Plant-Based Vaccines in Combat against Coronavirus Diseases
Abstract
:1. Introduction
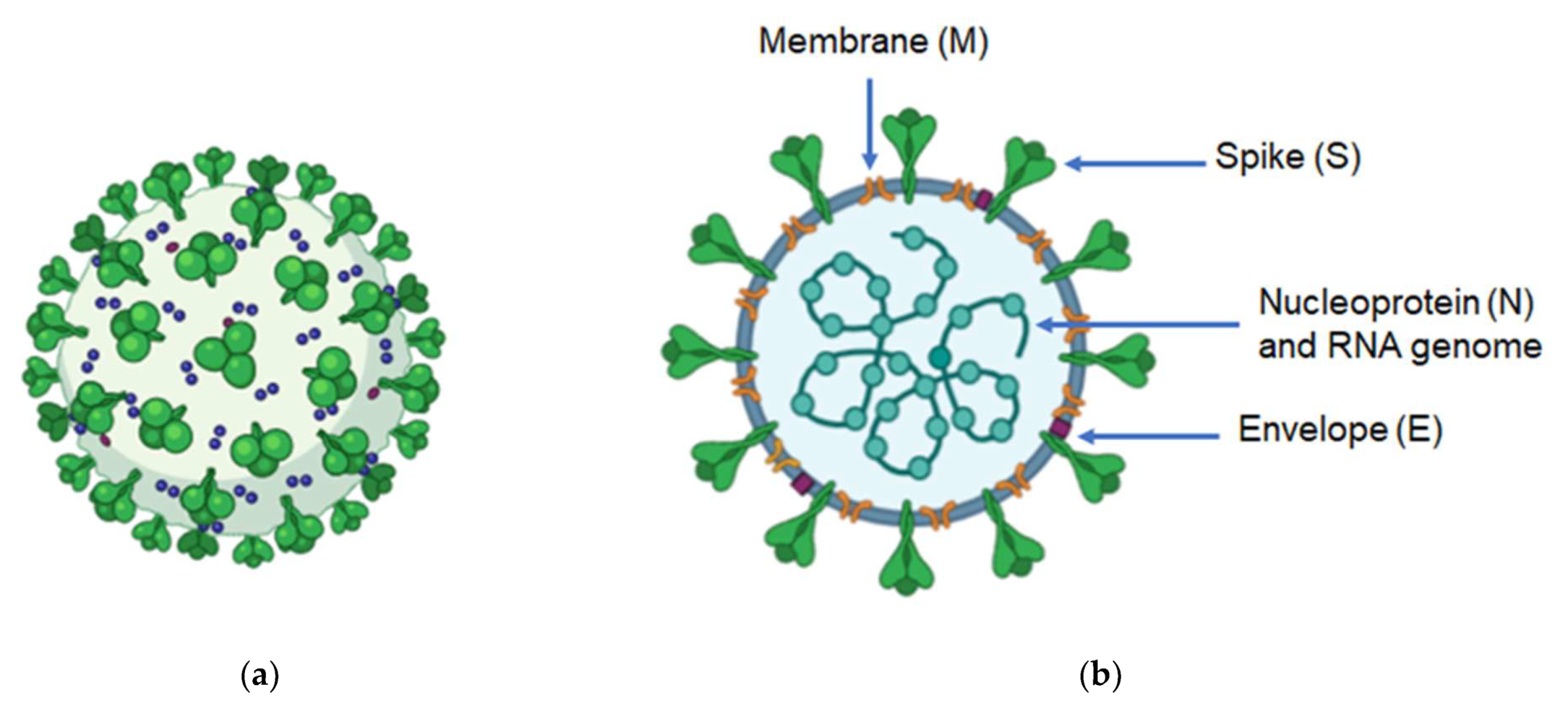
2. Genome and Structure of Coronaviruses
3. Treatment and Prophylaxis against COVID-19
4. Challenges in Developing CoV Vaccines
4.1. Approaches for Anti-CoV Vaccines
| Vaccine Platform | Advantages | Disadvantages | Developers (Vaccines) | Reference |
|---|---|---|---|---|
| Live attenuated | Mimic natural infection Long-lasting immunity Process used for several licensed human vaccines | Possibility to reverse to natural form Contraindicated in immunocompromised individuals | Codagenix (COVI-VAC) Meissa Vaccines, Inc. (MV-014-212) | [53,54,55,56,57,58,59,60,61] |
| Inactivated | No replication of the inactivated pathogen High stability Process used for several licensed human vaccines | Antigen and/or epitope integrity needs to be confirmed Multiple booster doses are required to obtain long-term protection | Sinovac (CoronaVac) Sinopharm (BIBP-CorV) Osaka University (CovidVax) | [39,55] |
| Subunit | Non-infectious Safe | Reduced immunogenicity, adjuvants are often needed | Novavax (NVX-CoV2373) Sanofi Pasteur (VAT00008) Instituto Finlay de Vacunas (Soberana 02) Kentucky Bioprocessing (KBP-201) | [36,57,58] |
| VLPs | Multimeric presentation of antigen Safe No viral replication | Purification can be a limiting factor | Astrazeneca (Vaxzevria) CanSino Biologics Inc. (Convidicea) Gamaleya Research Institute (Sputnik V) Janssen Pharmaceuticals (Ad26.COV2-S) | [59,60,61,62] |
| DNA | Induce both humoral and cell-mediated immune responses Highly scalable | Low immunogenicity | Zydus Cadila (ZyCoV-D) Inovio Pharmaceuticals (INO-4800) | [59,60,61] |
| RNA and modRNA | Safe Low-cost and highly scalable | Limited experimental information Instability | Pfizer/BioNTech (BNT162b2) Moderna (mRNA-1273) CureVac AG (CVnCoV) Imperial College London | [61,62,63] |
