Four Faces of Cell-Surface HLA Class-I: Their Antigenic and Immunogenic Divergence Generating Novel Targets for Vaccines
Abstract
:1. Introduction
1.1. Preamble
1.2. Polymorphism of Human Leukocyte Antigens (HLA)
- Centromere_HLA-II_HLA-B--HLA-C--HLA-E--HLA-A--HLA-G—HLA-F-Telomere
1.3. Natural Cell-Surface HLA Molecules Are Glycosylated
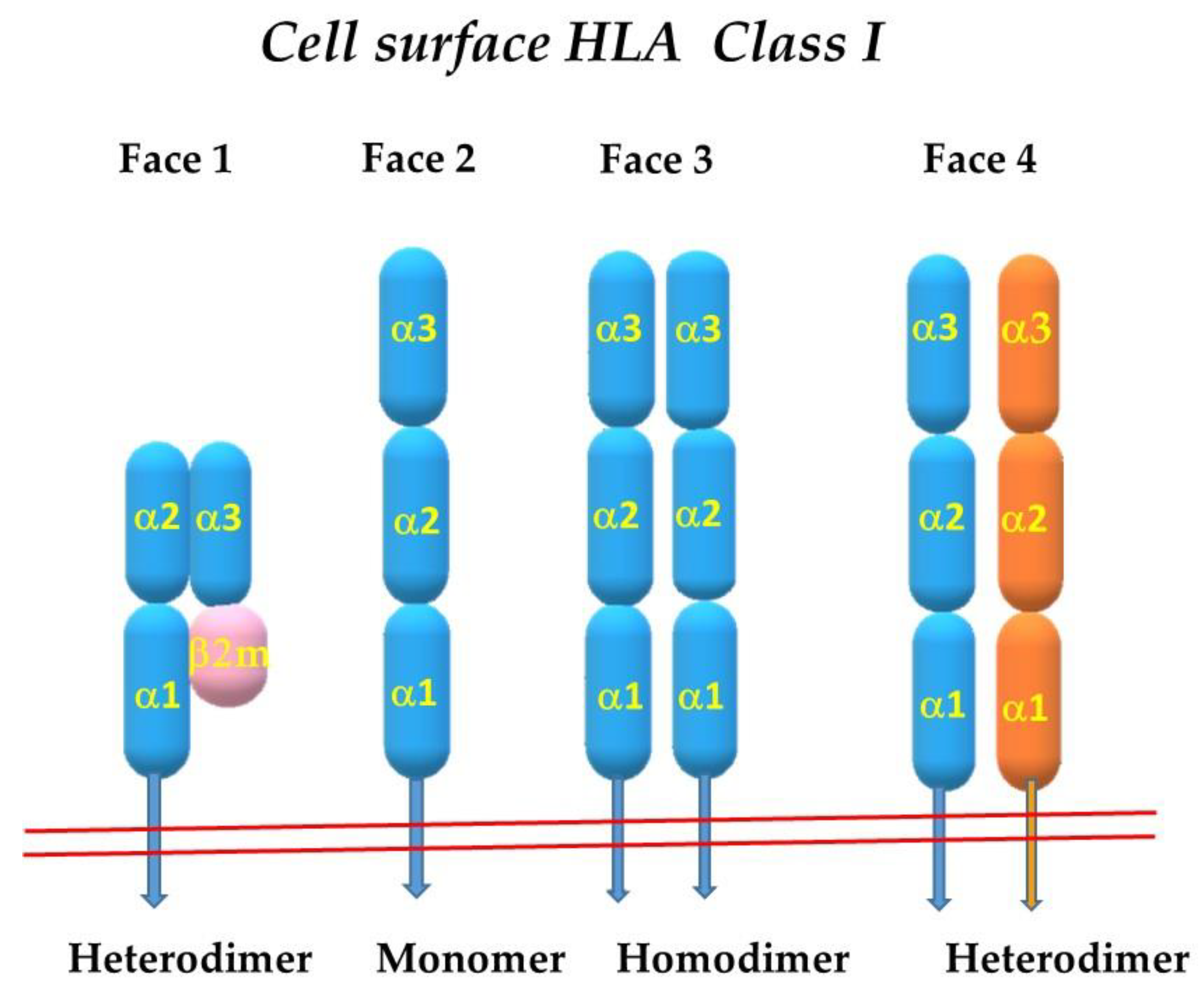
2. The Four Faces of the HLA Class-I Molecules
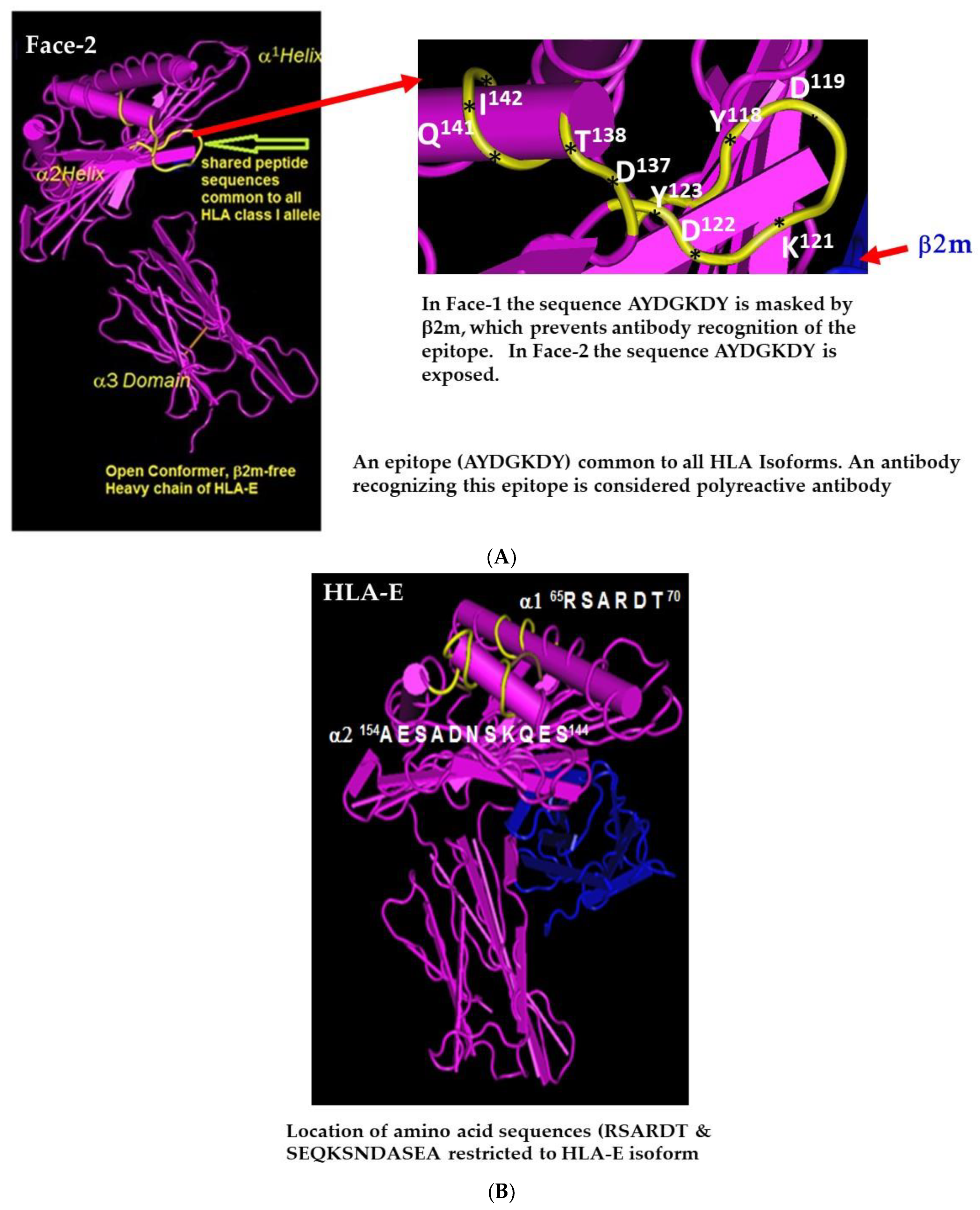
2.1. Face-2: The Open Conformers
2.2. The mAb LA45 Recognizes Only Face-2 but Not Face-1
2.3. Novel Cold Trypsin Assay Eliminated Face-2 from Cell Surface
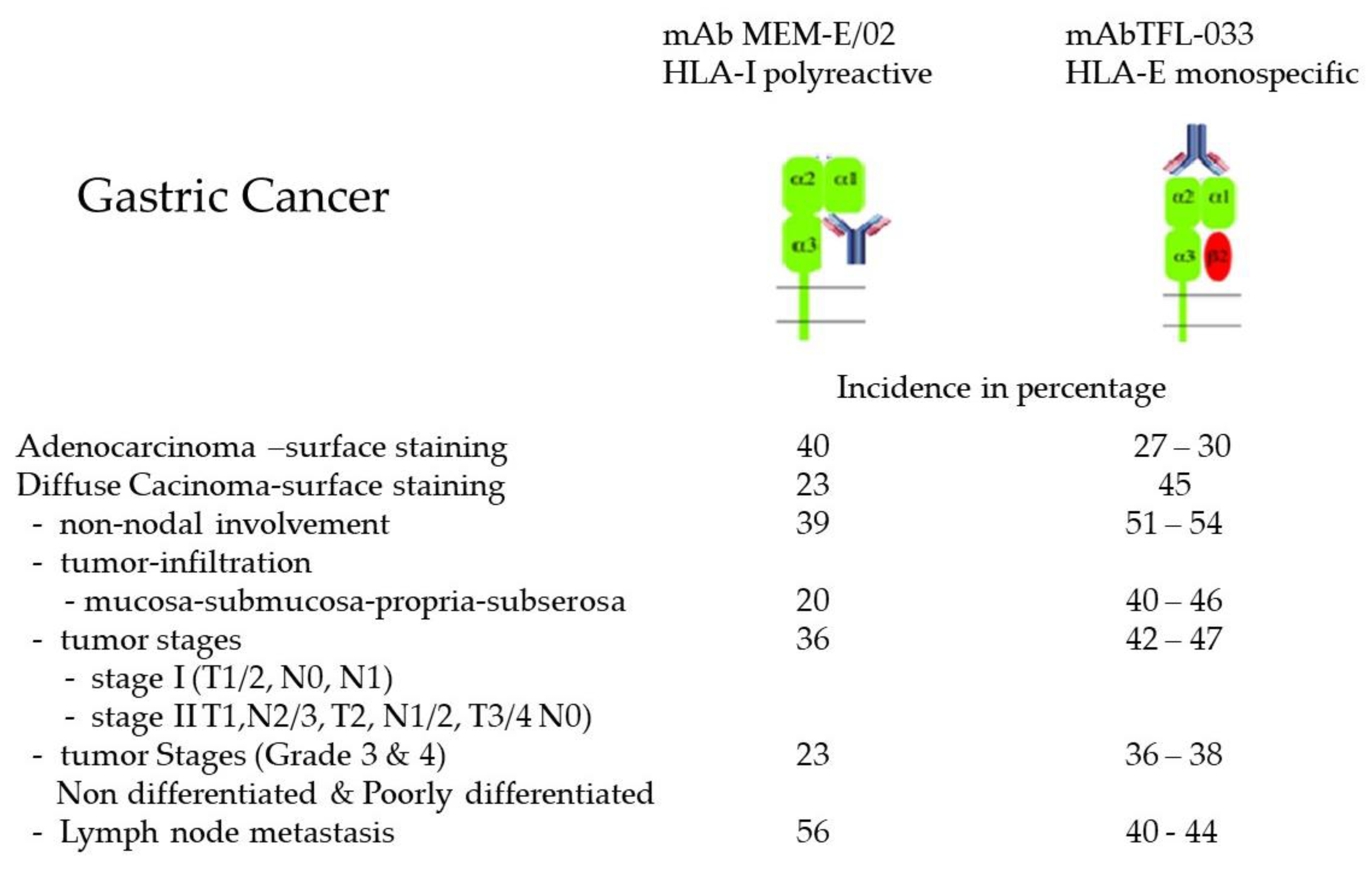
2.4. HLA-I Polyreactive mAbs Binding to Cryptic Sequences Exposed on Face-2
2.5. Admixture of Face-2 with Face-1 on Beadsets Hampers Specific Monitoring Pathogenic Anti-Face-1 HLA Antibodies in Transplant Patients
2.6. Overexpression of Face-2 in Malignant Cells
2.7. Homodimerization of HLA-I Face-2: The Face-3
2.8. Face-3 of HLA-B27
2.9. Face-3 of HLA-G
2.10. Heterodimerization of Two Different HLA-I Heavy Chains: Face-4
3. Significance of the Faces of HLA-I
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Param, P. Genomic Organization of the MHC: Structure, Origin and Function. Immunol. Rev. 1999, 167, 1–379. [Google Scholar]
- Arosa, F.A.; Esgalhado, A.J.; Padrão, C.A.; Cardoso, E.M. Divide, Conquer, and Sense: CD8 + CD28-T Cells in Perspective. Front. Immunol. 2017, 7, 665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufman, J.; Skjoedt, K.; Salomonsen, J. The MHC molecules of Non-mammalian Verebrates. Immunol. Rev. 1990, 113, 83–117. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Dijkstra, M. Major Histocompatibility Complex (MHC) Disease Resistance in Fish. Cells 2019, 8, 378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parnes, J.R. Molecular biology and function of CD4 and CD8. Adv. lmmunol. 1989, 44, 265–311. [Google Scholar]
- Townsend, A.; Bodmer, H. Antigen recognition by class I-restricted T lymphocytes. Annu. Rev. lmmunol. 1989, 7, 601–624. [Google Scholar] [CrossRef]
- Demaria, S.; DeVito-Haynes, L.D.; Salter, R.D.; Burlingham, W.J.; Bushkin, Y. Peptide-conformed beta2m-free class I heavy chains are intermediates in generation of soluble HLA by the membrane-bound metalloproteinase. Hum. Immunol. 1999, 60, 1216–1226. [Google Scholar] [CrossRef]
- Rizzo, R.; Trentini, A.; Bortolotti, D.; Manfrinato, M.C.; Rotola, A.; Castellazzi, M.; Melchiorri, L.; Di Luca, D.; Dallocchio, F.; Fainardi, E.; et al. Matrix metalloproteinase-2 (MMP-2) generates soluble HLA-G1 by cell surface proteolytic shedding. Mol. Cell. Biochem. 2013, 381, 243–255. [Google Scholar] [CrossRef]
- Parham, P.; Lomen, C.; Ei Lawlor, D.A.; Ways, J.P.; Holmes, N.; Coppin, H.L.; Salter, R.D.; Wan, A.M.; Ennis, P.D. Nature of polymorphism in HLA-A, -B, and -C molecules. Proc. Natl. Acad. Sci. USA 1988, 85, 4005–4009. [Google Scholar] [CrossRef] [Green Version]
- Neefjes, J.J.; De Bruijn, M.L.; Boog, C.J.; Nieland, J.D.; Boes, J.; Melief, C.J.; Ploegh, H.L. N-linked glycan modification on antigen-presenting cells restores an allospecific cytotoxic T cell response. J. Exp. Med. 1990, 171, 583–588. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, J.A.; Santos-Aguado, J.; Mentzer, S.J.; Strominger, J.L.; Burakoff, S.J.; Biro, P.A. Site-directed mutagenesis of class I HLA genes. Role of glycosylation in surface expression and functional recognition. J. Exp. Med. 1987, 166, 1329–1350. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Halliwell, J.A.; Hayhurst, J.D.; Flicek, P.; Parham, P.; Marsh, S.G. The IPD and IMGT/HLA database: Allele variant databases. Nucleic Acids Res. 2015, 43, 423–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salzberger, W.; Garcia-Beltran, W.F.; Dugan, H.; Gubbala, S.; Simoneau, C.; Gressens, S.B.; Jost, S.; Altfeld, M. Influence of Glycosylation Inhibition on the Binding of KIR3DL1 to HLA-B*57:01. PLoS ONE 2015, 10, e0145324. [Google Scholar] [CrossRef] [PubMed]
- Néel, D.; Merlu, B.; Turpin, E.; Rabourdin-Combe, C.; Mach, B.; Goussault, Y.; Charron, D.J. Characterization of N-linked oligosaccharides of an HLA DR molecule expressed in different cell lines. Biochem. J. 1987, 244, 433–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merlu, B.; Neel, D.; Goussault, Y.; Charron, D.J. Glycosylation of human leukocyte locus A molecules is dependent on the cell type. Eur. J. Biochem. 1989, 181, 755–760. [Google Scholar] [CrossRef]
- Wilkinson, G.W.G.; Aicheler, R.J.; Wang, E.C.Y. Cytomegaloviruses: From Molecular Pathogenesis to Intervention; Reddehasse, M., Ed.; Caister Academic Press: Wymondham, Norfolk, UK, 2013; pp. 172–190. [Google Scholar]
- Yoneyama, M.S.; Tobisawa, Y.; Hatakeyama, S.; Sato, M.; Tone, K.; Tatara, Y.; Kakizaki, I.; Funyu, T.; Fukuda, M.; Hoshi, S.; et al. A mechanism for evation of CTL immunity by altered O-glycsylation of HLA-I. J. Biochem. 2017, 161, 479–492. [Google Scholar]
- Bergeron, J.J.; Zapun, A.; Ou, W.J.; Hemming, R.; Parlati, F.; Cameron, P.H.; Thomas, D.Y. The role of the lectin calnexin in conformation independent binding to N-linked glycoproteins and quality control. Adv. Exp. Med. Biol. 1998, 435, 105–116. [Google Scholar]
- Trombetta, E.S.; Helenius, A. Lectins as chaperones in glycoprotein folding. Curr. Opin. Struct. Biol. 1998, 8, 587–592. [Google Scholar] [CrossRef]
