Bursal-Derived BP7 Induces the miRNA Molecular Basis of Chicken Macrophages and Promotes the Differentiation of B Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptide Synthesis, Cells and Animals
2.2. MTT Assay
2.3. Determination of Cytokine Expression in HD11 Cells Treated with BP7
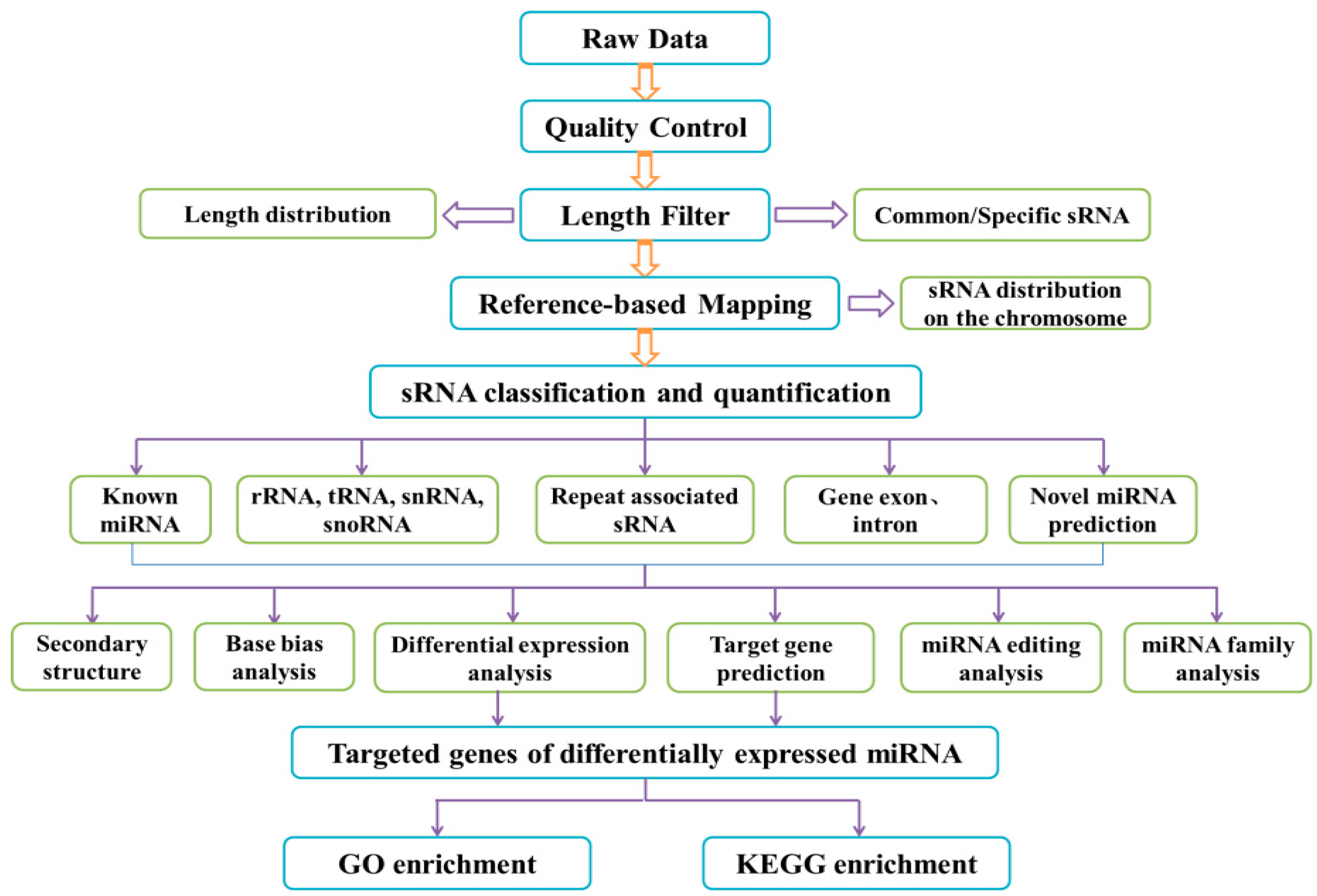
2.4. Preparation of Sequencing Samples and High Throughput miRNA Sequencing
2.5. Mice Immunization Model
2.6. Data Analysis
3. Results
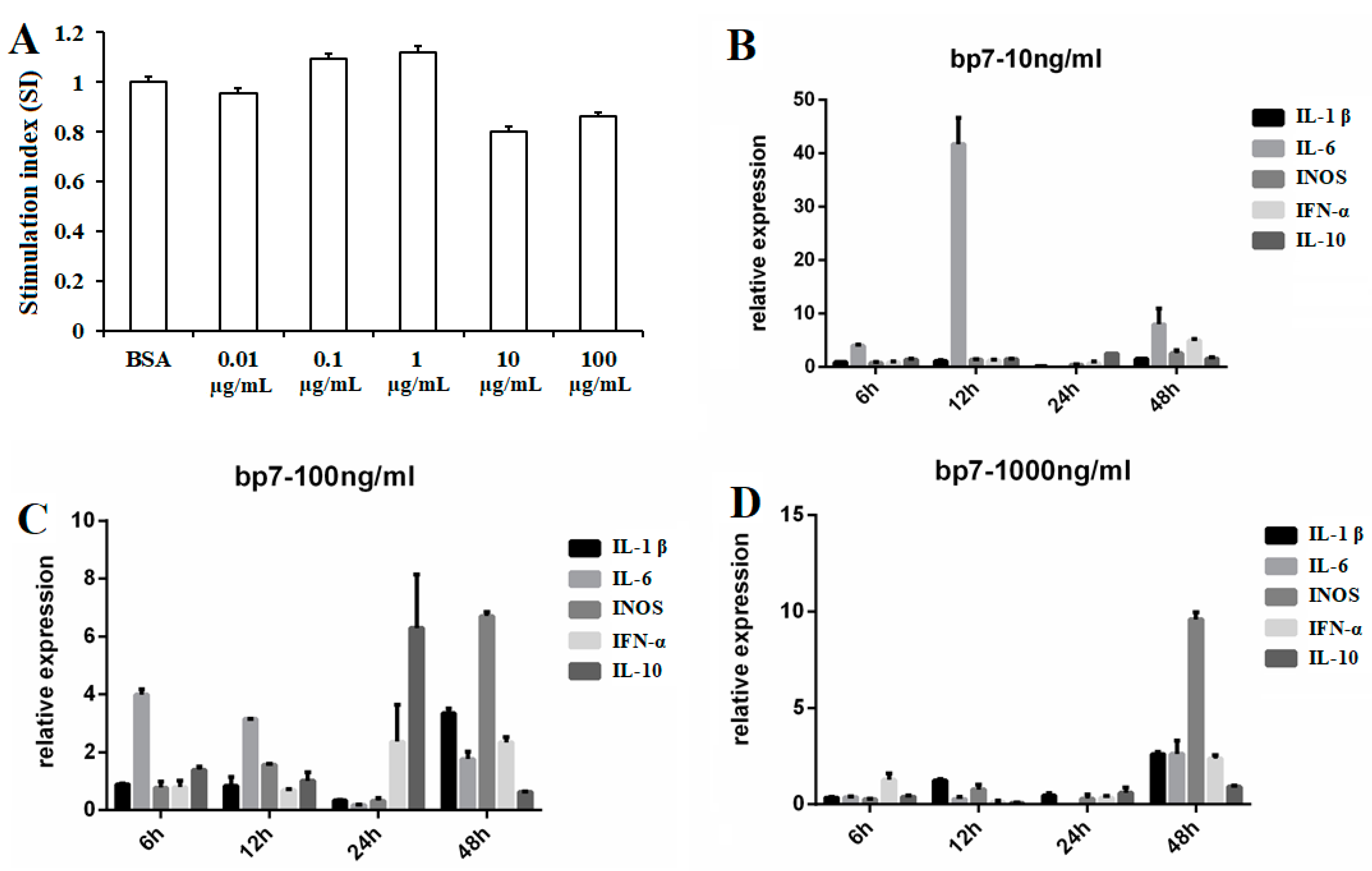
3.1. The Viabilities of HD11 Cells with BP7 Treatment
3.2. The Expressions of the Cytokine in HD11 Cells with BP7 Stimulation
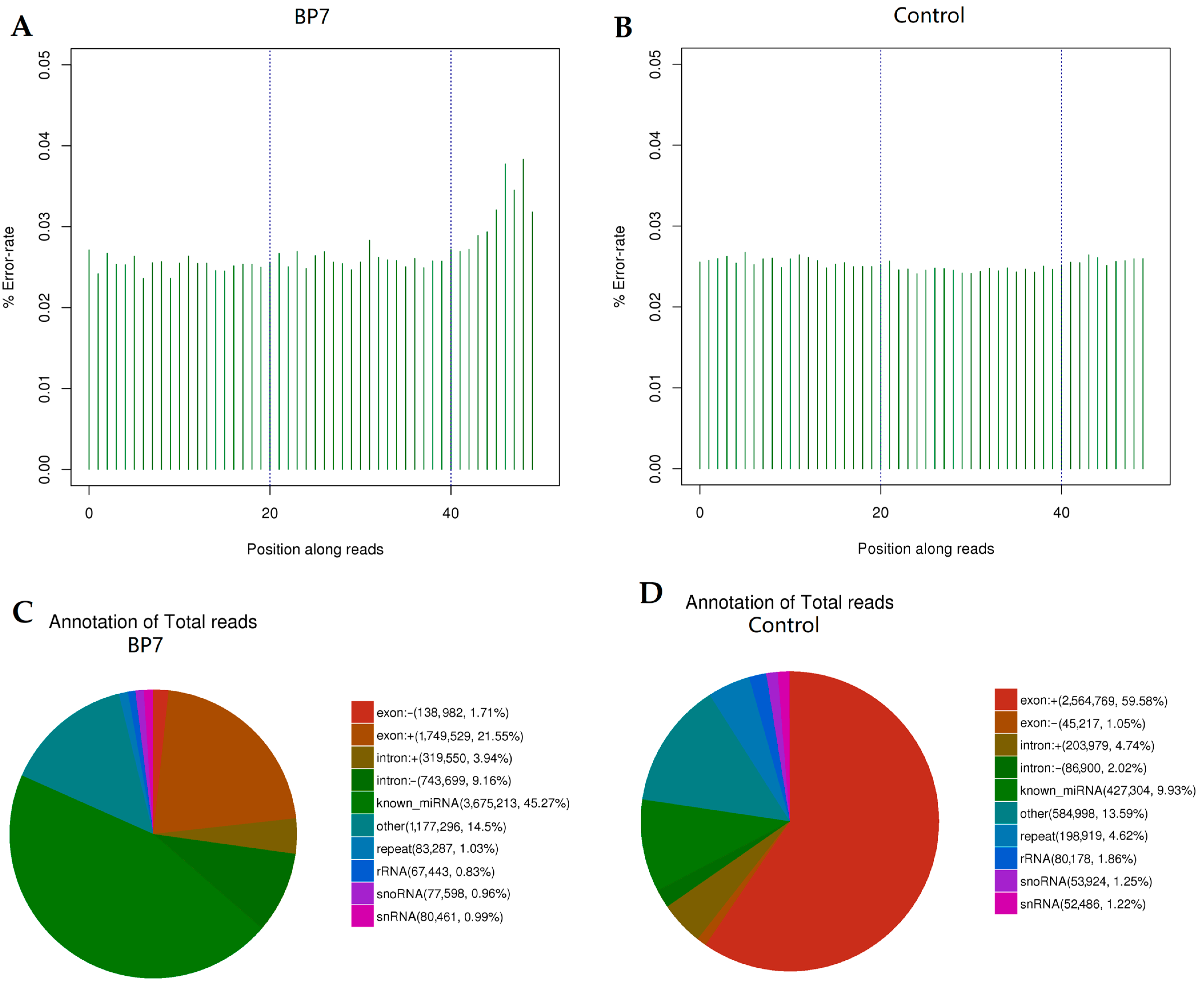
3.3. Library Construction Analysis
3.4. miRNA Expression Profile Analysis
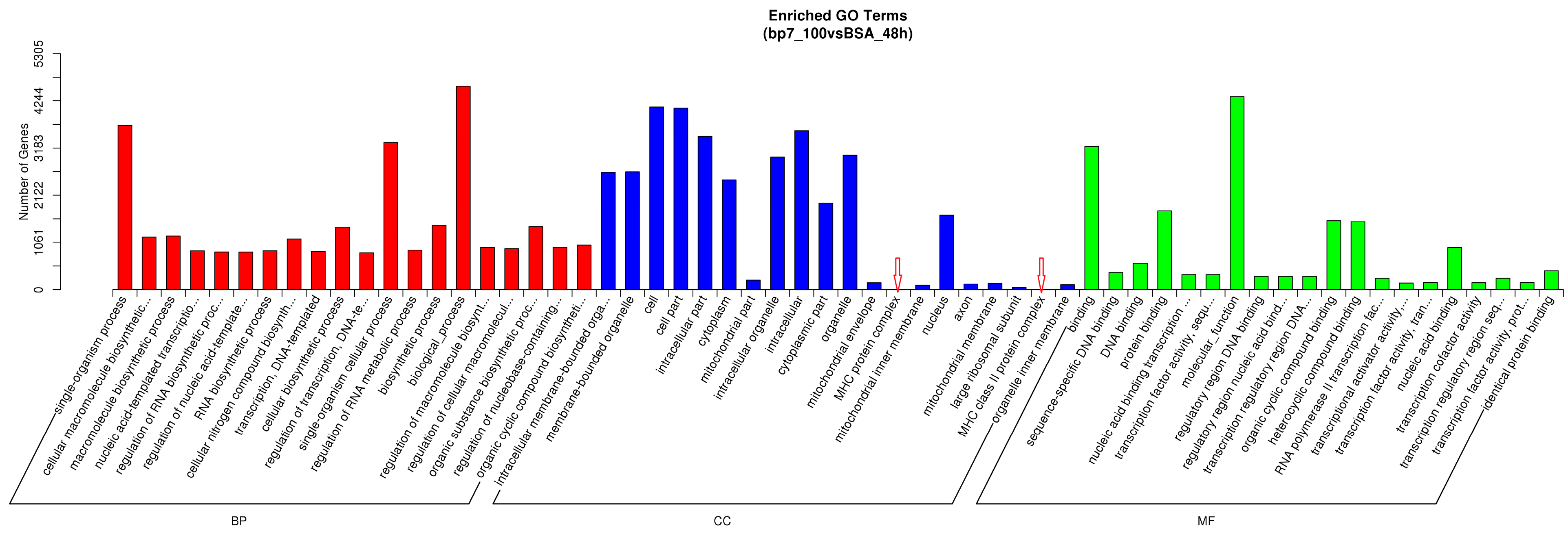
3.5. The Biological Function Analysis in HD11 Cells with BP7 Treatment
3.6. The Enriched Pathway in HD11 Cells with BP7 Treatment
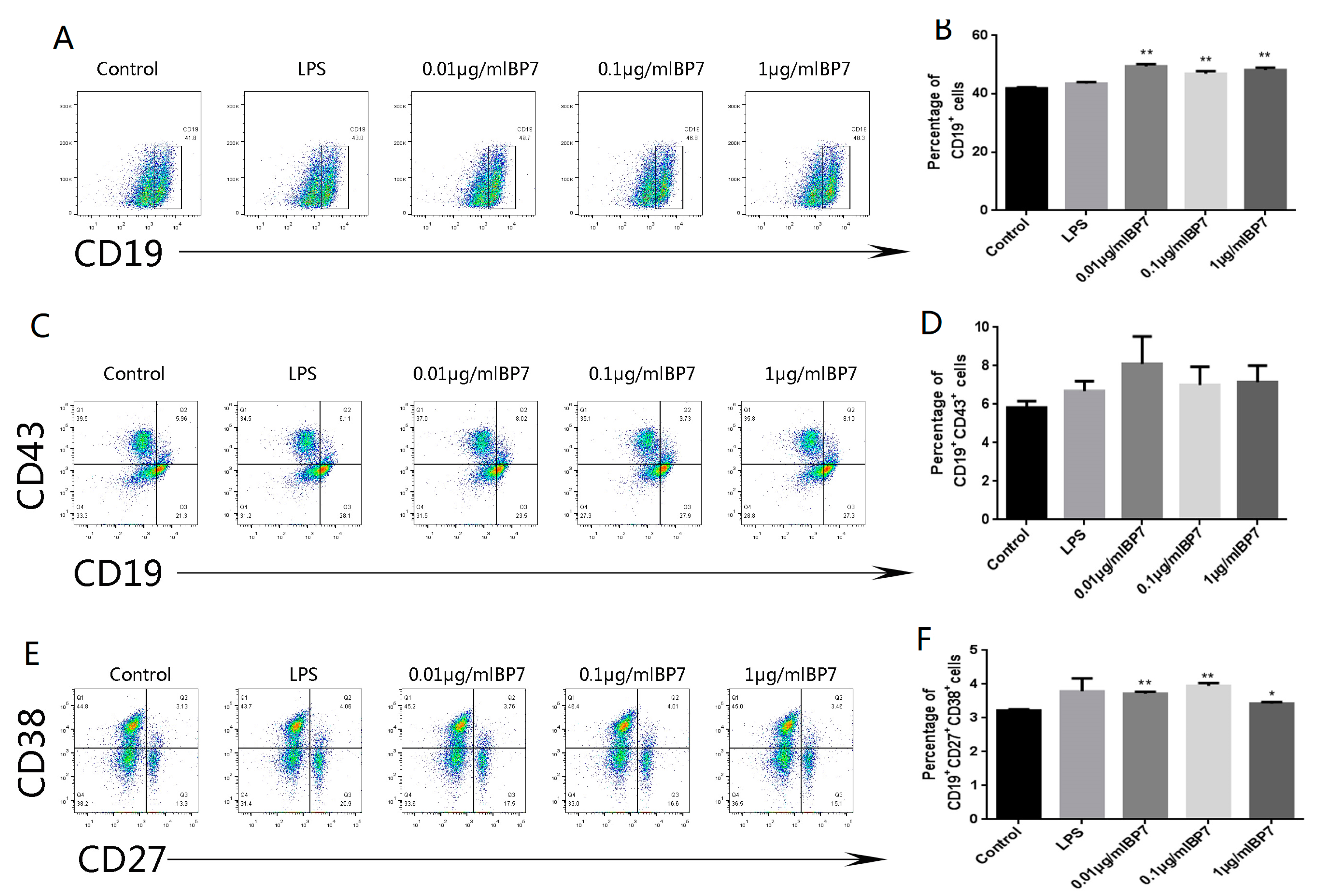
3.7. BP7 Induced B Cell Differentiation In Vitro
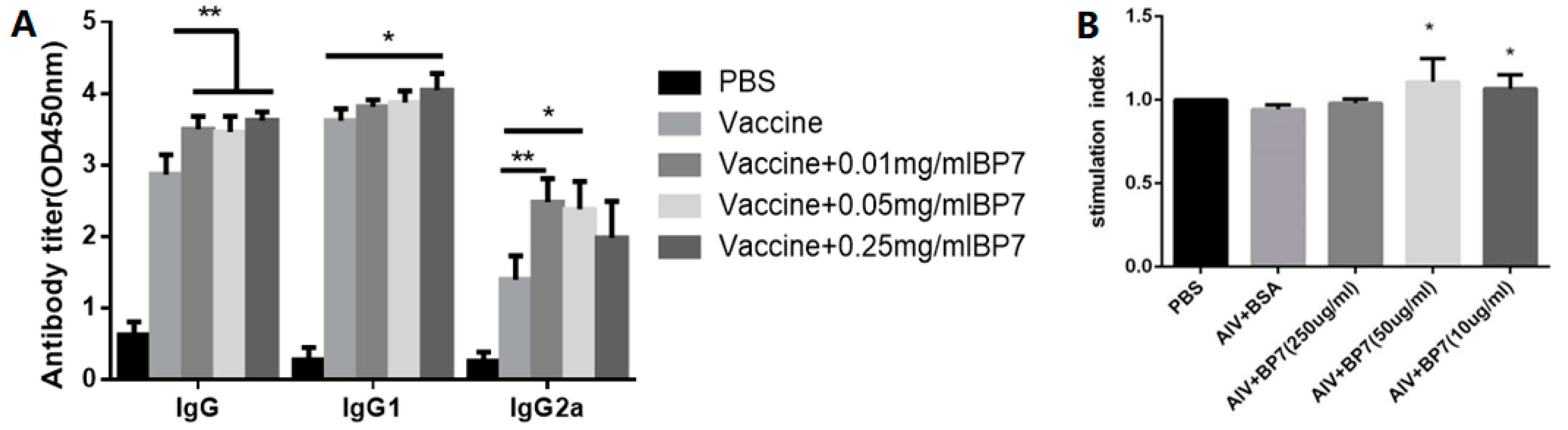
3.8. BP7 Induced B Cell Differentiation In Vivo
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ekino, S.; Sonoda, K. New insight into the origin of IgG-bearing cells in the bursa of Fabricius. Int. Rev. Cell Mol. Biol. 2014, 312, 101–137. [Google Scholar]
- Gitlin, A.D.; Nussenzweig, M.C. Immunology: Fifty years of B lymphocytes. Nature 2015, 517, 139–141. [Google Scholar] [CrossRef]
- Schat, K.A. The Importance of the Bursa of Fabricius, B Cells and T Cells for the Pathogenesis of Marek’s Disease: A Review. Viruses 2022, 14, 2015. [Google Scholar] [CrossRef] [PubMed]