4.2. Molecular Farming for Recombinant Protein Expression
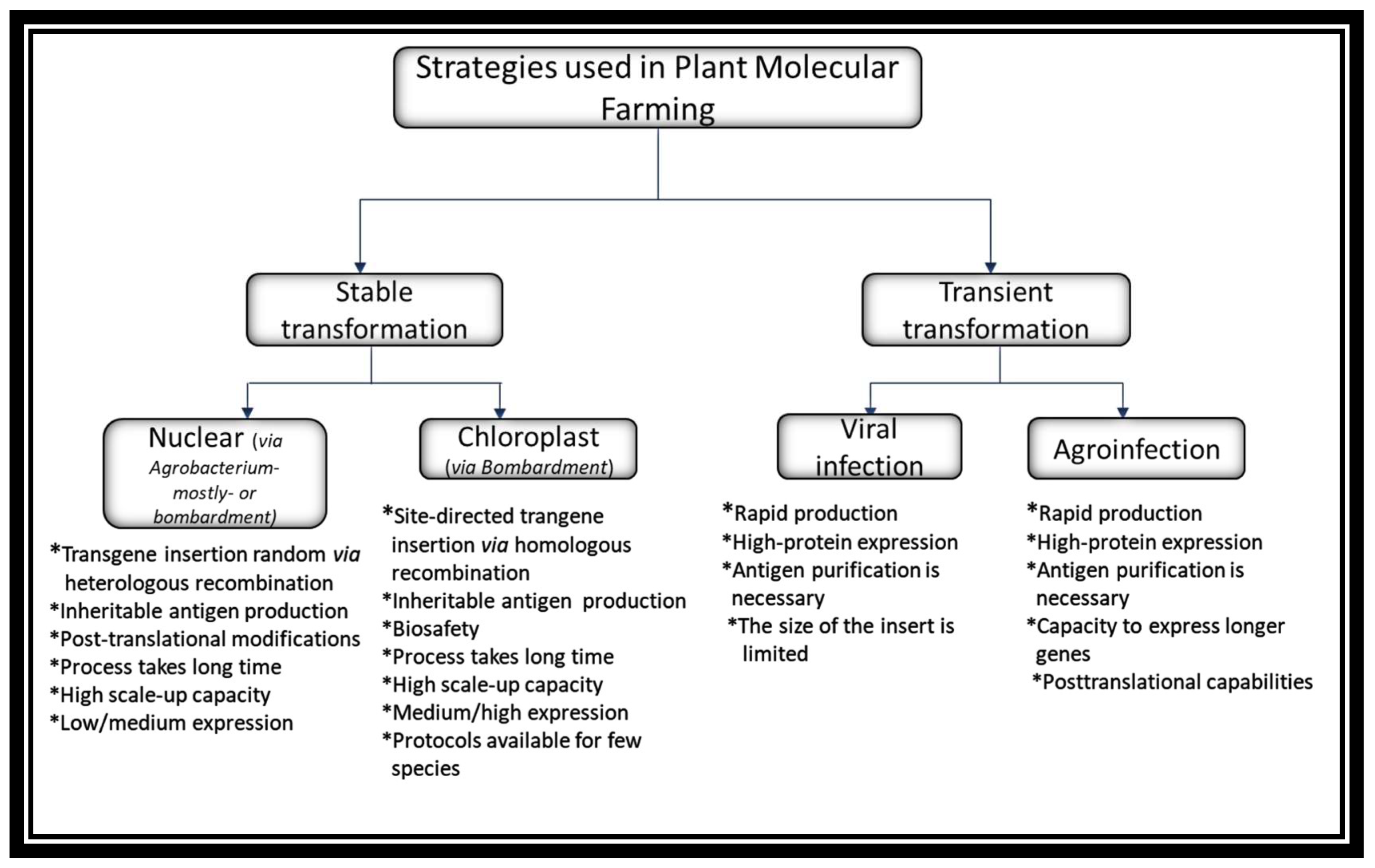
Plant Transformation Approaches
4.3. Plant-Made Vaccines against COVID-19 and SARS
5. Future Directions for Anti-CoV Plant-Made Vaccines
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef]
- Fehr, A.R.; Perlman, S. Coronaviruses: An Overview of Their Replication and Pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef]
- Snijder, E.J.; van der Meer, Y.; Zevenhoven-Dobbe, J.; Onderwater, J.J.M.; van der Meulen, J.; Koerten, H.K.; Mommaas, A.M. Ultrastructure and Origin of Membrane Vesicles Associated with the Severe Acute Respiratory Syndrome Coronavirus Replication Complex. J. Virol. 2006, 80, 5927–5940. [Google Scholar] [CrossRef] [PubMed]
- Cauchemez, S.; Van Kerkhove, M.D.; Riley, S.; Donnelly, C.A.; Fraser, C.; Ferguson, N.M. Transmission scenarios for middle east respiratory syndrome coronavirus (MERS-CoV) and how to tell them apart. Euro. Surveill. 2013, 18, 20503. [Google Scholar] [CrossRef]
- Ludwig, S.; Zarbock, A. Coronaviruses and SARS-CoV-2: A Brief Overview. Anesth. Analg. 2020, 30, 93–96. [Google Scholar] [CrossRef]
- Tian, X.; Li, C.; Huang, A.; Xia, S.; Lu, S.; Shi, Z.; Lu, L.; Jiang, S.; Yang, Z.; Wu, Y.; et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. Emerg. Microbes Infect. 2020, 9, 382–385. [Google Scholar] [CrossRef]
- Drexler, J.F.; Corman, V.M.; Drosten, C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014, 101, 45–56. [Google Scholar] [CrossRef]
- Prompetchara, E.; Ketloy, C.; Palaga, T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020, 38, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Weekly Epidemiological Update on COVID-19—30 November 2021. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---30-November-2021 (accessed on 30 November 2021).
