Computational Design of a Chimeric Vaccine against Plesiomonas shigelloides Using Pan-Genome and Reverse Vaccinology
Abstract
1. Introduction
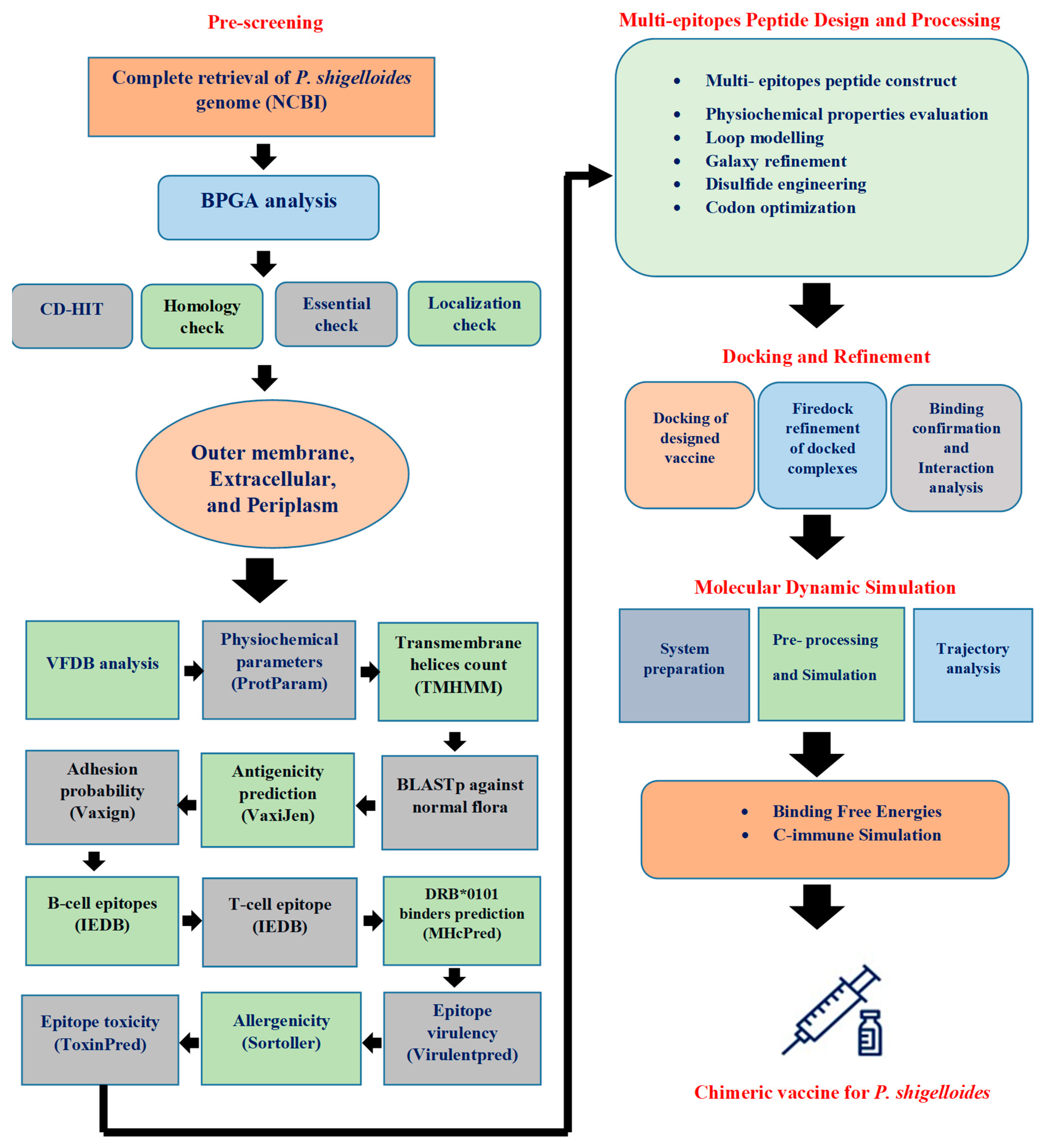
2. Research Methodology
2.1. Pre-Screening Phase
2.1.1. Complete Retrieval of P. shigelloides Genome
2.1.2. Screening Phase