- Saito, Y.; Ihara, Y.; Leach, M.R.; Cohen-Doyle, M.F.; Williams, D.B. Calreticulin functions in vitro as a molecular chaperone for both glycosylated and non-glycosylated proteins. EMBO J. 1999, 18, 6718–6729. [Google Scholar] [CrossRef] [Green Version]
- Wormald, M.R.; Dwek, R.A. Glycoproteins: Glycan presentation and protein-fold stability. Structure 1999, 7, R155–R160. [Google Scholar] [CrossRef] [Green Version]
- Dustin, M.L.; Golan, D.E.; Zhu, D.M.; Miller, J.M.; Meier, W.; Davies, E.A.; van der Merwe, P.A. Low affinity interaction of human or rat T cell adhesion molecule CD2 with its ligand aligns adhering membranes to achieve high physiological affinity. J. Biol. Chem. 1997, 272, 30889–30898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryan, S.O.; Bonomo, J.A.; Zhao, F.; Cobb, B.A. MHC-II glycosylation modulates Bacteroides fragilis carbohydrate antigen presentation. J. Exp. Med. 2011, 208, 1041–1053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, B.J.; Yu, L.G.; Rhodes, J.M. Altered glycosylation in inflammatory bowel disease: A possible role in cancer development. Glycoconj. J. 2001, 18, 851–858. [Google Scholar] [CrossRef]
- Bodger, K.; Halfvarson, J.; Dodson, A.R.; Campbell, F.; Wilson, S.; Lee, R.; Lindberg, E.; Järnerot, G.; Tysk, C.; Rhodes, J.M. Altered colonic glycoprotein expression in unaffected monozygotic twins of inflammatory bowel disease patients. Gut 2006, 55, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Barrera, C.; Espejo, R.; Reyes, V.E. Differential glycosylation of MHC class II molecules on gastric epithelial cells: Implications in local immune responses. Hum. Immunol. 2002, 63, 384–393. [Google Scholar] [CrossRef]
- Susal, C.; Ovens, J.; Mahmoud, K.; Döhler, B.; Scherer, S.; Ruhenstroth, A.; Hien Tran, T.; Heinold, A.; Opelz, G. No association of kidney graft loss with human leukocyte antigen antibodies detected exclusively by sensitive Luminex single-antigen testing: A Collaborative Transplant Study report. Transplantation 2011, 91, 883–887. [Google Scholar] [CrossRef]
- Poli, F.; Benazzi, E.; Innocente, A.; Nocco, A.; Cagni, N.; Gianatti, A.; Fiocchi, R.; Scalamogna, M. Heart transplantation with donor-specific antibodies directed toward denatured HLA A*02:01: A case report. Hum. Immunol. 2011, 72, 1045–1048. [Google Scholar] [CrossRef]
- Pereira, S.; Perkins, S.; Lee, J.H.; Shumway, W.; LeFor, W.; Lopez-Cepero, M.; Wong, C.; Connolly, A.; Tan, J.C.; Grumet, F.C. Donor-specific antibody against denatured HLA-A1: Clinically nonsignificant? Hum. Immunol. 2011, 72, 492–498. [Google Scholar] [CrossRef]
- Salvade, I.; Aubert, V.; Venetz, J.P.; Gohayan, D.; Saouli, A.C.; Matter, M.M.; Rotman, S.; Pantaleo, G.; Pascual, M. Clinically-relevant threshold of preformed donor-specific anti-HLA antibodies in kidney transplantation. Human Immunol. 2016, 77, 483–489. [Google Scholar] [CrossRef]
- Courant, M.; Visentin, J.; Linares, G.; Dubois, V.; Lepreux, S.; Guidicelli, G.; Thaunat, O.; Merville, P.L.; Taupin, J.-L. The disappointing contribution of antihuman leukocyte antigen donor-specific antibodies characteristics for predicting allograft loss. Nephrol. Dial. Transpl. 2018, 33, 1853–1863. [Google Scholar] [CrossRef]
- Kamburova, E.G.; Wisse, B.W.; Joosten, I.; Allebes, W.A.; van der Meer, A.; Hilbrands, L.B.; Baas, M.C.; Spierings, E.; Hack, C.E.; van Reekum, F.E.; et al. Pretransplant C3d-Fixing Donor-Specific Anti-HLA Antibodies are not associated with Increased Risk for Kidney Graft Failure. J. Am. Soc. Nephrol. 2018, 29, 2279–2285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCaughan, J.; Xu, Q.; Tinckam, K. Detecting donor-specific antibodies: The importance of sorting the wheat from the chaff. Hepatobiliary Surg. Nutr. 2019, 8, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.S.; Glassy, M.C.; Quaranta, V.; Ng, A.K.; Ferrone, S. Effect of tunicamycin on the assembly and antigenicity of HLA antigens: Analysis with monoclonal antibodies. Scand. J. Immunol. 1981, 14, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Fellous, M.; Kamoun, M.; Wiels, J.; Dausset, J.; Clements, G.; Zeuthen, J.; Klein, G. Induction of HLA expression in Daudi cells after cell fusion. lmmunogenetics 1977, 5, 423. [Google Scholar]
- Hyman, R.; Stallings, V. Characterization of a TL-variant ofa homozygous TL ÷ mouse lymphoma. Immunogenetics 1976, 3, 75. [Google Scholar] [CrossRef]
- Hyman, R.; Stallings, V. Analysis of hybrids between a H-2 + TL-lymphoma and an H-2 ÷ TL + lymphoma and its H-2-TL-variant subline. Immunogenetics 1977, 4, 171. [Google Scholar] [CrossRef]
- Krangel, M.S.; Orr, H.T.; Strominger, J.L. Assembly and maturation of HLA-A and HLA-B antigens in vivo. Cell 1979, 18, 979–991. [Google Scholar] [CrossRef]
- Lancet, D.; Parham, P.; Strominger, J.L. Heavy chain ofHLA-A and HLA-B antigens is conformationally labile: A possible role for beta2microglobulin. Proc. Natl. Acad. Sci. USA 1979, 76, 3844–3848. [Google Scholar] [CrossRef] [Green Version]
- Ladasky, J.J.; Shum, B.P.; Canavez, F.; Seuanez, H.N.; Parham, P. Residue 3 of β2-microglobulin affects binding of class I MHC molecules by the W6/32 antibody. Immunogenetics 1999, 49, 312–320. [Google Scholar] [CrossRef]
