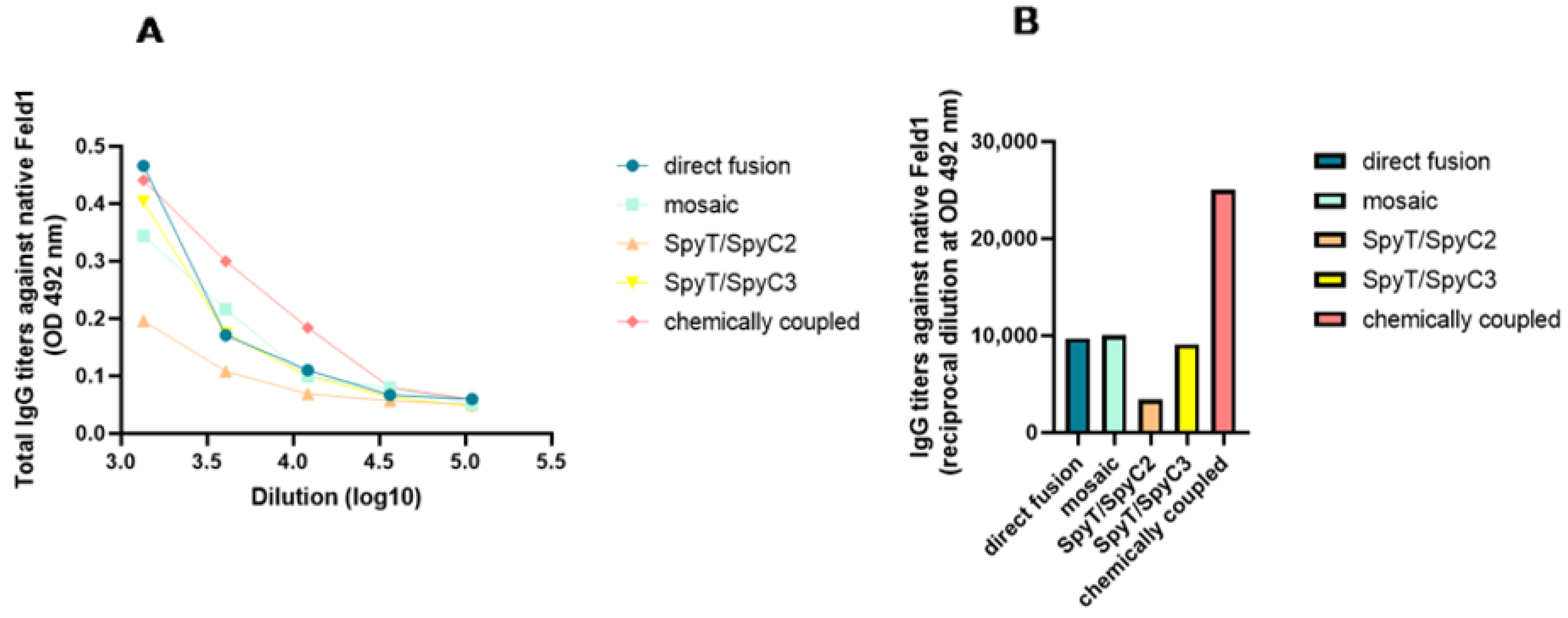
3.3.4. Native Fel d 1 Recognition
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balke, I.; Zeltins, A. Use of plant viruses and virus-like particles for the creation of novel vaccines. Adv. Drug Deliv. Rev. 2019, 145, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Zeltins, A. Viral Nanoparticles: Principles of Construction and Characterization. In Viral Nanotechnology; Khudyakov, Y.E., Pumpens, P., Eds.; CRC Press: Boca Raton, FL, USA, 2016; pp. 93–119. [Google Scholar]
- Lebel, M.E.; Chartrand, K.; Leclerc, D.; Lamarre, A. Plant Viruses as Nanoparticle-Based Vaccines and Adjuvants. Vaccines 2015, 3, 620–637. [Google Scholar] [CrossRef] [PubMed]
- Benne, N.; van Duijn, J.; Kuiper, J.; Jiskoot, W.; Slütter, B. Orchestrating immune responses: How size, shape and rigidity affect the immunogenicity of particulate vaccines. J. Control. Release 2016, 234, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Moyer, T.J.; Zmolek, A.C.; Irvine, D.J. Beyond antigens and adjuvants: Formulating future vaccines. J. Clin. Investig. 2016, 126, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Citiulo, F.; Crosatti, C.; Cattivelli, L.; Biselli, C. Frontiers in the Standardization of the Plant Platform for High Scale Production of Vaccines. Plants 2021, 10, 1828. [Google Scholar] [CrossRef]
- Tariq, H.; Batool, S.; Asif, S.; Ali, M.; Abbasi, B.H. Virus-Like Particles: Revolutionary Platforms for Developing Vaccines against Emerging Infectious Diseases. Front. Microbiol. 2022, 12, 4137. [Google Scholar] [CrossRef]
- Mohsen, M.O.; Balke, I.; Zinkhan, S.; Zeltina, V.; Liu, X.; Chang, X.; Krenger, P.S.; Plattner, K.; Gharailoo, Z.; Vogt, A.S.; et al. A scalable and highly immunogenic virus-like particle-based vaccine against SARS-CoV-2. Allergy 2022, 77, 243–257. [Google Scholar] [CrossRef]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697. [Google Scholar] [CrossRef]
- Brune, K.D.; Leneghan, D.; Brian, I.J.; Ishizuka, A.; Bachmann, M.F.; Draper, S.; Biswas, S.; Howarth, M. Plug-and-Display: Decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Sci. Rep. 2016, 6, 19234. [Google Scholar] [CrossRef]
- Marini, A.; Zhou, Y.; Li, Y.; Taylor, I.; Leneghan, D.B.; Jin, J.; Zaric, M.; Mekhaiel, D.; Long, C.A.; Miura, K.; et al. A Universal Plug-and-Display Vaccine Carrier Based on HBsAg VLP to Maximize Effective Antibody Response. Front. Immunol. 2019, 10, 2931. [Google Scholar] [CrossRef]
- Fredsgaard, L.; Goksøyr, L.; Thrane, S.; Aves, K.-L.; Theander, T.; Sander, A. Head-to-Head Comparison of Modular Vaccines Developed Using Different Capsid Virus-Like Particle Backbones and Antigen Conjugation Systems. Vaccines 2021, 9, 539. [Google Scholar] [CrossRef] [PubMed]