- Ciriaco, E.; Pinera, P.P.; Diaz-Esnal, B.; Laura, R. Age-related changes in the avian primary lymphoid organs (thymus and bursa of Fabricius). Microsc. Res. Tech. 2003, 62, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Toivanen, P.; Toivanen, A.; Good, R.A. Ontogeny of bursal function in chicken. I. Embryonic stem cell for humoral immunity. J. Immunol. 1972, 109, 1058–1070. [Google Scholar] [PubMed]
- Glick, B. Historical perspective: The bursa of Fabricius and its influence on B-cell development, past and present. Vet. Immunol. Immunopathol. 1991, 30, 3–12. [Google Scholar] [CrossRef]
- Ratcliffe, M.J. Antibodies, immunoglobulin genes and the bursa of Fabricius in chicken B cell development. Dev. Comp. Immunol. 2006, 30, 101–118. [Google Scholar] [CrossRef]
- Funk, P.E.; Palmer, J.L. Dynamic control of B lymphocyte development in the bursa of fabricius. Arch. Immunol. Exp. 2003, 51, 389–398. [Google Scholar]
- Audhya, T.; Kroon, D.; Heavner, G.; Viamontes, G.; Goldstein, G. Tripeptide structure of bursin, a selective B-cell-differentiating hormone of the bursa of fabricius. Science 1986, 231, 997–999. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, T.; Zhou, J.; Chen, L.; Shi, S.; Wang, X.; Zhang, M.; Wang, C.; Liao, C. Bursal peptide BP-IV as a novel immunoadjuvant enhances the protective efficacy of an epitope peptide vaccine containing T and B cell epitopes of the H9N2 avian influenza virus. Microb. Pathog. 2021, 158, 105095. [Google Scholar] [CrossRef]