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar] [PubMed]
- Ziebuhr, J. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 2005, 287, 57–94. [Google Scholar] [CrossRef]
- De Diego, M.L.; Nieto-Torres, J.L.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Castaño-Rodriguez, C.; Fernandez-Delgado, R.; Usera, F.; Enjuanes, L. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014, 194, 124–137. [Google Scholar] [CrossRef]
- Nal, B.; Chan, C.; Kien, F.; Siu, L.; Tse, J.; Chu, K.; Kam, J.; Staropoli, I.; Crescenzo-Chaigne, B.; Escriou, N.; et al. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S, M and E. J. Gen. Virol. 2005, 86, 1423–1434. [Google Scholar] [CrossRef] [PubMed]
- Neuman, B.W.; Kiss, G.; Kunding, A.H.; Bhella, D.; Baksh, M.F.; Connelly, S.; Droese, B.; Klaus, J.P.; Makino, S.; Sawicki, S.G.; et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011, 174, 11–22. [Google Scholar] [CrossRef]
- De Diego, M.L.; Álvarez, E.; Almazán, F.; Rejas, M.T.; Lamirande, E.; Roberts, A.; Shieh, W.-J.; Zaki, S.R.; Subbarao, K.; Enjuanes, L. A Severe Acute Respiratory Syndrome Coronavirus That Lacks the E Gene Is Attenuated In Vitro and In Vivo. J. Virol. 2007, 81, 1701–1713. [Google Scholar] [CrossRef] [PubMed]
- Luan, J.; Lu, Y.; Jin, X.; Zhang, L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem. Biophys. Res. Commun. 2020, 526, 165–169. [Google Scholar] [CrossRef] [PubMed]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.J.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Sheahan, T.P.; Sims, A.C.; Graham, R.L.; Menachery, V.D.; Gralinski, L.E.; Case, J.B.; Leist, S.R.; Pyrc, K.; Feng, J.Y.; Trantcheva, I.; et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017, 9, eaal3653. [Google Scholar] [CrossRef] [PubMed]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef]
- Dong, L.; Hu, S.; Gao, J. Discovering drugs to treat coronavirus disease 2019 (COVID-19). Drug Discov. Ther. 2020, 14, 58–60. [Google Scholar] [CrossRef]
- Amawi, H.; Abu Deiab, G.I.; Aljabali, A.A.A.; Dua, K.; Tambuwala, M.M. COVID-19 pandemic: An overview of epidemiology, pathogenesis, diagnostics and potential vaccines and therapeutics. Ther. Deliv. 2020, 11, 245–268. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Yao, Y.; Yeung, M.-L.; Deng, W.; Bao, L.; Jia, L.; Li, F.; Xiao, C.; Gao, H.; Yu, P.; et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015, 212, 1904–1913. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.M.; Cheng, V.C.C.; Hung, I.F.N.; Wong, M.M.L.; Chan, K.H.; Chan, K.S.; Kao, R.Y.T.; Poon, L.L.M.; Wong, C.L.P.; Guan, Y.; et al. Role of lopinavir/ritonavir in the treatment of SARS: Initial virological and clinical findings. Thorax 2004, 59, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Khamitov, R.A.; Loginova, S.; Shchukina, V.N.; Borisevich, S.V.; Maksimov, V.A.; Shuster, A. Antiviral activity of arbidol and its derivatives against the pathogen of severe acute respiratory syndrome in the cell cultures. Vopr. Virusol. 2008, 53, 9–13. (In Russian) [Google Scholar] [PubMed]
- Xia, S.; Zhu, Y.; Liu, M.; Lan, Q.; Xu, W.; Wu, Y.; Ying, T.; Liu, S.; Shi, Z.; Jiang, S.; et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell. Mol. Immunol. 2020, 17, 765–767. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Yi, L.; Chen, J.; Qu, X.; Qing, T.; Rao, X.; Jiang, P.; Hu, J.; Xiong, Z.; Nie, Y.; et al. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem. Biophys. Res. Commun. 2004, 319, 746–752. [Google Scholar] [CrossRef]
- Zhu, Y.; Yu, D.; Yan, H.; Chong, H.; He, Y. Design of Potent Membrane Fusion Inhibitors against SARS-CoV-2, an Emerging Coronavirus with High Fusogenic Activity. J. Virol. 2020, 94, e00635-20. [Google Scholar] [CrossRef]