- Chevillard, C.; Amen, A.; Besson, S.; Hannani, D.; Bally, I.; Dettling, V.; Gout, E.; Moreau, C.J.; Buisson, M.; Gallet, S.; et al. Elicitation of potent SARS-CoV-2 neutralizing antibody responses through immunization with a versatile adenovirus-inspired multimerization platform. Mol. Ther. 2022. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhou, H.; Wang, W.; Tan, W.; Fu, Y.-X.; Zhu, M. A novel method for synthetic vaccine construction based on protein assembly. Sci. Rep. 2014, 4, 7266. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, B.; Messaoudi, K.; Jacomet, F.; Michaud, E.; Fauquert, J.L.; Caillaud, D.; Evrard, B. An update on molecular cat allergens: Fel d 1 and what else? Chapter 1: Fel d 1, the major cat allergen. Allergy Asthma Clin. Immunol. 2018, 14, 14. [Google Scholar] [CrossRef]
- Zeltins, A.; West, J.; Zabel, F.; El Turabi, A.; Balke, I.; Haas, S.; Maudrich, M.; Storni, F.; Engeroff, P.; Jennings, G.T.; et al. Incorporation of tetanus-epitope into virus-like particles achieves vaccine responses even in older recipients in models of psoriasis, Alzheimer’s and cat allergy. npj Vaccines 2017, 2, 30. [Google Scholar] [CrossRef]
- Thoms, F.; Haas, S.; Erhart, A.; Nett, C.S.; Rüfenacht, S.; Graf, N.; Strods, A.; Patil, G.; Leenadevi, T.; Fontaine, M.C.; et al. Immunization of Cats against Fel d 1 Results in Reduced Allergic Symptoms of Owners. Viruses 2020, 12, 288. [Google Scholar] [CrossRef]
- Kalnciema, I.; Skrastina, D.; Ose, V.; Pumpens, P.; Zeltins, A. Potato Virus Y-Like Particles as a New Carrier for the Presentation of Foreign Protein Stretches. Mol. Biotechnol. 2011, 52, 129–139. [Google Scholar] [CrossRef]
- Zeltins, A.; Turks, M.; Skrastina, D.; Lugiņina, J.; Kalnciema, I.; Balke, I.; Bizdēna, Ē.; Skrivelis, V. Synthesis and Immunological Evaluation of Virus-Like Particle-Milbemycin A3/A4 Conjugates. Antibiotics 2017, 6, 18. [Google Scholar] [CrossRef]
- Zinkhan, S.; Ogrina, A.; Balke, I.; Reseviča, G.; Zeltins, A.; de Brot, S.; Lipp, C.; Chang, X.; Zha, L.; Vogel, M.; et al. The impact of size on particle drainage dynamics and antibody response. J. Control. Release 2021, 331, 296–308. [Google Scholar] [CrossRef]
- Li, L.; Fierer, J.O.; Rapoport, T.A.; Howarth, M. Structural Analysis and Optimization of the Covalent Association between SpyCatcher and a Peptide Tag. J. Mol. Biol. 2014, 426, 309–317. [Google Scholar] [CrossRef]