- Feng, X.L.; Zong, M.M.; Zhou, G.F.; Zheng, Y.; Yu, Y.N.; Cao, R.B.; Chen, P.Y.; Yang, M. The Functions and Mechanism of a New Oligopeptide BP9 from Avian Bursa on Antibody Responses, Immature B Cell, and Autophagy. J. Immunol. Res. 2019, 2019, 1574383. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.D.; Zhou, B.; Cao, R.B.; Feng, X.L.; Ma, Z.Y.; Chen, P.Y. BP5 regulated B cell development promoting anti-oxidant defence. Amino Acids 2014, 46, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Li, D.Y.; Xue, M.Y.; Geng, Z.R.; Chen, P.Y. The suppressive effects of Bursopentine (BP5) on oxidative stress and NF-kB activation in lipopolysaccharide-activated murine peritoneal macrophages. Cell. Physiol. Biochem. 2012, 29, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhou, J.; Li, S.; Cai, K.; Guo, X.; Liao, C.; Wang, C. Bursal Hexapeptide, A Potential Immunomodulator, Inhibits Tumor Cells Proliferation via p53 Signaling Pathway. Anticancer Agents Med. Chem. 2018, 18, 1582–1588. [Google Scholar] [CrossRef]
- Zhou, G.F.; Liu, Q.T.; Zhou, B.; Qiu, Y.F.; Liu, X.D.; Ma, Z.Y.; Feng, X.L.; Cao, R.B.; Chen, P.Y. The potential molecular effects of bursal septpeptide II on immune induction and antitumor activity. J. Vet. Sci. 2015, 16, 325–331. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.L.; Liu, Q.T.; Cao, R.B.; Zhou, B.; Li de, Y.; Zhang, Y.P.; Liu, K.; Liu, X.D.; Wei, J.C.; Qiu, Y.F.; et al. Gene expression profiling of hybridoma cells after bursal-derived bioactive factor BP5 treatment. Amino Acids 2012, 43, 2443–2456. [Google Scholar] [CrossRef]