2.1.3. Bacterial Pan-Genome Analysis
2.1.4. Cd-Hit Analysis (Cluster Data at High Identity with Tolerance)
2.1.5. Subcellular Localization
2.1.6. Vaccine Candidate’s Prioritization Phase
2.1.7. Antigenicity, Allergenicity, and Adhesion Probability Prediction
2.1.8. Immune Cell Epitopes Prediction
2.1.9. MHcPred Analysis
2.2. Multi-Epitopes Peptide Construct
Disulfide Engineering and Codon Optimization
2.3. Molecular Docking
2.4. Molecular Dynamics Simulation (MDS) Analysis
2.4.1. Binding Free Energies Estimation
2.4.2. Vaccine Immune Simulation
3. Results
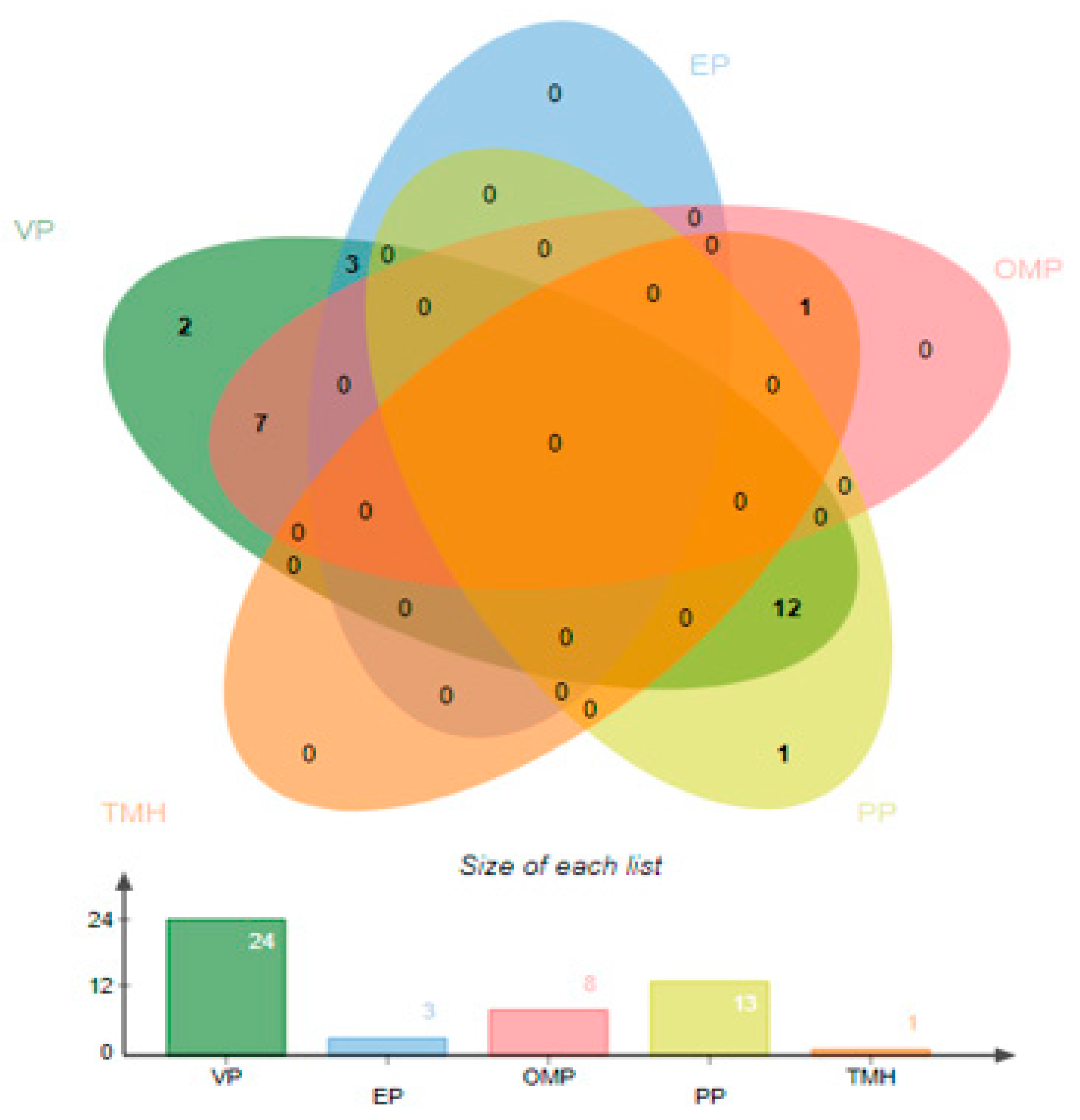
3.1. Retrieval of P. shigelloides Proteomics, Pan-Proteomics and Redundency Check