- Vanpatten, S.; He, M.; Altiti, A.; Cheng, K.F.; Ghanem, M.H.; Al-Abed, Y. Evidence supporting the use of peptides and peptidomimetics as potential SARS-CoV-2 (COVID-19) therapeutics. Future Med. Chem. 2020, 12, 1647–1656. [Google Scholar] [CrossRef]
- Savarino, A.; Boelaert, J.R.; Cassone, A.; Majori, G.; Cauda, R. Effects of chloroquine on viral infections: An old drug against today’s diseases? Lancet Infect. Dis. 2003, 3, 722–727. [Google Scholar] [CrossRef]
- Duan, J.; Yan, X.; Guo, X.; Cao, W.; Han, W.; Qi, C.; Feng, J.; Yang, D.; Gao, G.; Jin, G. A human SARS-CoV neutralizing antibody against epitope on S2 protein. Biochem. Biophys. Res. Commun. 2005, 333, 186–193. [Google Scholar] [CrossRef]
- Pascal, K.E.; Coleman, C.M.; Mujica, A.O.; Kamat, V.; Badithe, A.; Fairhurst, J.; Hunt, C.; Strein, J.; Berrebi, A.; Sisk, J.M.; et al. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc. Natl. Acad. Sci. USA 2015, 112, 8738–8743. [Google Scholar] [CrossRef]
- Zhu, Z.; Chakraborti, S.; He, Y.; Roberts, A.; Sheahan, T.; Xiao, D.; Hensley, L.E.; Prabakaran, P.; Rockx, B.; Sidorov, I.A.; et al. Potent cross-reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc. Natl. Acad. Sci. USA 2007, 104, 12123–12128. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wong, R.; Soo, Y.O.Y.; Wong, W.S.; Lee, C.K.; Ng, M.H.L.; Chan, P.; Wong, K.C.; Leung, C.B.; Cheng, G. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005, 24, 44–46. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Y.M.; Xu, J.Y.; Cao, B. Potential antiviral therapeutics for 2019 Novel Coronavirus. Chinese J. Tuberc. Respir. Dis. 2020, 43, E002. (In Chinese) [Google Scholar] [CrossRef]
- Weiss, S.R.; Navas-Martin, S. Coronavirus Pathogenesis and the Emerging Pathogen Severe Acute Respiratory Syndrome Coronavirus. Microbiol. Mol. Biol. Rev. 2005, 69, 635–664. [Google Scholar] [CrossRef]
- Afrough, B.; Dowall, S.; Hewson, R. Emerging viruses and current strategies for vaccine intervention. Clin. Exp. Immunol. 2019, 196, 157–166. [Google Scholar] [CrossRef]
- Gouglas, D.; Thanh Le, T.; Henderson, K.; Kaloudis, A.; Danielsen, T.; Hammersland, N.C.; Robinson, J.M.; Heaton, P.M.; Røttingen, J.A. Estimating the cost of vaccine development against epidemic infectious diseases: A cost minimisation study. Lancet Glob. Health 2018, 6, e1386–e1396. [Google Scholar] [CrossRef]
- Hunter, D.J. Developing Covid-19 vaccines at pandemic speed. N. Engl. J. Med. 2020, 31, 1969–1973. [Google Scholar]
- Diamond, M.S.; Pierson, T.C. The Challenges of Vaccine Development against a New Virus during a Pandemic. Cell Host Microbe 2020, 27, 699–703. [Google Scholar] [CrossRef]
- Dai, L.; Zheng, T.; Xu, K.; Han, Y.; Xu, L.; Huang, E.; An, Y.; Cheng, Y.; Li, S.; Liu, M.; et al. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell. 2020, 182, 722–733. [Google Scholar] [CrossRef] [PubMed]
- Dagotto, G.; Yu, J.; Barouch, D.H. Approaches and Challenges in SARS-CoV-2 Vaccine Development. Cell Host Microbe 2020, 28, 364–370. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Quadeer, A.A.; McKay, M.R. Preliminary identification of potential vaccine targets for the COVID-19 Coronavirus (SARS-CoV-2) Based on SARS-CoV Immunological Studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef]
- Liang, Z.; Zhu, H.; Wang, X.; Jing, B.; Li, Z.; Xia, X.; Sun, H.; Yang, Y.; Zhang, W.; Shi, L.; et al. Adjuvants for coronavirus vaccines. Front. Immunol. 2020, 11, 2896–2905. [Google Scholar] [CrossRef]
- Coleman, C.M.; Liu, Y.V.; Mu, H.; Taylor, J.K.; Massare, M.; Flyer, D.C.; Smith, G.E.; Frieman, M.B. Purified coronavirus spike protein nanoparticles induce coronavirus neutralizing antibodies in mice. Vaccine. 2014, 32, 3169–3174. [Google Scholar] [CrossRef]