- Liu, S.; Zhuang, Q.; Wang, S.; Jiang, W.; Jin, J.; Peng, C.; Hou, G.; Li, J.; Yu, J.; Yu, X.; et al. Control of avian influenza in China: Strategies and lessons. Transbound. Emerg. Dis. 2020, 67, 1463–1471. [Google Scholar] [CrossRef]
- Yu, Y.N.; Zheng, Y.; Hao, S.S.; Zhang, Z.; Cai, J.X.; Zong, M.M.; Feng, X.L.; Liu, Q.T. The molecular evolutionary characteristics of new isolated H9N2 AIV from East China and the function of vimentin on virus replication in MDCK cells. Virol. J. 2020, 17, 78. [Google Scholar] [CrossRef]
- Song, W.; Qin, K. Human-infecting influenza A (H9N2) virus: A forgotten potential pandemic strain? Zoonoses Public Health 2020, 67, 203–212. [Google Scholar] [CrossRef]
- Dong, J.; Zhou, Y.; Pu, J.; Liu, L. Status and Challenges for Vaccination against Avian H9N2 Influenza Virus in China. Life 2022, 12, 1326. [Google Scholar] [CrossRef]
- Alqazlan, N.; Astill, J.; Raj, S.; Sharif, S. Strategies for enhancing immunity against avian influenza virus in chickens: A review. Avian Pathol. 2022, 51, 211–235. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Jin, Y.; Zhang, S.; Xu, J.; Zhang, J. microRNA, a Subtle Indicator of Human Cytomegalovirus against Host Immune Cells. Vaccines 2022, 10, 144. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Wei, Y. Modulators of MicroRNA Function in the Immune System. Int. J. Mol. Sci. 2020, 21, 2357. [Google Scholar] [CrossRef] [PubMed]