- Kalnciema, I.; Balke, I.; Skrastina, D.; Ose, V.; Zeltins, A. Potato Virus M-Like Nanoparticles: Construction and Characterization. Mol. Biotechnol. 2015, 57, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Peyret, H.; Ponndorf, D.; Meshcheriakova, Y.; Richardson, J.; Lomonossoff, G.P. Covalent protein display on Hepatitis B core-like particles in plants through the in vivo use of the SpyTag/SpyCatcher system. Sci. Rep. 2020, 10, 17095. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Pan, C.; Sun, P.; Peng, Z.; Feng, E.; Wu, J.; Wang, H.; Zhu, L. Orthogonal modular biosynthesis of nanoscale conjugate vaccines for vaccination against infection. Nano Res. 2021, 15, 1645–1653. [Google Scholar] [CrossRef] [PubMed]
- Stander, J.; Chabeda, A.; Rybicki, E.P.; Meyers, A.E. A Plant-Produced Virus-Like Particle Displaying Envelope Protein Domain III Elicits an Immune Response Against West Nile Virus in Mice. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Rioux, G.; Babin, C.; Majeau, N.; Leclerc, D. Engineering of Papaya Mosaic Virus (PapMV) Nanoparticles through Fusion of the HA11 Peptide to Several Putative Surface-Exposed Sites. PLoS ONE 2012, 7, e31925. [Google Scholar] [CrossRef]
- Thérien, A.; Bédard, M.; Carignan, D.; Rioux, G.; Gauthier-Landry, L.; Laliberté-Gagné, M.-È.; Bolduc, M.; Savard, P.; Leclerc, D. A versatile papaya mosaic virus (PapMV) vaccine platform based on sortase-mediated antigen coupling. J. Nanobiotechnol. 2017, 15, 54. [Google Scholar] [CrossRef]
- Andersson, T.N.; Ekman, G.J.; Grönlund, H.; Buentke, E.; Eriksson, T.L.J.; Scheynius, A.; Van Hage-Hamsten, M.; Gafvelin, G. A novel adjuvant-allergen complex, CBP-rFel d 1, induces up-regulation of CD86 expression and enhances cytokine release by human dendritic cells in vitro. Immunology 2004, 113, 253–259. [Google Scholar] [CrossRef]
- Jegerlehner, A.; Maurer, P.; Bessa, J.; Hinton, H.J.; Kopf, M.; Bachmann, M.F. TLR9 Signaling in B Cells Determines Class Switch Recombination to IgG2a. J. Immunol. 2007, 178, 2415–2420. [Google Scholar] [CrossRef]
- Klein, J.S.; Bjorkman, P.J. Few and Far Between: How HIV May Be Evading Antibody Avidity. PLOS Pathog. 2010, 6, e1000908. [Google Scholar] [CrossRef]
- Vorup-Jensen, T. On the roles of polyvalent binding in immune recognition: Perspectives in the nanoscience of immunology and the immune response to nanomedicines. Adv. Drug Deliv. Rev. 2012, 64, 1759–1781. [Google Scholar] [CrossRef]
- Dobaño, C.; Sanz, H.; Sorgho, H.; Dosoo, D.; Mpina, M.; Ubillos, I.; Aguilar, R.; Ford, T.; Díez-Padrisa, N.; Williams, N.A.; et al. Concentration and avidity of antibodies to different circumsporozoite epitopes correlate with RTS,S/AS01E malaria vaccine efficacy. Nat. Commun. 2019, 10, 2174. [Google Scholar] [CrossRef] [PubMed]