3.2. Subcellular Localization
3.3. Virulence Proteins Analysis and Transmembrane Helices Analysis
3.4. Physiochemical Properties of Proteins
3.5. Human and Normal Flora Homology, Antigenicity, Allergenicity, and Adhesion Probability Analysis
3.6. Immune Epitopes Prediction
3.7. Antigenicity, Allergenicity, Solubility, and Toxicity Analysis of Predicted Epitopes
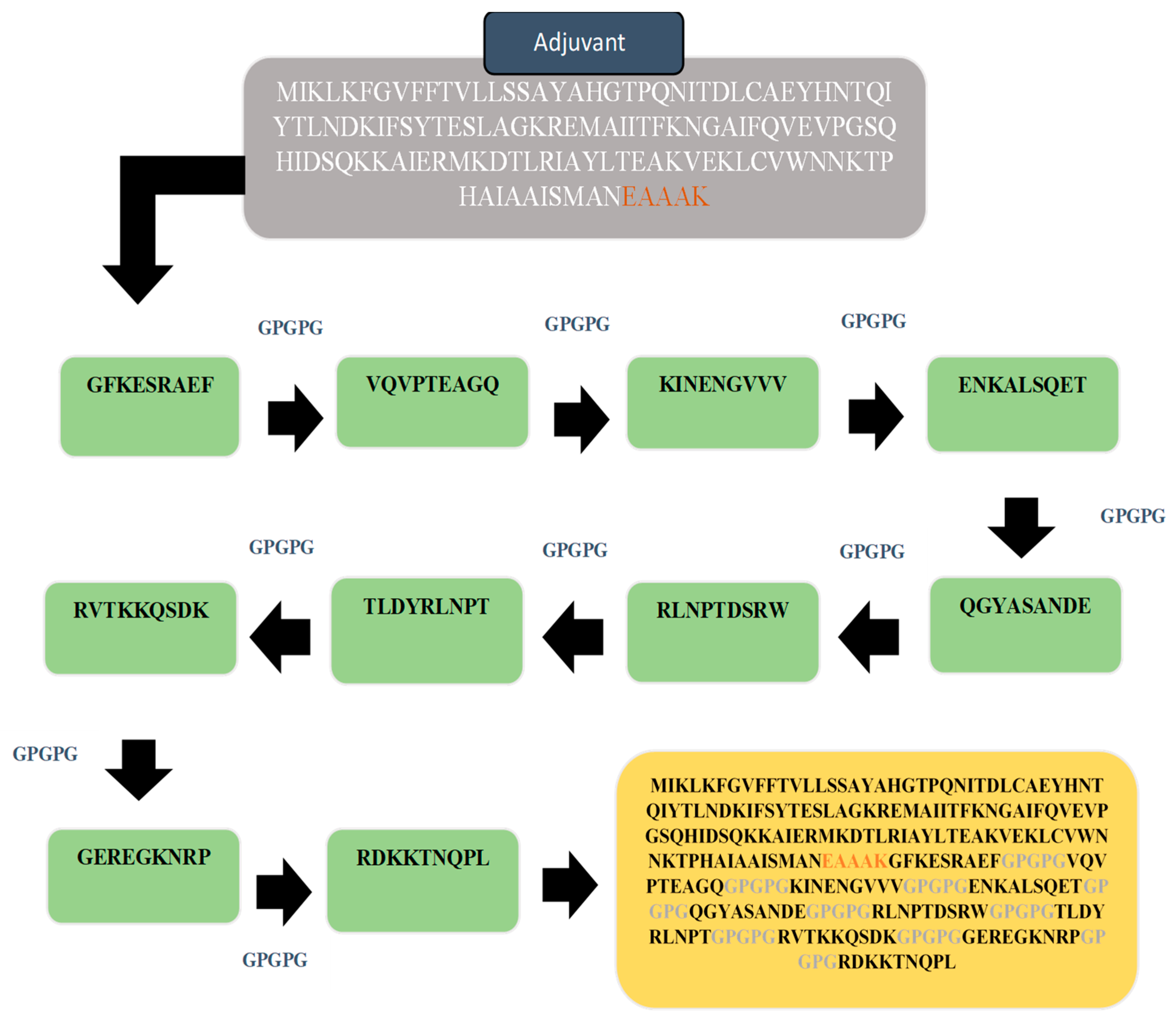
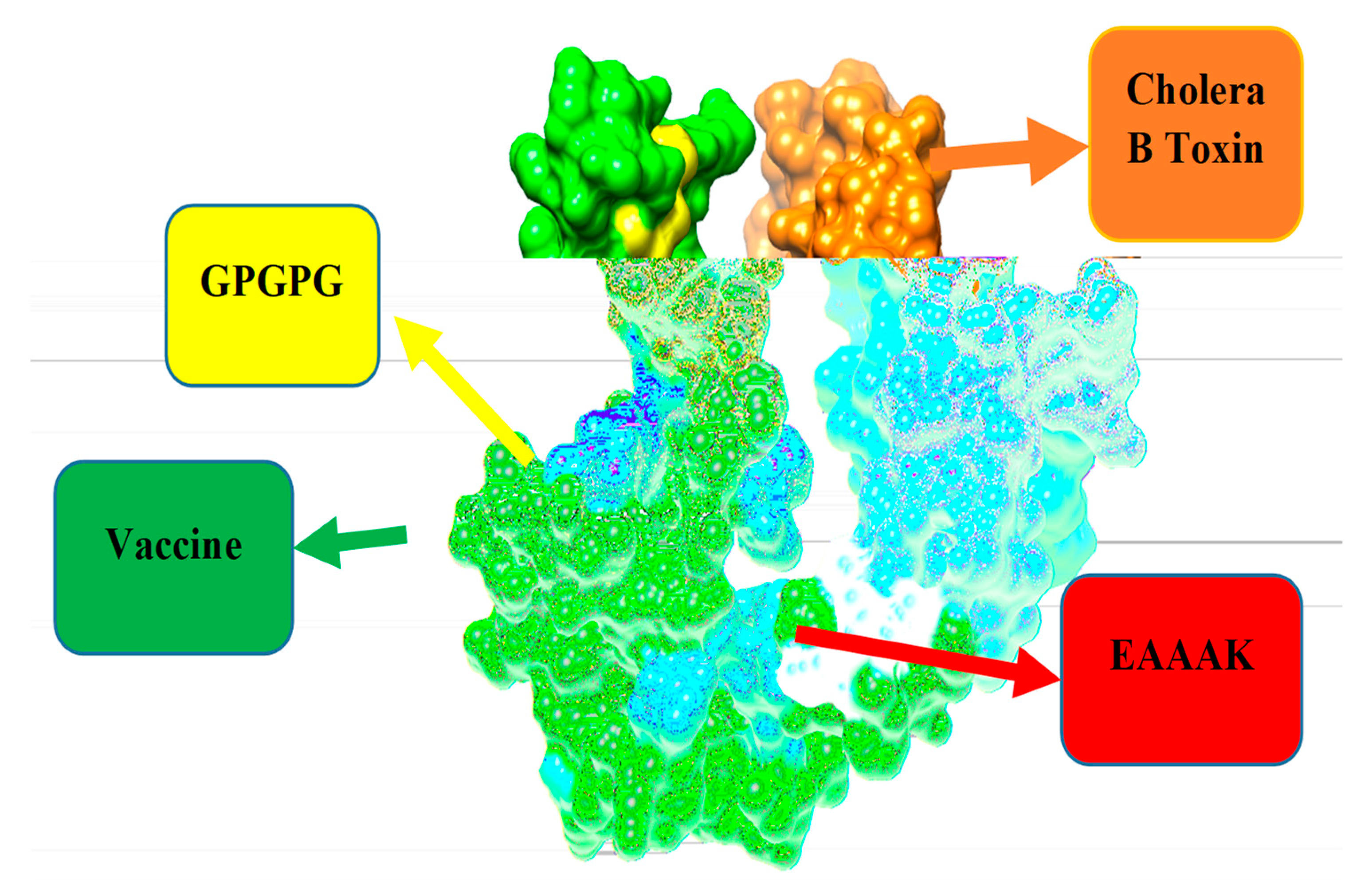
3.8. Multi-Epitopes Vaccine Construct
D Structure of Vaccine, Loop Modeling, and Refinement
3.9. Disulfide Engineering and Codon Optimization
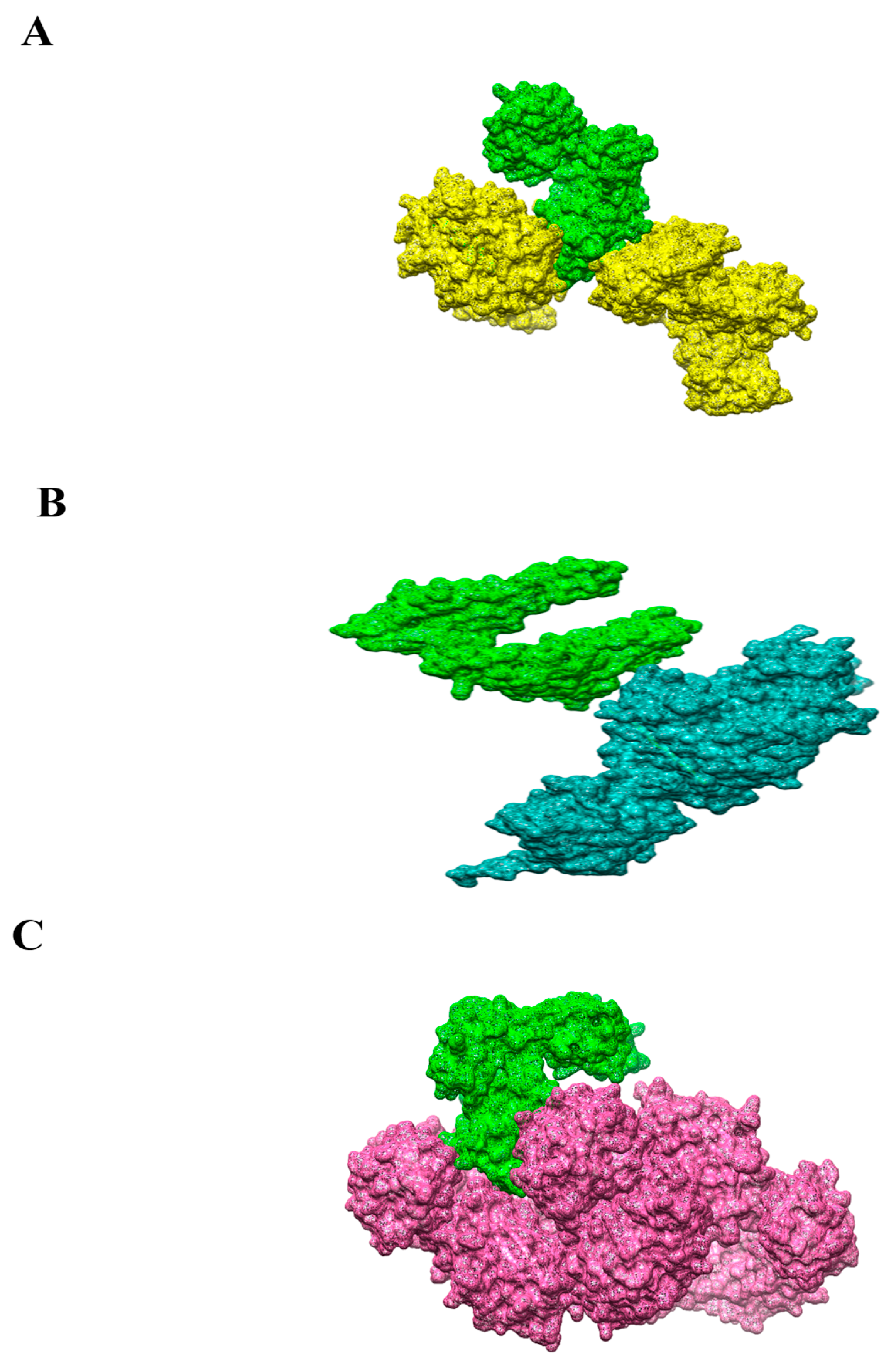