- Lan, J.; Deng, Y.; Chen, H.; Lu, G.; Wang, W.; Guo, X.; Lu, Z.; Gao, G.F.; Tan, W. Tailoring subunit vaccine immunity with adjuvant combinations and delivery routes using the Middle East respiratory coronavirus (MERS-CoV) receptor-binding domain as an antigen. PLoS ONE 2014, 9, e112602. [Google Scholar] [CrossRef]
- Moderbacher, C.R.; Ramirez, S.I.; Dan, J.M.; Grifoni, A.; Hastie, K.M.; Weiskopf, D.; Belanger, S.; Abbott, R.K.; Kim, C.; Choi, J.; et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell 2020, 183, 996–1012. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Li, K.; Rowe, D.K.; Diaz, M.C.H.; Griffin, E.F.; Beavis, A.C.; Johnson, S.K.; Padykula, I.; Jones, C.A.; Briggs, K.; et al. Protection of K18-hACE2 mice and ferrets against SARS-CoV-2 challenge by a single-dose mucosal immunization with a parainfluenza virus 5-based COVID-19 vaccine. Sci. Adv. 2021, 7, eabi5246. [Google Scholar] [CrossRef] [PubMed]
- Corbett, K.S.; Flynn, B.; Foulds, K.E.; Francica, J.R.; Boyoglu-Barnum, S.; Werner, A.P.; Flach, B.; O’Connell, S.; Bock, K.W.; Minai, M.; et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N. Engl. J. Med. 2020, 383, 1544–1555. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, Y.; Huang, B.; Deng, W.; Quan, Y.; Wang, W.; Xu, W.; Zhao, Y.; Li, N.; Zhang, J.; et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell 2020, 182, 713–721.e9. [Google Scholar] [CrossRef]
- Mercado, N.B.; Zahn, R.; Wegmann, F.; Loos, C.; Chandrashekar, A.; Yu, J.; Liu, J.; Peter, L.; McMahan, K.; Tostanoski, L.H.; et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature 2020, 586, 583–588. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Lambe, T.; Spencer, A.; Belij-Rammerstorfer, S.; Purushotham, J.N.; Port, J.R.; Avanzato, V.A.; Bushmaker, T.; Flaxman, A.; Ulaszewska, M.; et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 2020, 586, 578–582. [Google Scholar] [CrossRef]
- Yu, J.; Tostanosk, L.H.; Peter, L.; Mercad, N.B.; McMahan, K.; Mahrokhia, S.H.; Nkolol, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Hajj Hussein, I.; Chams, N.; Chams, S.; El Sayegh, S.; Badran, R.; Raad, M.; Gerges-Geagea, A.; Leone, A.; Jurjus, A. Vaccines Through Centuries: Major Cornerstones of Global Health. Front. Public Health 2015, 3, 269. [Google Scholar] [CrossRef] [PubMed]
- Pulendran, B.; Ahmed, R. Immunological mechanisms of vaccination. Nat. Immunol. 2011, 12, 509–517. [Google Scholar] [CrossRef]
- Vetter, V.; Denizer, G.; Friedland, L.R.; Krishnan, J.; Shapiro, M. Understanding modern-day vaccines: What you need to know. Ann. Med. 2018, 50, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Gillim-Ross, L.; Subbarao, K. Emerging respiratory viruses: Challenges and vaccine strategies. Clin. Microbiol. Rev. 2006, 19, 614–636. [Google Scholar] [CrossRef]
- Droppa-Almeida, D.; Franceschi, E.; Padilha, F.F. Immune-informatic analysis and design of peptide vaccine from multi-epitopes against Corynebacterium pseudotuberculosis. Bioinform. Biol. Insights 2018, 12, 25–29. [Google Scholar] [CrossRef]
- Rappuoli, R.; Pizza, M.; Del Giudice, G.; De Gregorio, E. Vaccines, new opportunities for a new society. Proc. Natl. Acad. Sci. USA 2014, 111, 12288–12293. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.F.; Zhou, J.; Chen, S.; Cai, L.L.; Bao, Q.Y.; Zheng, F.Y.; Lu, J.Q.; Padmanabha, J.; Hengst, K.; Malcolm, K.; et al. HPV6b virus like particles are potent immunogens without adjuvant in man. Vaccine 2000, 18, 1051–1058. [Google Scholar] [CrossRef]
- García-Sastre, A.; Mena, I. Novel vaccine strategies against emerging viruses. Curr. Opin. Virol. 2013, 3, 210–216. [Google Scholar] [CrossRef]