- Klasing, K.C. Avian macrophages: Regulators of local and systemic immune responses. Poult. Sci. 1998, 77, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.L.; Zheng, Y.; Zong, M.M.; Hao, S.S.; Zhou, G.F.; Cao, R.B.; Chen, P.Y.; Liu, Q.T. The immunomodulatory functions and molecular mechanism of a new bursal heptapeptide (BP7) in immune responses and immature B cells. Veter Res. 2019, 50, 64. [Google Scholar] [CrossRef]
- Li, C.; Huang, X.; Cai, J.; Lu, A.; Hao, S.; Zhang, Z.; Sun, H.; Feng, X. The Immunomodulatory Functions of Various CpG Oligodeoxynucleotideson CEF Cells and H9N2 Subtype Avian Influenza Virus Vaccination. Vaccines 2022, 10, 616. [Google Scholar] [CrossRef]
- Klatte-Schulz, F.; Thiele, K.; Scheibel, M.; Duda, G.N.; Wildemann, B. Subacromial Bursa: A Neglected Tissue Is Gaining More and More Attention in Clinical and Experimental Research. Cells 2022, 11, 663. [Google Scholar] [CrossRef]
- Cline, T.D.; Beck, D.; Bianchini, E. Influenza virus replication in macrophages: Balancing protection and pathogenesis. J. Gen. Virol. 2017, 98, 2401–2412. [Google Scholar] [CrossRef]
- Balzar, S. Self-Centered Function of Adaptive Immunity in Regulation of Immune Responses and in Tolerance. J. Immunol. Res. 2021, 2021, 7507459. [Google Scholar] [CrossRef]
- Vazquez, M.I.; Catalan-Dibene, J.; Zlotnik, A. B cells responses and cytokine production are regulated by their immune microenvironment. Cytokine 2015, 74, 318–326. [Google Scholar] [CrossRef]
- Kajikhina, K.; Tsuneto, M.; Melchers, F. B-Lymphopoiesis in Fetal Liver, Guided by Chemokines. Adv. Immunol. 2016, 132, 71–89. [Google Scholar] [PubMed]