- Visciano, M.L.; Tagliamonte, M.; Tornesello, M.L.; Buonaguro, F.M.; Buonaguro, L. Effects of adjuvants on IgG subclasses elicited by virus-like Particles. J. Transl. Med. 2012, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Rostamian, M.; Sohrabi, S.; Kavosifard, H.; Niknam, H.M. Lower levels of IgG1 in comparison with IgG2a are associated with protective immunity against Leishmania tropica infection in BALB/c mice. J. Microbiol. Immunol. Infect. 2017, 50, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.C.; Roesti, E.S.; El-Turabi, A.; Bachmann, M.F. Type of RNA Packed in VLPs Impacts IgG Class Switching—Implications for an Influenza Vaccine Design. Vaccines 2019, 7, 47. [Google Scholar] [CrossRef]
- Huber, V.C.; McKeon, R.M.; Brackin, M.N.; Miller, L.A.; Keating, R.; Brown, S.A.; Makarova, N.; Perez, D.; MacDonald, G.H.; McCullers, J.A. Distinct Contributions of Vaccine-Induced Immunoglobulin G1 (IgG1) and IgG2a Antibodies to Protective Immunity against Influenza. Clin. Vaccine Immunol. 2006, 13, 981–990. [Google Scholar] [CrossRef]
- Snapper, C.M.; Paul, W.E. Interferon-γ and B Cell Stimulatory Factor-1 Reciprocally Regulate Ig Isotype Production. Science 1987, 236, 944–947. [Google Scholar] [CrossRef]
- Hocart, M.J.; MacKenzie, J.S.; Stewart, G.A. The Immunoglobulin G Subclass Responses of Mice to Influenza A Virus: The Effect of Mouse Strain, and the Neutralizing Abilities of Individual Protein A-purified Subclass Antibodies. J. Gen. Virol. 1989, 70, 2439–2448. [Google Scholar] [CrossRef]
- Markine-Goriaynoff, D.; Van Der Logt, J.T.; Truyens, C.; Nguyen, T.D.; Heessen, F.W.A.; Bigaignon, G.; Carlier, Y.; Coutelier, J.-P. IFN-γ-independent IgG2a production in mice infected with viruses and parasites. Int. Immunol. 2000, 12, 223–230. [Google Scholar] [CrossRef]
- Ichikawa, K.; Iwasaki, E.; Baba, M.; Chapman, M. High prevalence of sensitization to cat allergen among Japanese children with asthma, living without cats. Clin. Exp. Allergy 1999, 29, 754–761. [Google Scholar] [CrossRef]
- Reddy, S.T.; Van Der Vlies, A.J.; Simeoni, E.; Angeli, V.; Randolph, G.J.; O’Neil, C.P.; Lee, L.K.; Swartz, M.A.; Hubbell, J.A. Exploiting lymphatic transport and complement activation in nanoparticle vaccines. Nat. Biotechnol. 2007, 25, 1159–1164. [Google Scholar] [CrossRef]
- Caldeira, J.C.; Perrine, M.; Pericle, F.; Cavallo, F. Virus-Like Particles as an Immunogenic Platform for Cancer Vaccines. Viruses 2020, 12, 488. [Google Scholar] [CrossRef] [PubMed]