- Wallis, J.; Shenton, D.P.; Carlisle, R.C. Novel approaches for the design, delivery and administration of vaccine technologies. Clin. Exp. Immunol. 2019, 196, 189–204. [Google Scholar] [CrossRef]
- Maslow, J.N. Vaccine development for emerging virulent infectious diseases. Vaccine 2017, 35, 5437–5443. [Google Scholar] [CrossRef] [PubMed]
- Roncati, L.; Corsi, L. Nucleoside-modified messenger RNA COVID-19 vaccine platform. J. Med. Virol. 2021, 93, 4054–4057. [Google Scholar] [CrossRef] [PubMed]
- Obembe, O.O.; Popoola, J.O.; Leelavathi, S.; Reddy, S.V. Advances in plant molecular farming. Biotechnol. Adv. 2011, 29, 210–222. [Google Scholar] [CrossRef]
- Twyman, R.M.; Stoger, E.; Schillberg, S.; Christou, P.; Fischer, R. Molecular farming in plants: Host systems and expression technology. Trends Biotechnol. 2003, 21, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Lienard, D. Pharming and transgenic plants. Biotechnol. Annu. Rev. 2007, 13, 115–147. [Google Scholar]
- Giddings, G. Transgenic plants as protein factories. Curr. Opin. Biotechnol. 2001, 12, 450–454. [Google Scholar] [CrossRef]
- Fischer, R.; Emans, N. Molecular farming of pharmaceutical proteins. Transgenic Res. 2000, 9, 279–299. [Google Scholar] [CrossRef]
- Tekoah, Y.; Shulman, A.; Kizhner, T.; Ruderfer, I.; Fux, L.; Nataf, Y.; Bartfeld, D.; Ariel, T.; Gingis-Velitski, S.; Hanania, U.; et al. Large-scale production of pharmaceutical proteins in plant cell culture-the protalix experience. Plant Biotechnol. J. 2015, 13, 1199–1208. [Google Scholar] [CrossRef]
- Naderi, S.; Baratali, F. Overview of Plant-based Vaccines. Res. J. Fish. Hydrobiol. 2015, 10, 275–289. [Google Scholar]
- Ward, B.J.; Makarkov, A.; Séguin, A.; Pillet, S.; Trépanier, S.; Dhaliwall, J.; Libman, M.D.; Vesikari, T.; Landry, N. Efficacy, immunogenicity, and safety of a plant-derived, quadrivalent, virus-like particle influenza vaccine in adults (18–64 years) and older adults (≥65 years): Two multicentre, randomised phase 3 trials. Lancet 2020, 396, 1491–1503. [Google Scholar] [CrossRef]
- Shanmugaraj, B.; Bulaon, C.J.I.; Phoolcharoen, W. Plant molecular farming: A viable platform for recombinant biopharmaceutical production. Plants 2020, 9, 842. [Google Scholar] [CrossRef]
- Sartaj Sohrab, S.; Suhail, M.; Kamal, M.A.; Husen, A.; Azhar, E.I. Recent Development and Future Prospects of Plant-Based Vaccines. Curr. Drug Metab. 2017, 18, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.B.; Park, J.S.; Park, Y.I.; Song, I.J.; Lee, H.J.; Cho, H.S.; Jeon, J.H.; Kim, H.S. Development of systems for the production of plant-derived biopharmaceuticals. Plants 2020, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Rosales-Mendoza, S.; Márquez-Escobar, V.A.; González-Ortega, O.; Nieto-Gómez, R.; Arévalo-Villalobos, J.I. What does plant-based vaccine technology offer to the fight against COVID-19? Vaccines 2020, 8, 183. [Google Scholar] [CrossRef]
- Horn, M.E.; Woodard, S.L.; Howard, J.A. Plant molecular farming: Systems and products. Plant Cell Rep. 2004, 22, 711–720. [Google Scholar] [CrossRef]
- Laere, E.; Ling, A.P.K.; Wong, Y.P.; Koh, R.Y.; Mohd Lila, M.A.; Hussein, S. Plant-based vaccines: Production and challenges. J. Bot. 2016, 2016, 4928637. [Google Scholar] [CrossRef]
- Gleba, Y.; Klimyuk, V.; Marillonnet, S. Magnifection—A new platform for expressing recombinant vaccines in plants. Vaccine 2005, 23, 2042–2048. [Google Scholar] [CrossRef] [PubMed]
- Fujiki, M.; Kaczmarczyk, J.F.; Yusibov, V.; Rabindran, S. Development of a new cucumber mosaic virus-based plant expression vector with truncated 3a movement protein. Virology 2008, 381, 136–142. [Google Scholar] [CrossRef]
- COVID-19 Vaccine Tracker and Landscape. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 30 November 2021).