- Parham, P.; Barnstable, C.J.; Bodmer, W.F. Use of a monoclonal antibody fW6/32) in structural studies of HLA-A, B, C antigens. J. Immunol. 1979, 123, 342–349. [Google Scholar]
- Nössner, E.; Parham, P. Species-specific differences in chaperone interaction of human and mouse major histocompatibility complex class I molecules. J. Exp. Med. 1995, 181, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Bernabeu, C.; van de Rijn, M.; Lerch, P.G.; Terhorst, C.P. β2-microglobulin from serum associates with MHC class I antigens on the surface of cultured cells. Nature 1984, 308, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Allen, H.J.; Fraser, J.; Flyer, D.; Calvin, S.; Flavell, R. B2-microglobulin is not required for cell surface expression of the routine class I histocompatibility antigen H-2D b or of a truncated H-2D b. Proc. Natl. Acad. Sci. USA 1986, 83, 7447–7451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitiello, A.; Potter, T.A.; Sherman, L. The role of β2-microglobulin in peptide binding by class I molecules. Science 1990, 25, 1423–1426. [Google Scholar] [CrossRef]
- Bix, M.; Raulet, D.H. Functionally conformed free class I heavy chains exist on the surface of β2-microglobulin negative cells. J. Exp. Med. 1992, 176, 829–834. [Google Scholar] [CrossRef] [Green Version]
- Raulet, D.H. MHC class I deficient mice. Adv. Immunol. 1994, 55, 381–421. [Google Scholar]
- Sege, K.; Rusk, L.; Peterson, P.A. Role of β2-microglobulin in the intracellular processing of HLA antigens. Biochemistry 1981, 20, 4523–4530. [Google Scholar] [CrossRef]
- Rock, K.L.; Gamble, S.; Rothstein, L.; Gramm, C.; Benacerraf, B. Dissociation of beta 2microglobulin leads to the accumulation of a substantial pool of inactive class I MHC heavy chains on the cell surface. Cell 1991, 65, 611–620. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Taniguchi, M.; Chen, C.W.; Ozawa, M.; Kaneku, H.; El-Awar, N.; Cai, J.; Terasaki, P.I. HLA-E monoclonal antibodies recognize shared peptide sequences on classical HLA class Ia: Relevance to human natural HLA antibodies. Mol. Immunol. 2010, 47, 121–131. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Pham, T.; El-Awar, N.; Kaneku, H.; Terasaki, P.I. Anti-HLA-E mAb 3D12 mimics MEM-E/02 in binding to HLA-B and HLA-C alleles: Web-tools validate the immunogenic epitopes of HLA-E recognized by the Abs. Mol. Immunol. 2011, 48, 423–430. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Terasaki, P.I.; Pham, T.; Jucaud, V.; Kawakita, S. Therapeutic preparations of IVIg contain naturally occurring anti-HLA-E Abs that react with HLA-Ia (HLA-A/-B/-Cw) alleles. Blood 2013, 121, 2013–2028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravindranath, M.H.; Zhu, D.; Pham, T.; Jucaud, V.; Hopfield, J.; Kawakita, S.; Terasaki, P.I. Anti-HLA-E monoclonal Abs reacting with HLA-la and lb alleles like IVIg as potential IVIg-immunomimetics: An evolving therapeutic concept. Clinical Transplant. 2013, 2013, 293–305. [Google Scholar]
- Ravindranath, M.H. HLA Class Ia and Ib Polyreactive Anti-HLA-E IgG2a MAbs (TFL-006 and TFL-007) Suppress Anti-HLA IgG Production by CD19+ B-cells and Proliferation of CD4+ T-cells While Upregulating Tregs. J. Immunol. Res. 2017, 2017, 3475926. [Google Scholar] [CrossRef] [Green Version]
- Ravindranath, M.H.; El Hilali, F. Monospecific and Polyreactive MAbs against Human Leukocyte Antigen-E: Diagnostic and Therapeutic Relevance. In Monoclonal Antibodies; Resaei, N., Ed.; Intech Open: London, UK, 2021; 38p. [Google Scholar]
- Ravindranath, M.H.; Filippone, E.J.; Amato-Menker, C.J.; Arosa, F.A.; Das, B.; Ou, Y.; Norin, A.J. Antibodies to Cryptic Epitopes on HLA Class I and Class II Heavy Chains Bound to Single Antigen Beads: Clinically Relevant? Transpl. Immunol. 2021, 69, 101482. [Google Scholar] [CrossRef] [PubMed]
- Raine, T.; Brown, D.; Bowness, P.; Hill Gaston, J.S.; Moffett, A.; Trowsdale, J.; Allen, R.L. Consistent patterns of expression of HLA class I free heavy chains in healthy individuals and raised expression in spondyloarthropathy patients point to physiological and pathological roles. Rheumatology 2006, 45, 1338–1344. [Google Scholar] [CrossRef] [Green Version]
- Schnabl, E.; Stockinger, H.; Majdic, O.; Gaugitsch, H.; Lindley, I.J.; Maurer, D.; Hajek-Rosenmayr, A.; Knapp, W. Activated human T lymphocytes express MHC class I heavy chains not associated with beta 2-microglobulin. J. Exp. Med. 1990, 171, 1431–1442. [Google Scholar] [CrossRef]
- Bjorkman, P.J.; Saper, M.A.; Samraoui, B.; Bennett, W.S.; Strominger, J.L.; Wiley, D.C. Structure of the human class I histocompatibility antigen, HLA-A2. Nature 1987, 329, 506. [Google Scholar] [CrossRef]
- Madrigal, J.A.; Belich, M.P.; Benjamin, R.J.; Little, A.M.; Hildebrand, W.H.; Mann, D.L.; Parham, P. Molecular definition of a polymorphic antigen (LA45) of free HLA-A and -B heavy chains found on the surfaces of activated B and T cells. J. Exp. Med. 1991, 174, 1085–1095. [Google Scholar] [CrossRef] [Green Version]
- Demaria, S.; Schwab, R.; Bushkin, Y. The origin and fate of beta 2m-free MHC class I molecules induced on activated T cells. Cell Immunol. 1992, 142, 103–113. [Google Scholar] [CrossRef]
- Aiuti, A.; Forte, P.; Simeoni, L.; Lino, M.; Pozzi, L.; Fattorossi, A.; Giacomini, P.; Ginelli, E.; Beretta, A.; Siccardi, A.; et al. Membrane expression of HLA-Cw4 free chains in activated T cells of transgenic mice. Immunogenetics 1995, 42, 368–375. [Google Scholar] [CrossRef]