- Pitek, A.S.; Wen, A.M.; Shukla, S.; Steinmetz, N.F. The Protein Corona of Plant Virus Nanoparticles Influences their Dispersion Properties, Cellular Interactions, and In Vivo Fates. Small 2016, 12, 1758–1769. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dintzis, H.M.; Dintzis, R.Z.; Vogelstein, B. Molecular determinants of immunogenicity: The immunon model of immune response. Proc. Natl. Acad. Sci. USA 1976, 73, 3671–3675. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Peng, Y.; Lin, L.; Pan, X.; Fang, M.; Zhao, Y.; Bao, K.; Li, R.; Han, J.; Chen, J.; et al. A pathogen-like antigen-based vaccine confers immune protection against SARS-CoV-2 in non-human primates. Cell Rep. Med. 2021, 2, 100448. [Google Scholar] [CrossRef]
- Benne, C.; Harmsen, M.; van der Graaff, W.; Verheul, A.; Snippe, H.; Kraaijeveld, C. Influenza virus neutralizing antibodies and IgG isotype profiles after immunization of mice with influenza A subunit vaccine using various adjuvants. Vaccine 1997, 15, 1039–1044. [Google Scholar] [CrossRef]
- Gupta, C.L.; Akhtar, S.; Waye, A.; Pandey, N.R.; Pathak, N.; Bajpai, P. Cross talk between Leishmania donovani CpG DNA and Toll-like receptor 9: An immunoinformatics approach. Biochem. Biophys. Res. Commun. 2015, 459, 424–429. [Google Scholar] [CrossRef]
- Chang, X.; Krenger, P.; Krueger, C.C.; Zha, L.; Han, J.; Yermanos, A.; Roongta, S.; Mohsen, M.O.; Oxenius, A.; Vogel, M.; et al. TLR7 Signaling shapes and maintains antibody diversity upon virus-like particle immunization. Front. Immunol. 2022, 12, 827256. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogrina, A.; Skrastina, D.; Balke, I.; Kalnciema, I.; Jansons, J.; Bachmann, M.F.; Zeltins, A. Comparison of Bacterial Expression Systems Based on Potato Virus Y-like Particles for Vaccine Generation. Vaccines 2022, 10, 485. https://doi.org/10.3390/vaccines10040485
Ogrina A, Skrastina D, Balke I, Kalnciema I, Jansons J, Bachmann MF, Zeltins A. Comparison of Bacterial Expression Systems Based on Potato Virus Y-like Particles for Vaccine Generation. Vaccines. 2022; 10(4):485. https://doi.org/10.3390/vaccines10040485
Chicago/Turabian StyleOgrina, Anete, Dace Skrastina, Ina Balke, Ieva Kalnciema, Juris Jansons, Martin F. Bachmann, and Andris Zeltins. 2022. "Comparison of Bacterial Expression Systems Based on Potato Virus Y-like Particles for Vaccine Generation" Vaccines 10, no. 4: 485. https://doi.org/10.3390/vaccines10040485
APA StyleOgrina, A., Skrastina, D., Balke, I., Kalnciema, I., Jansons, J., Bachmann, M. F., & Zeltins, A. (2022). Comparison of Bacterial Expression Systems Based on Potato Virus Y-like Particles for Vaccine Generation. Vaccines, 10(4), 485. https://doi.org/10.3390/vaccines10040485