- Li, H.Y.; Ramalingam, S.; Chye, M.L. Accumulation of recombinant SARS-CoV spike protein in plant cytosol and chloroplasts indicate potential for development of plant-derived oral vaccines. Exp. Biol. Med. 2006, 231, 1346–1352. [Google Scholar] [CrossRef]
- Zheng, N.; Xia, R.; Yang, C.; Yin, B.; Li, Y.; Duan, C.; Liang, L.; Guo, H.; Xie, Q. Boosted expression of the SARS-CoV nucleocapsid protein in tobacco and its immunogenicity in mice. Vaccine 2009, 27, 5001–5007. [Google Scholar] [CrossRef] [PubMed]
- Pogrebnyak, N.; Golovkin, M.; Andrianov, V.; Spitsin, S.; Smirnov, Y.; Egolf, R.; Koprowski, H. Severe Acute Respiratory Syndrome (SARS) S Protein Production in Plants. Proc. Natl. Acad. Sci. USA 2005, 102, 9062–9067. [Google Scholar] [CrossRef]
- Ward, B.J.; Gobeil, P.; Séguin, A.; Atkins, J.; Boulay, I.; Charbonneau, P.Y.; Couture, M.; D’Aoust, M.A.; Dhaliwall, J.; Finkle, C.; et al. Phase 1 randomized trial of a plant-derived virus-like particle vaccine for COVID-19. Nat. Med. 2021, 27, 1071–1078. [Google Scholar] [CrossRef]
- Kumar, A.U.; Kadiresen, K.; Gan, W.C.; Ling, A.P.K. Current updates and research on plant-based vaccines for coronavirus disease 2019. Clin. Exp. Vaccine Res. 2021, 10, 13–23. [Google Scholar] [CrossRef]
- Maharjan, P.M.; Choe, S. Plant-based COVID-19 vaccines: Current status, design, and development strategies of candidate vaccines. Vaccines 2021, 9, 992. [Google Scholar] [CrossRef] [PubMed]
- Shanmugaraj, B.; Siriwattananon, K.; Malla, A.; Phoolcharoen, W. Potential for developing plant-derived candidate vaccines and biologics against emerging coronavirus infections. Pathogens 2021, 10, 1051. [Google Scholar] [CrossRef]
- Yusibov, V.; Kushnir, N.; Streatfield, S.J. Antibody Production in Plants and Green Algae. Annu. Rev. Plant Biol. 2016, 67, 669–701. [Google Scholar] [CrossRef]
- Rybicki, E.P. Plant molecular farming of virus-like nanoparticles as vaccines and reagents. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1587. [Google Scholar] [CrossRef] [PubMed]
- Blokhina, E.A.; Kuprianov, V.V.; Stepanova, L.A.; Tsybalova, L.M.; Kiselev, O.I.; Ravin, N.V.; Skryabin, K.G. A molecular assembly system for presentation of antigens on the surface of HBc virus-like particles. Virology 2013, 435, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Pumpens, P.; Grens, E. The true story and advantages of the famous Hepatitis B virus core particles. Mol. Biol. 2016, 50, 489–509. [Google Scholar] [CrossRef]
- Stephen, S.L.; Beales, L.; Peyret, H.; Roe, A.; Stonehouse, N.J.; Rowlands, D.J. Recombinant expression of tandem-HBc virus-like particles (VLPs). Methods Mol. Biol. 2018, 1776, 97–123. [Google Scholar] [CrossRef]
- Thuenemann, E.C.; Meyers, A.E.; Verwey, J.; Rybicki, E.P.; Lomonossoff, G.P. A method for rapid production of heteromultimeric protein complexes in plants: Assembly of protective bluetongue virus-like particles. Plant Biotechnol. J. 2013, 11, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Zahmanova, G.; Mazalovska, M.; Takova, K.; Toneva, V.; Minkov, I.; Peyret, H.; Lomonossoff, G. Efficient production of chimeric hepatitis b virus-like particles bearing an epitope of hepatitis e virus capsid by transient expression in Nicotiana benthamiana. Life 2021, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lai, H.; Sun, H.; Chen, Q. Virus-like particles that display Zika virus envelope protein domain III induce potent neutralizing immune responses in mice. Sci. Rep. 2017, 7, 7679. [Google Scholar] [CrossRef]
- Ravin, N.V.; Kotlyarov, R.Y.; Mardanova, E.S.; Kuprianov, V.V.; Migunov, A.I.; Stepanova, L.A.; Tsybalova, L.M.; Kiselev, O.I.; Skryabin, K.G. Plant-produced recombinant influenza vaccine based on virus-like HBc particles carrying an extracellular domain of M2 protein. Biochemistry 2012, 77, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Bandurska, K.; Brodzik, R.; Spitsin, S.; Kohl, T.; Portocarrero, C.; Smirnov, Y.; Pogrebnyak, N.; Sirko, A.; Koprowski, H.; Golovkin, M. Plant-produced hepatitis B core protein chimera carrying anthrax protective antigen domain-4. Hybridoma 2008, 27, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Kesik-Brodacka, M.; Lipiec, A.; Kozak Ljunggren, M.; Jedlina, L.; Miedzinska, K.; Mikolajczak, M.; Plucienniczak, A.; Legocki, A.B.; Wedrychowicz, H. Immune response of rats vaccinated orally with various plant-expressed recombinant cysteine proteinase constructs when challenged with Fasciola hepatica metacercariae. PLoS Negl. Trop. Dis. 2017, 11, e0005451. [Google Scholar] [CrossRef]
- Qian, B.; Shen, H.; Liang, W.; Guo, X.; Zhang, C.; Wang, Y.; Li, G.; Wu, A.; Cao, K.; Zhang, D. Immunogenicity of recombinant hepatitis B virus surface antigen fused with preS1 epitopes expressed in rice seeds. Transgenic Res. 2008, 17, 621–631. [Google Scholar] [CrossRef]