- Arosa, F.A.; Santos, S.G.; Powis, S.J. Open conformers: The hidden Face of MHC-I molecules. Trends Immunol. 2007, 28, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Terasaki, P.I.; Anderson, N.; Lachmann, N.; Schönemann, C. Intact HLA not beta2m-free HC-specific HLA class I antibodies are predictive of graft failure. Transplantation 2009, 88, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Otten, H.G.; Verhaar, M.C.; Borst, H.P.E.; van Eck, M.; van Ginkel, W.G.J.; Hené, R.J.; van Zuilen, A.D. The significance of pretransplant donor-specific antibodies reactive with intact or denatured human leucocyte antigen in kidney transplantation. Clin. Exp. Immunol. 2013, 173, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Visentin, J.; Guidicelli, G.; Moreau, J.F.; Lee, J.H.; Taupin, J.L. Deciphering allogeneic antibody response against native and denatured HLA epitopes in organ transplantation. Eur. J. Immunol. 2015, 45, 2111–2121. [Google Scholar] [CrossRef] [PubMed]
- Visentin, J.G.; Guidicelli, T.; Nong, J.-F.; Moreau, J.-F.; Merville, P.; Couzi, L.; Lee, J.-H.; Taupin, J.-L. Evaluation of the iBeads assay as a tool for identifying class I HLA antibodies. Hum. Immunol. 2015, 76, 651–656. [Google Scholar] [CrossRef]
- Michel, K.; Santella, R.; Steers, J.; Sahajpal, A.; Downey, F.X.; Thohan, V.; Oaks, M. Many de novo donor-specific antibodies recognize β2-microglobulin-free, but not intact HLA heterodimers. HLA 2016, 87, 356–366. [Google Scholar] [CrossRef] [Green Version]
- Ravindranath, M.H.; Kaneku, H.; El-Awar, N.; Morales-Buenrostro, L.E.; Terasaki, P.I. Abs to HLA-E in alloimmunized males: Pattern of HLA-Ia reactivity of anti-HLA-E-positive sera. J. Immunol. 2010, 185, 1935–1948. [Google Scholar] [CrossRef] [Green Version]
- Ravindranath, M.H.; Flippone, E.J.; Mahowald, G.; Callender, C.; Babu, A.; Saidman, S.; Ferrone, S. Significance of the intraindividual variability of HLA IgG antibodies in renal disease patients observed with different SABs monitored with two different secondary antibodies on a Luminex platform. Immunol. Res. 2018, 66, 584–604. [Google Scholar] [CrossRef] [Green Version]
- Ravindranath, M.H.; Ravindranath, N.M.; Amato-Menker, C.J. Luminex Multiplex Bead Assay monitoring HLA IgG antibodies in sensitized pre- and post-transplant patients: Clonality of the Detection Antibody Impacts Specificity and Sensitivity. Appl. Sci. 2021, 11, 6430. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Jucaud, V.; Ferrone, S. Monitoring native HLA-I trimer specific antibodies in Luminex multiplex single antigen bead assay: Evaluation of beadsets from different manufacturers. J. Immunol. Methods 2017, 450, 73–80. [Google Scholar] [CrossRef]
- Jucaud, V.; Ravindranath, M.H.; Terasaki, P.I. Conformational Variants of the Individual HLA-I Antigens on Luminex Single Antigen Beads Used in Monitoring HLA Antibodies: Problems and Solutions. Transplantation 2017, 101, 764–777. [Google Scholar] [CrossRef]
- Algarra, I.; Garcia-Lora, A.; Cabrera, T.; Ruiz-Cabello, F.; Garrido, F. The selection of tumor variants with altered expression of classical and nonclassical MHC class I molecules: Implications for tumor immune escape. Cancer Immunol. Immunother. 2004, 53, 904–910. [Google Scholar] [CrossRef] [PubMed]
- Stam, N.J.; Spits, H.; Ploegh, H.L. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J. Immunol. 1986, 137, 2299–2307. [Google Scholar] [PubMed]
- Grassi, F.; Meneveri, R.; Gullberg, M.; Lopalco, L.; Rossi, G.B.; Lanza, P.; De Santis, C.; Bratsand, G.; Butto, S.; Ginelli, E.; et al. Human immunodeficiency virus type 1 gpl20 mimics a hidden monomorphic epitope borne by class I major histocompatibility complex heavy chains. J. Exp. Med. 1990, 174, 53–62. [Google Scholar] [CrossRef]
- Marozzi, A.; Meneveri, R.; Bunone, G.; De Santis, C.; Lopalco, L.; Beretta, A.A.; Agresti, A.; Siccardi, A.G.; Della Valle, G.; Ginelli, E. Expression of beta 2m-free HLA class I heavy chains in neuroblastoma cell lines. Scand. J. Immunol. 1993, 37, 661–667. [Google Scholar]
- Martayan, A.; Fiscella, M.; Setini, A.; Ciccarelli, G.; Gambari, R.; Feriotto, G.; Beretta, A.; Siccardi, A.G.; Appella, E.; Giacomini, P. Conformation and surface expression of free HLA-CW1 heavy chains in the absence of beta 2-microglobulin. Hum. Immunol. 1997, 53, 23–33. [Google Scholar] [CrossRef]
- Giacomini, P.; Beretta, A.; Nicotra, M.R.; Ciccarelli, G.; Martayan, A.; Cerboni, C.; Lopalco, L.; Bini, D.; Delfino, L.; Ferrara, G.B.; et al. HLA-C heavy chains free of beta2-microglobulin: Distribution in normal tissues and neoplastic lesions of non-lymphoid origin and interferon-y responsiveness. Tissue Antigens 1997, 50, 555–566. [Google Scholar] [CrossRef]
- Lee, N.; Goodlett, D.R.; Ishitani, A.; Marquardt, H.; Geraghty, D.E. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA Class I signal sequences. J. Immunol. 1998, 160, 4951–4960. [Google Scholar]
- Sensi, M.; Pietra, G.; Molla, A.; Nicolini, G.; Vegetti, C.; Bersani, I.; Millo, E.; Weiss, E.; Moretta, L.; Mingari, M.C.; et al. Peptides with dual binding specificity for HLA-A2 and HLA-E are encoded by alternatively spliced isoforms of the antioxidant enzyme peroxiredoxin 5. Int. Immunol. 2009, 21, 257–268. [Google Scholar] [CrossRef] [Green Version]