- Zhao, Q.; Elson, C.O. Adaptive immune education by gut microbiota antigens. Immunology 2018, 154, 28–37. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Li, H.; Wang, M.; Chu, E.; Wei, N.; Lin, J.; Hu, Y.; Dai, J.; Chen, A.; Zheng, H.; et al. mTOR participates in the formation, maintenance, and function of memory CD8+T cells regulated by glycometabolism. Biochem. Pharm. 2022, 204, 115197. [Google Scholar] [CrossRef] [PubMed]
- Klarquist, J.; Cross, E.W.; Thompson, S.B.; Willett, B.; Aldridge, D.L.; Caffrey-Carr, A.K.; Xu, Z.; Hunter, C.A.; Getahun, A.; Kedl, R.M. B cells promote CD8 T cell primary and memory responses to subunit vaccines. Cell Rep. 2021, 36, 109591. [Google Scholar] [CrossRef]
- Malhotra, S.; Kovats, S.; Zhang, W.; Coggeshall, K.M. B cell antigen receptor endocytosis and antigen presentation to T cells require Vav and dynamin. J. Biol. Chem. 2009, 284, 24088–24097. [Google Scholar] [CrossRef]
- Hoogeboom, R.; Tolar, P. Molecular Mechanisms of B Cell Antigen Gathering and Endocytosis. B Cell Recept. Signal. 2016, 393, 45–63. [Google Scholar]
- Wang, J.; Wan, Z.; Liu, W. Understanding of B Cell Receptor Signaling through a Photo-Activatable Antigen Presentation System. B Cell Recept. Signal. 2018, 1707, 225–234. [Google Scholar]


| Primer Names | Primer Sequences |
|---|---|
| IL-1β-F | ACCCGCTTCATCTTCTACCG |
| IL-1β-R | TCAGCGCCCACTTAGCTTG |
| IL-6-F | AGGACGAGATGTGCAAGAAGTTC |
| IL-6-R | TTGGGCAGGTTGAGGTTGTT |
| IL-10-F | CGCTGTCACCGCTTCTTCA |
| IL-10-R | CGTCTCCTTGATCTGCTTGATG |
| INOS-F | AGGCCAAACATCCTGGAGGTC |
| INOS-R | TCATAGAGACGCTGCTGCCAG |
| IFN-α-F | GGACATGGCTCCCACACTAC |
| IFN-α-R | GGCTGCTGAGGATTTTGAAGA |
| β-actin-F | AGACATCAGGGTGTGATGGTTGGT |
| β-actin-R | TGGTGACAATACCGTGTTCAATGG |