- Matić, S.; Rinaldi, R.; Masenga, V.; Noris, E. Efficient production of chimeric Human papillomavirus 16 L1 protein bearing the M2e influenza epitope in Nicotiana benthamiana plants. BMC Biotechnol. 2011, 11, 106. [Google Scholar] [CrossRef]
- Mohammadzadeh, S.; Roohvand, F.; Memarnejadian, A.; Jafari, A.; Ajdary, S.; Salmanian, A.H.; Ehsani, P. Co-expression of hepatitis C virus polytope-HBsAg and p19-silencing suppressor protein in tobacco leaves. Pharm. Biol. 2016, 54, 465–473. [Google Scholar] [CrossRef]
- Balke, I.; Zeltins, A. Use of plant viruses and virus-like particles for the creation of novel vaccines. Adv. Drug Deliv. Rev. 2019, 145, 119–129. [Google Scholar] [CrossRef]
- Ibrahim, A.; Odon, V.; Kormelink, R. Plant viruses in plant molecular pharming: Toward the use of enveloped viruses. Front. Plant Sci. 2019, 10, 803. [Google Scholar] [CrossRef]
- Ríos-Huerta, R.; Monreal-Escalante, E.; Govea-Alonso, D.O.; Angulo, C.; Rosales-Mendoza, S. Expression of an immunogenic LTB-based chimeric protein targeting Zaire ebolavirus epitopes from GP1 in plant cells. Plant Cell Rep. 2017, 36, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Randall, T.D. Bronchus-Associated Lymphoid Tissue (BALT). In Structure and Function, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2010; Volume 107. [Google Scholar]
- Pabst, R. Mucosal vaccination by the intranasal route. Nose-associated lymphoid tissue (NALT)-Structure, function and species differences. Vaccine 2015, 33, 4406–4413. [Google Scholar] [CrossRef]
- Vela Ramirez, J.E.; Sharpe, L.A.; Peppas, N.A. Current state and challenges in developing oral vaccines. Adv. Drug Deliv. Rev. 2017, 114, 116–131. [Google Scholar] [CrossRef] [PubMed]
- Rezende, R.M.; Weiner, H.L. History and mechanisms of oral tolerance. Semin. Immunol. 2017, 30, 3–11. [Google Scholar] [CrossRef]
- Pniewski, T.; Milczarek, M.; Wojas-Turek, J.; Pajtasz-Piasecka, E.; Wietrzyk, J.; Czyż, M. Plant lyophilisate carrying S-HBsAg as an oral booster vaccine against HBV. Vaccine 2018, 36, 6070–6076. [Google Scholar] [CrossRef] [PubMed]
| Approach | Expressed Antigen/ Administration Route | Relevant Results | Reference |
|---|---|---|---|
| Nuclear and chloroplast transformation | 1-658 amino acids of SARS-CoV-1 S protein | The antigen was successfully expressed in transgenic tobacco and lettuce as well as in transplastomic tobacco. | [80] |
| Transient expression | Recombinant SARS-CoV-1 nucleocapsid (rN) protein/intraperitoneal | p19 protein enhanced the transient expression of rN up to a concentration of 79 µg per g fresh leaf weight, which induced in mice high levels of IgG1 and IgG2a. | [81] |
| Nuclear expression | N-terminal fragment of SARS-CoV S protein (S1)/oral | S1 protein was expressed in tomato and low-nicotine tobacco plants, which induced specific IgA and IgG responses in mice. | [82] |
| Transient expression | Full-length S glycoprotein of SARS-CoV-2/intramuscular | CoVLP alone or adjuvanted with either CpG1018 or AS03 suggests that the candidate vaccine is well-tolerated and immunogenic. Its immunogenicity, particularly at low doses, is radically enhanced by the presence of an adjuvant. | [83] |
| Transient expression | Protein subunit vaccine based on the SARS-CoV-2 receptor-binding domain (RBD)/intramuscular | The vaccine showed a positive result on stimulation of immune responses in pre-clinical trials. Clinical trials results not yet published. | [84] |
| Transient expression | Subunit vaccine combining antigens derived from the SARS-CoV-2 spike protein fused to LicKM/intramuscular | In pre-clinical trials the vaccine (IBIO-201) stimulated the immune response producing high titers of neutralizing antibodies. | [85] |
| Transient expression | Subunit vaccine from SARS-CoV-2 spike protein/injection (no more data provided) | The vaccine was able to induce antigen-specific IgG and neutralizing responses as well as cellular immunity in animals. | [86] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortega-Berlanga, B.; Pniewski, T. Plant-Based Vaccines in Combat against Coronavirus Diseases. Vaccines 2022, 10, 138. https://doi.org/10.3390/vaccines10020138
Ortega-Berlanga B, Pniewski T. Plant-Based Vaccines in Combat against Coronavirus Diseases. Vaccines. 2022; 10(2):138. https://doi.org/10.3390/vaccines10020138
Chicago/Turabian StyleOrtega-Berlanga, Benita, and Tomasz Pniewski. 2022. "Plant-Based Vaccines in Combat against Coronavirus Diseases" Vaccines 10, no. 2: 138. https://doi.org/10.3390/vaccines10020138
APA StyleOrtega-Berlanga, B., & Pniewski, T. (2022). Plant-Based Vaccines in Combat against Coronavirus Diseases. Vaccines, 10(2), 138. https://doi.org/10.3390/vaccines10020138