- de Kruijf, E.M.; Sajet, A.; van Nes, G.H.; Natanov, R.; Putter, H.; Smit, V.T.H.B.M.; Jan Liefers, G.; van den Elsen, P.J.; van de Velde, C.J.H.; Kuppen, P.J.K. HLA-E and HLA-G expression in classical HLA class Inegative tumors is of prognostic value for clinical outcome of early breast cancer patients. J. Immunol. 2010, 185, 7452–7459. [Google Scholar] [CrossRef] [Green Version]
- Goncalves, M.A.G.; Le Discorde, M.; Simoes, R.T.; Rabreau, M.; Soares, E.G.; Donadi, E.A.; Carosella, E.D. Classical and non-classical HLA molecules and p16(INK4a) expression in precursors lesions and invasive cervical cancer. Eur. J. Obstet. Gynecol. Reprod. Biol. 2008, 141, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Marin, R.; Ruiz-Cabello, F.; Pedrinaci, S.; Méndez, M.; Jiménez, P.; Geraghty, D.E.; Garrido, F. Analysis of HLA-E expression in human tumors. Immunogenetics 2003, 54, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Derre, L.; Corvaisier, M.; Charreau, B.; Moreau, A.; Godefroy, M.; Moreau-Aubry, A.; Francine Jotereau, F.; Gervois, N. Expression and release of HLA-E by melanoma cells and melanocytes: Potential impact on the response of cytotoxic effector cells. J. Immunol. 2006, 177, 3100–3107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coupel, S.; Moreau, A.; Hamidou, M.; Horejsi, V.; Soulillou, J.-P.; Béatrice Charreau, B. Expression and release of soluble HLA-E is an immunoregulatory feature of endothelial cell activation. Blood 2007, 109, 2806–2814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wischhusen, J.; Friese, M.A.; Mittelbronn, M.; Meyermann, R.; Weller, M. HLA-E protects glioma cells from NKG2Dmediated immune responses in vitro: Implications for immune escape in vivo. J. Neuropathol. Exp. Neurol. 2005, 64, 523–528. [Google Scholar] [CrossRef]
- Mittelbronn, M.; Simon, P.; Löffler, C.; Capper, D.; Bunz, B.; Harter, P.; Schlaszus, H.; Schleich, A.; Tabatabai, G.; Goeppert, B.; et al. Elevated HLA-E levels in human glioblastomas but not in grade I to III astrocytomas correlate with infiltrating CD81 cells. J. Neuroimmunol. 2007, 189, 50–58. [Google Scholar] [CrossRef]
- Levy, E.M.; Sycz, G.; Arriaga, J.M.; Barrio, M.M.; von Euw, E.M.; Morales, S.B.; González, M.; Mordoh, J.; Bianchini, M. Cetuximab-mediated cellular cytotoxicity is inhibited by HLA-E membrane expression in colon cancer cells. Innate Immun. 2009, 15, 91–100. [Google Scholar] [CrossRef]
- Kren, L.; Muckova, K.; Lzicarova, E.; Sova, M.; Vybihal, V.; Svoboda, T.; Fadrus, P.; Smrcka, M.; Slaby, O.; Lakomy, R.; et al. Production of immune-modulatory nonclassical molecules HLA-G and HLA-E by tumor infiltrating ameboid microglia=macrophages in glioblastomas: A role in innate immunity? J. Neuroimmunol. 2010, 220, 131–135. [Google Scholar] [CrossRef]
- Kren, L.; Slaby, O.; Muckova, K.; Lzicarova, E.; Sova, M.; Vybihal, V.; Svoboda, T.; Fadrus, P.; Lakomy, R.; Vanhara, P.; et al. Expression of immune-modulatory molecules HLA-G and HLA-E by tumor cells in glioblastomas: An unexpected prognostic significance? Neuropathology 2011, 31, 129–134. [Google Scholar] [CrossRef]
- Silva, T.G.; Crispim, J.C.; Miranda, F.A.; Hassumi, M.K.; de Mello, J.M.Y.; Simões, R.T.; Souto, F.; Soares, E.G.; Donadi, E.A.; Soares, C.P. Expression of the nonclassical HLA-G and HLA-E molecules in laryngeal lesions as biomarkers of tumor invasiveness. Histol. Histopathol. 2011, 26, 1487–1497. [Google Scholar]
- Levy, E.M.; Bianchini, M.; Von Euw, E.M.; Barrio, M.M.; Bravo, A.I.; Furman, D.; Domenichini, E.; Macagno, C.; Pinsky, V.; Zucchini, C.; et al. Human leukocyte antigen-E protein is overexpressed in primary human colorectal cancer. Int. J. Oncol. 2008, 32, 633–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benevolo, M.; Mottolese, M.; Tremante, E.; Rollo, F.; Diodoro, M.G.; Ercolani, C.; Sperduti, I.; Monaco, E.L.; Cosimelli, M.; Giacomini, P. High expression of HLA-E in colorectal carcinoma is associated with a favorable prognosis. J. Transl. Med. 2011, 9, 184–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bossard, C.; Bezieau, S.; Matysiak-Budnik, T.; Volteau, C.; Laboisse, C.L.; Jotereau, F.; Mosnier, J.-F. HLA-E=beta2 microglobulin overexpression in colorectal cancer is associated with recruitment of inhibitory immune cells and tumor progression. Int. J. Cancer 2012, 131, 855–863. [Google Scholar] [CrossRef] [PubMed]
- Hanak, L.; Slaby, O.; Lauerova, L.; Kren, L.; Nenutil, R.; Michalek, J. Expression pattern of HLA class I antigens in renal cell carcinoma and primary cell line cultures: Methodological implications for immunotherapy. Med. Sci. Monit. 2009, 15, CR638–CR643. [Google Scholar]
- Kren, L.; Valkovsky, I.; Dolezel, J.; Capak, I.; Pacik, D.; Poprach, A.; Lakomy, R.; Redova, M.; Fabian, P.; Krenova, Z.; et al. HLA-G and HLA-E specific mRNAs connote opposite prognostic significance in renal cell carcinoma. Diagn. Pathol. 2012, 7, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, S.J.; Duan, L.-N.; Luo, Y.; Suo, T.-L.; Lu, C.-R.; Tang, J. Expression of NKG2D and NKG2A with their ligands MHC-I A=B and HLA-E in acute leukemia patients and its significance. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2011, 19, 312–316. [Google Scholar]