| Sample | Concentration (ng/µL) | Total RNA (μg) | RIN | Detection |
|---|---|---|---|---|
| BP7 | 334 | 10.688 | 10.00 | A |
| Control | 63 | 2.016 | 10.00 | A |
| GO Accession | Description | Term Type | Over_Represented_p-Value | DEG Item |
|---|---|---|---|---|
| Immune-related GO terms | ||||
| GO:0002683 | negative regulation of immune system process | BP | 0.0032905 | 99 |
| GO:0002376 | immune system process | BP | 0.0072332 | 488 |
| GO:0045824 | negative regulation of innate immune response | BP | 0.030752 | 10 |
| MHC and antigen presentation-related GO terms | ||||
| GO:0042611 | MHC protein complex | CC | 4.71 × 10−5 | 9 |
| GO:0042613 | MHC class II protein complex | CC | 0.00014287 | 8 |
| GO:0042287 | MHC protein binding | MF | 0.040487 | 7 |
| GO:0002483 | antigen processing and presentation of endogenous peptide antigen | BP | 0.046177 | 4 |
| B cell-related GO terms | ||||
| GO:0050869 | negative regulation of B cell activation | BP | 0.014968 | 12 |
| GO:0045578 | negative regulation of B cell differentiation | BP | 0.038157 | 3 |
| GO:0001922 | B-1 B cell homeostasis | BP | 0.039526 | 3 |
| Cytokine-related GO terms | ||||
| GO:0001816 | cytokine production | BP | 0.0028395 | 148 |
| GO:0071346 | cellular response to interferon-gamma | BP | 0.016662 | 31 |
| GO:0034341 | response to interferon-gamma | BP | 0.029455 | 32 |
| GO:0070098 | chemokine-mediated signaling pathway | BP | 0.0014371 | 29 |
| GO:0048020 | CCR chemokine receptor binding | MF | 0.0049025 | 15 |
| ID | Term | Rich Factor | Number | q Value |