- Sasaki, T.; Ravindranath, M.H.; Terasaki, P.I.; Freitas, M.C.; Kawakita, S.; Jucaud, V. Gastric cancer progression may involve a shift in HLA-E profile from an intact heterodimer to β2m-free monomer. Int. J. Cancer 2014, 134, 1558–1570. [Google Scholar] [CrossRef]
- Zhu, D.; Ravindranath, M.H.; Terasaki, P.I.; Miyazaki, T.; Pham, T.; Jucaud, V. Suppression of allo-human leucocyteantigen (HLA) Abs secreted by B memory cells in vitro: Intravenous immunoglobulin (IVIg) versus a monoclonal anti-HLA-E IgG that mimics HLA-I reactivities of IVIg. Clin. Exp. Immunol. 2014, 177, 464–477. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Terasaki, P.I.; Pham, T.; Jucaud, V.; Kawakita, S. Suppression of blastogenesis and proliferation of activated CD4(+) T-cells: Intravenous immunoglobulin (IVIg) versus novel anti-human leucocyte antigen (HLA)-E mAbs mimicking HLA-I reactivity of IVIg. Clin. Exp. Immunol. 2014, 178, 154–177. [Google Scholar] [CrossRef]
- Brewerton, D.A.; Caffrey, M.; Hart, F.D.; James, D.C.O.; Nichols, A.; Sturrock, R.D. Ankylosing spondylitis and HL-A27. Lancet 1973, 1, 904. [Google Scholar] [CrossRef]
- Wakefield, D.; Clarke, D.P. Recent Developments in HLA B27 Anterior Uveitis. Front. Immunol. 2020, 11, 608134. [Google Scholar] [CrossRef] [PubMed]
- Dangoria, N.S.; DeLay, M.L.; Kingsbury, D.J.; Meaar, M.P.; Uchanska-Ziegler, B.; Ziegler, A.; Colbert, R.A. HLA-B27 misfolding is associated with aberrant intermolecular disulfide bond formation (dimerization) in the endoplasmic reticulum. J. Biol. Chem. 2002, 277, 23459–23468. [Google Scholar] [CrossRef] [Green Version]
- Mear, J.P.; Schreiber, K.L.; Münz, C.; Zhu, X.; Stevanović, S.; Rammensee, H.G.; Rowland-Jones, S.L.; Colbert, R.A. Misfolding of HLA-B27 as a result of its B pocket suggests a novel mechanism for its role in susceptibility to spondyloarthropathies. J. Immunol. 1999, 163, 6665–6670. [Google Scholar] [PubMed]
- Allen, R.L.; O’Callaghan, C.A.; McMichael, A.J.; Bowness, P. Cutting Edge: HLA-B27 Can Form a Novel b2-Microglobulin-Free Heavy Chain Homodimer Structure. J. Immunol. 1999, 162, 5045–5048. [Google Scholar] [PubMed]
- Khare, S.D.; Hansen, J.; Luthra, H.S.; David, C.S. HLA-B27 heavy chains contribute to spontaneous inflammatory disease in B27/human b2-microglobulin (2m) double transgenic mice with disrupted mouse b2m. J. Clin. Investig. 1996, 98, 2746. [Google Scholar] [CrossRef]
- Khare, S.D.; Bull, M.J.; Hansen, J.; Luthra, H.S.; David, C.S. Spontaneous inflammatory Disease in HLA-B27 transgenic mice is independent of MHC class II molecules: A direct role for B27 heavy chains and not B27-derived peptides. J. Immunol. 1998, 60, 101. [Google Scholar]
- Bird, L.A.; Peh, C.A.; Kollnberger, S.; Elliott, T.; McMichael, A.J.; Bowness, P. Lymphoblastoid cells express HLA-B27 homodimers both intracellularly and at the cell surface following endosomal recycling. Eur. J. Immunol. 2003, 33, 748–759. [Google Scholar] [CrossRef]
- Kollnberger, S.; Bird, L.; Sun, M.Y.; Retiere, C.; Braud, V.M.; McMichael, A.J.; Bowness, P. Cell-surface expression and immune receptor recognition of HLA-B27 homodimers. Arthritis Rheumatol. 2002, 46, 972–982. [Google Scholar] [CrossRef]
- Turner, M.J.; Delay, M.L.; Bai, S.; Klenk, E.; Colbert, R.A. HLA-B27 up-regulation causes accumulation of misfolded heavy chains and correlates with the magnitude of the unfolded protein response in transgenic rats: Implications for the pathogenesis of spondylarthritis-like disease. Arthritis Rheumatol. 2007, 6, 215–223. [Google Scholar] [CrossRef]
- Antoniou, A.N.; Ford, S.; Taurog, J.D.; Butcher, G.W.; Powis, S.J. Formation of HLA-B27 homodimers and their relationship to assembly kinetics. J. Biol. Chem. 2004, 27, 8895–8902. [Google Scholar] [CrossRef] [Green Version]
- Kollnberger, S.; Bird, L.A.; Roddis, M.; Hacquard-Bouder, C.; Kubagawa, H.; Bodmer, H.C.; Breban, M.; McMichael, A.J.; Bowness, P. HLA-B27 heavy chain homodimers are expressed in HLA-B27 transgenic rodent models of spondyloarthritis and are ligands for paired Ig-like receptors. J. Immunol. 2004, 173, 1699–1710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, W.C.; Chen, C.J.; Yen, J.H.; Ou, T.T.; Tsai, J.J.; Liu, C.S.; Liu, H.W. Free HLA class I heavy chain-carrying monocytes—a potential role in the pathogenesis of spondyloarthro pathies. J. Rheumatol. 2002, 29, 966–972. [Google Scholar] [PubMed]
- Santos, S.G.; Lynch, S.; Campbell, E.C.; Antoniou, A.N.; Powis, S.J. Induction of HLA-B27 heavy chain homodimer formation after activation in dendritic cells. Arthritis Res. Ther. 2008, 10, R100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuon, W.; Holzhutter, H.G.; Appel, H.; Grolms, M.; Kollnberger, S.; Traeder, A.; Henklein, P.; Weiss, E.; Thiel, A.; Lauster, R.; et al. Identification of HLA-B27-restricted peptides from the Chlamydia trachomatis proteome with possible relevance to HLA-B27-associated diseases. J. Immunol. 2001, 167, 4738–4746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenart, I.; Guiliano, D.B.; Burn, G.; Campbell, E.C.; Morley, K.D.; Fussell, H.; Powis, S.J.; Antoniou, A.N. The MHC Class I heavy chain structurally conserved cysteines 101 and 164 participate in HLA-B27 dimer formation. Antioxid. Redox Signal. 2012, 16, 33–43. [Google Scholar] [CrossRef]
- Boyson, J.E.; Erskine, R.; Whitman, M.C.; Chiu, M.; Lau, J.M.; Koopman, L.A.; Valter, M.M.; Angelisova, P.; Horejsi, V.; Stroming, J.L. Disulfide bond-mediated dimerization of HLA-G on the cell surface. Proc. Natl. Acad. Sci. USA 2002, 99, 16180–16185. [Google Scholar] [CrossRef] [Green Version]
- Gonen-Gross, T.; Achdout, H.; Gazit, R.; Hanna, J.; Mizrahi, S.; Markel, G.; Goldman-Wohl, D.; Yagel, S.; Horejsí, V.; Levy, O.; et al. Complexes of HLA-G protein on the cell surface are important for leukocyte Ig-like receptor-1 function. J. Immunol. 2003, 17, 1343–1351. [Google Scholar] [CrossRef]
- Wainwright, S.D.; Biro, P.A.; Holmes, C.H. HLA-F is a predominantly empty, intracellular, TAP-associated MHC class Ib protein with a restricted expression pattern. J. Immunol. 2000, 164, 319–328. [Google Scholar] [CrossRef]
- Lee, N.; Geraghty, D.E. HLA-F surface expression on B cell and monocyte cell lines is partially independent from tapasin and completely independent from TAP. J. Immunol. 2003, 171, 5264–5271. [Google Scholar] [CrossRef] [Green Version]
- Boyle, L.H.; Gillingham, A.K.; Munro, S.; Trowsdale, J. Selective export of HLA-F by its cytoplasmic tail. J. Immunol. 2006, 176, 6464–6472. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Ishitani, A.; Geraghty, D.E. HLA-F is a surface marker on activated lymphocytes. Eur. J. Immunol. 2010, 40, 2308–2318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodridge, J.P.; Burian, A.; Lee, N.; Geraghty, D.E. HLA-F complex without peptide binds to MHC class I protein in the open conformer form. J. Immunol. 2010, 184, 6199–6208. [Google Scholar] [CrossRef] [PubMed]
- Due, C.; Simonsen, M.; Olsson, L. The major histocompatibilitycomplex class I heavy chain as a structural subunit of the human cell membrane insulin receptor: Implications for the range of biological functions of histocompatibility antigens. Proc. Natl. Acad. Sci. USA 1986, 83, 6007–6011. [Google Scholar] [CrossRef] [Green Version]
- Ramalingam, T.S.; Chakrabarti, A.; Edidin, M. Interaction of class I human leukocyte antigen (HLA-I) molecules with insulin receptors and its effect on the insulin-signaling cascade. Mol. Biol. Cell 1997, 8, 2463–2474. [Google Scholar] [CrossRef] [Green Version]
- Schreiber, A.B.; Schlessinger, J.; Edidin, M. Interaction between major histocompatibility complex antigens and epidermal growth factor receptors on human cells. J. Cell Biol. 1984, 98, 725–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, A.; Yan, W.-H. The emerging roles of human leukocyte antigen-F in immune modulation and viral infection. Front. Immunol. 2019, 10, 964. [Google Scholar] [CrossRef]
- Lepin, E.J.; Bastin, J.M.; Allan, D.S.; Roncador, G.; Braud, V.M.; Mason, D.Y.; van der Merwe, P.A.; McMichael, A.J.; Bell, J.I.; Powis, S.H.; et al. Functional characterization of HLA-F and binding of HLA-F tetramers to ILT2 and ILT4 receptors. Eur. J. Immunol. 2000, 30, 3552–3561. [Google Scholar] [CrossRef]
- Burian, A.; Wang, K.L.; Finton, K.A.; Lee, N.; Ishitani, A.; Strong, R.K.; Geraghty, D.E. HLA-F and MHC-I Open Conformers Bind Natural Killer Cell Ig-Like Receptor KIR3DS1. PLoS ONE 2016, 11, e0163297. [Google Scholar] [CrossRef]
- Edgerly, C.H.; Weimer, E.T. The Past, Present, and Future of HLA Typing in Transplantation. Methods Mol. Biol. 2018, 1802, 1–10. [Google Scholar]
- Dunckley, H. Methods HLA typing by SSO and SSP methods. Mol. Biol. 2012, 882, 9–25. [Google Scholar]
- Ravindranath, M.H.; Filippone, E.J.; Devarajan, A.; Asgharzadeh, S. Enhancing Natural Killer and CD8+ T Cell-Mediated Anticancer Cytotoxicity and Proliferation of CD8+ T Cells with HLA-E Monospecific Monoclonal Antibodies. Monoclon. Antib. Immunodiagn. Immunother. 2019, 38, 38–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrido, F. MHC Class-I loss and Cancer. Adv. Exp. Med. Biol. 2019, 1151, 1–101. [Google Scholar] [PubMed]
- Ravindranath, M.H.; Morton, D.L. Role of gangliosid.des in active immunotherapy with melanoma vaccine. Int. Rev. Immunol. 1991, 7, 303–329. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, M.H.; Morton, D.L. Active Specific Immunotherapy with Vaccines. Section XVIII, Principles of Biotherapeutics. In Cancer Medicine, 4th ed.; Holland, J.F., Bast, R.C., Morton, D.L., Frei, E., III, Kufe, D.W., Weichselbaum, R.R., Eds.; Williams and Wilkins: Philadelphia, PS, USA; Waverly Co.: Boston, MA, USA, 1993; Volume 2. [Google Scholar]
- Ravindranath, M.H.; Morton, D.L.; Irie, R.F. An epitope common to gangliosides O-acetyl-GD3 and GD3 recognized by antibodies in melanoma patients after active specific immunotherapy. Cancer Res. 1989, 49, 3891–3897. [Google Scholar]
- Ravindranath, M.H.; Morton, D.L.; Irie, R.F. Attachment of monophosphoryl lipid A (MPL) to cells and liposomes augments antibody response to membrane-bound gangliosides. J. Autoimmun. 1994, 7, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Ravindranath, M.H.; Brazeau, S.M.; Morton, D.L. Efficacy of tumor cell vaccine after incorporating monophosphoryl lipid A (MPL) in tumor cell membranes containing tumor-associated ganglioside. Experientia 1994, 50, 648–653. [Google Scholar] [CrossRef]
- Klareskog, L.; Banck, G.; Forsgren, A.; Peterson, P.A. Binding of HLA antigen-containing liposomes to bacteria. Proc. Natl. Acad. Sci. USA 1978, 75, 6197–6201. [Google Scholar] [CrossRef] [Green Version]


| Updated on | HLA Class I Alleles | HLA Class II Alleles | |||||||