|---|---|---|---|---|
| gga04672 | Intestinal immune network for IgA production | 0.6875 | 22 | 0.9476629 |
| gga00051 | Fructose and mannose metabolism | 0.625 | 20 | 0.9476629 |
| gga04144 | Endocytosis | 0.4635417 | 89 | 0.9476629 |
| gga03010 | Ribosome | 0.4833333 | 58 | 0.9476629 |
| gga00563 | Glycosylphosphatidylinositol (GPI)-anchor biosynthesis | 0.625 | 15 | 0.9476629 |
| gga04514 | Cell adhesion molecules (CAMs) | 0.4732143 | 53 | 0.9476629 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, J.; Zhang, Z.; Li, C.; Hao, S.; Lu, A.; Huang, X.; Feng, X. Bursal-Derived BP7 Induces the miRNA Molecular Basis of Chicken Macrophages and Promotes the Differentiation of B Cells. Vaccines 2022, 10, 1960. https://doi.org/10.3390/vaccines10111960
Cai J, Zhang Z, Li C, Hao S, Lu A, Huang X, Feng X. Bursal-Derived BP7 Induces the miRNA Molecular Basis of Chicken Macrophages and Promotes the Differentiation of B Cells. Vaccines. 2022; 10(11):1960. https://doi.org/10.3390/vaccines10111960
Chicago/Turabian StyleCai, Jiaxi, Ze Zhang, Chenfei Li, Shanshan Hao, Anran Lu, Xiangyu Huang, and Xiuli Feng. 2022. "Bursal-Derived BP7 Induces the miRNA Molecular Basis of Chicken Macrophages and Promotes the Differentiation of B Cells" Vaccines 10, no. 11: 1960. https://doi.org/10.3390/vaccines10111960
APA StyleCai, J., Zhang, Z., Li, C., Hao, S., Lu, A., Huang, X., & Feng, X. (2022). Bursal-Derived BP7 Induces the miRNA Molecular Basis of Chicken Macrophages and Promotes the Differentiation of B Cells. Vaccines, 10(11), 1960. https://doi.org/10.3390/vaccines10111960