|---|---|---|---|---|---|---|---|---|---|
| α-Chain | α-Chain | β-Chain | |||||||
| Genes | Alleles | Proteins | Genes | Alleles | Proteins | Genes | Alleles | Proteins | |
| 14 October 2020 | A | 6766 | 4064 | DRA | 29 | 2 | DRB1 | 2949 | 2015 |
| 7 July 2021 | 6921 | 4156 | 29 | 2 | 3801 | 3801 | 2620 | ||
| 14 October 2020 | B | 7967 | 4962 | DRB2 | 1 | 0 | |||
| 7 July 2021 | 8181 | 5090 | 1 | 0 | |||||
| 14 October 2020 | C | 6621 | 3831 | DRB3 | 390 | 293 | |||
| 7 July 2021 | 6779 | 3927 | 404 | 301 | |||||
| 14 October 2020 | E | 271 | 110 | DRB4 | 194 | 129 | |||
| 7 July 2021 | 271 | 110 | 203 | 131 | |||||
| 14 October 2020 | F | 45 | 6 | DRB5 | 155 | 120 | |||
| 7 July 2021 | 45 | 6 | 163 | 125 | |||||
| 14 October 2020 | G | 82 | 22 | DRB6 | 0 | 0 | |||
| 7 July 2021 | 88 | 26 | 3 | 0 | |||||
| 14 October 2020 | α-chain pairs with β2-microglobulin | DRB7 | 2 | 0 | |||||
| 7 July 2021 | 2 | 0 | |||||||
| 14 October 2020 | DRB8 | 1 | 0 | ||||||
| 7 July 2021 | 1 | 0 | |||||||
| 14 October 2020 | DRB9 | 6 | 0 | ||||||
| 7 July 2021 | 6 | 0 | |||||||
| 14 October 2020 | DQA1 | 306 | 143 | DQA2 | 40 | 11 | |||
| 7 July 2021 | 343 | 172 | 40 | 11 | |||||
| 14 October 2020 | DQB1 | 1997 | 1303 | ||||||
| 7 July 2021 | 2033 | 1324 | |||||||
| 14 October 2020 | DPA1 | 258 | 107 | DPB1 | 1749 | 1106 | |||
| 7 July 2021 | 298 | 135 | 1862 | 1180 | |||||
| 14 October 2020 | DPA2 | 5 | 0 | DPB2 | 6 | 0 | |||
| 7 July 2021 | 5 | 2 | 6 | 3 | |||||
| 14 October 2020 | DMA | 7 | 4 | DMB | 13 | 7 | |||
| 7 July 2021 | 7 | 4 | 13 | 7 | |||||
| 14 October 2020 | DOA | 12 | 3 | DOB | 13 | 5 | |||
| 7 July 2021 | 12 | 3 | 13 | 5 | |||||
| α-chain pairs with β-chain | |||||||||
| Face-2 Peptide Sequences or Open Conformers of HLA-E | HLA Alleles | |||||
|---|---|---|---|---|---|---|
| HLA-Ia | HLA-Ib | Specificity | ||||
| [Number of Amino Acids] | A | B | Cw | F | G | |
| 47PRAPWMEQE55 [9] | 1 | 0 | 0 | 0 | 0 | A*3306 |
| 59EYWDRETR65 [8] | 5 | 0 | 0 | 0 | 0 | A-restricted |
| 65RSARDTA71 [6] | 0 | 0 | 0 | 0 | 0 | E-restricted |
| 90AGSHTLQW97 [8] | 1 | 10 | 48 | 0 | 0 | Polyspecific |
| 108RFLRGYE123 [7] | 24 | 0 | 0 | 0 | 0 | A-restricted |
| 115QFAYDGKDY123 [9] | 1 | 104 | 75 | 0 | 0 | Polyspecific |
| 117AYDGKDY123 [7] | 491 | 831 | 271 | 21 | 30 | Polyspecific |
| 126LNEDLRSWTA135 [10] | 239 | 219 | 261 | 21 | 30 | Polyspecific |
| 137DTAAQI142 [6] | 0 | 824 | 248 | 0 | 30 | Polyspecific |
| 137DTAAQIS143 [7] | 0 | 52 | 4 | 0 | 30 | Polyspecific |
| 143SEQKSNDASE152 [10] | 0 | 0 | 0 | 0 | 0 | E-restricted |
| 157RAYLED162 [6] | 0 | 1 | 0 | 0 | 0 | B*8201-restricted |
| 163TCVEWL168 [6] | 282 | 206 | 200 | 0 | 30 | Polyspecific |
| 182EPPKTHVT190 [8] | 0 | 0 | 19 | 0 | 0 | C-restricted |
| Peptide Sequence Exposed in the Absence of β2-Microglobuin | Number of Positive HLA-I Alleles | Prediction SCORES | Immunogenicity Rank | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Classical HLA-Ia | Non-Classical HLA-Ib | Specificity | Beta-Turn | Antigencity | Flexibility | Hydro Philicity | |||||
| [Total Number of Amino Acids] | A | B | Cw | F | G | Chou & Fasman (1978) | Kolaskar & Tangaonkar (1990) | Karplus & Schulz (1985) | Parker (1986) | ||
| 115QFAYDGKDY123 [9] | 1 | 104 | 75 | 0 | 0 | Polyspecific | 1.059 | 1.001 | 0.993 | 2.629/3.201 | 5 |
| 117AYDGKDY123 [7] | 491 | 831 | 271 | 21 | 30 | Polyspecific | 1.204 | 0.989 | 1.061 | 4.243 | 1 |
| Cysteine Position | ||||||
|---|---|---|---|---|---|---|
| Alleles of HLA-I Isoforms | 42 | 67 | 101 | 164 | 203 | 259 |
| B *1401/2/3/4/5/6 | C | C | C | C | ||
| B *1509/10/66 | C | C | C | C | ||
| B *2701/2–17/19–36 | C | C | C | C | ||
| B *3801/2/4/5/8–15 | C | C | C | C | ||
| B 41*3901/3–7/9/12/14/15/18/19/22/24/26–37/41 | C | C | C | C | ||
| B *7301 | C | C | C | C | ||
| B *7803 | C | C | C | C | ||
| almost All C* alleles | C | C | C | |||
| almost All A* alleles | C | C | C | |||
| HLA-E | C | C | C | C | ||
| HLA-F | C | C | C | C | ||
| HLA-G | C | C | C | C | C | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ravindranath, M.H.; Ravindranath, N.M.; Selvan, S.R.; Filippone, E.J.; Amato-Menker, C.J.; El Hilali, F. Four Faces of Cell-Surface HLA Class-I: Their Antigenic and Immunogenic Divergence Generating Novel Targets for Vaccines. Vaccines 2022, 10, 339. https://doi.org/10.3390/vaccines10020339
Ravindranath MH, Ravindranath NM, Selvan SR, Filippone EJ, Amato-Menker CJ, El Hilali F. Four Faces of Cell-Surface HLA Class-I: Their Antigenic and Immunogenic Divergence Generating Novel Targets for Vaccines. Vaccines. 2022; 10(2):339. https://doi.org/10.3390/vaccines10020339
Chicago/Turabian StyleRavindranath, Mepur H., Narendranath M. Ravindranath, Senthamil R. Selvan, Edward J. Filippone, Carly J. Amato-Menker, and Fatiha El Hilali. 2022. "Four Faces of Cell-Surface HLA Class-I: Their Antigenic and Immunogenic Divergence Generating Novel Targets for Vaccines" Vaccines 10, no. 2: 339. https://doi.org/10.3390/vaccines10020339
APA StyleRavindranath, M. H., Ravindranath, N. M., Selvan, S. R., Filippone, E. J., Amato-Menker, C. J., & El Hilali, F. (2022). Four Faces of Cell-Surface HLA Class-I: Their Antigenic and Immunogenic Divergence Generating Novel Targets for Vaccines. Vaccines, 10(2), 339. https://doi.org/10.3390/vaccines10020339