3.10. Molecular Docking
3.11. Refinement of Docked Complexes
3.12. Residues Wise Interaction Analysis of MHC-MHC- and TLR-4 to Vaccine
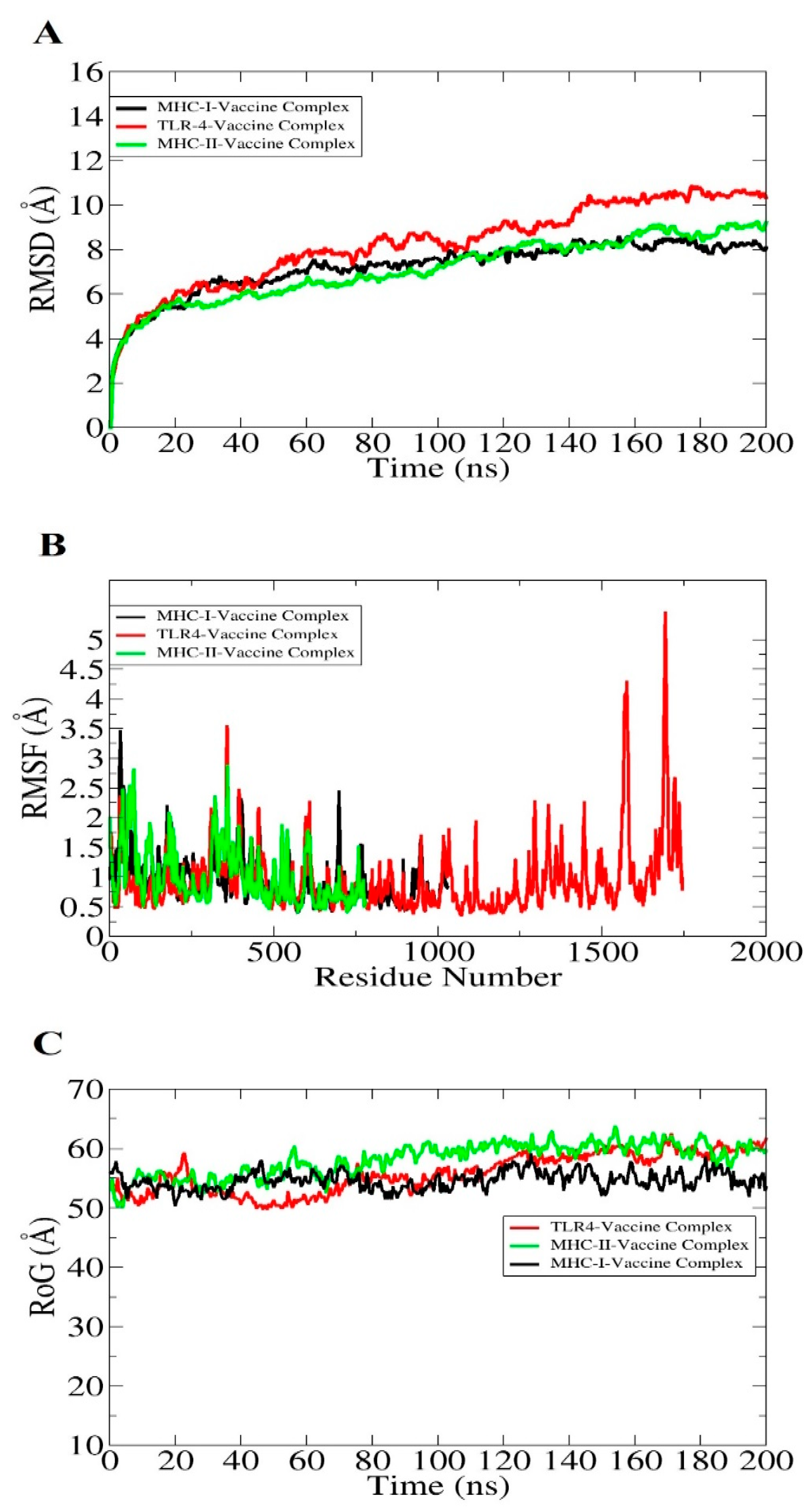
3.13. Molecular Dynamic Simulation
3.14. Estimation of Binding Free Energies of Vaccine Construct with MHC-I, MHC-II, and TLR-4
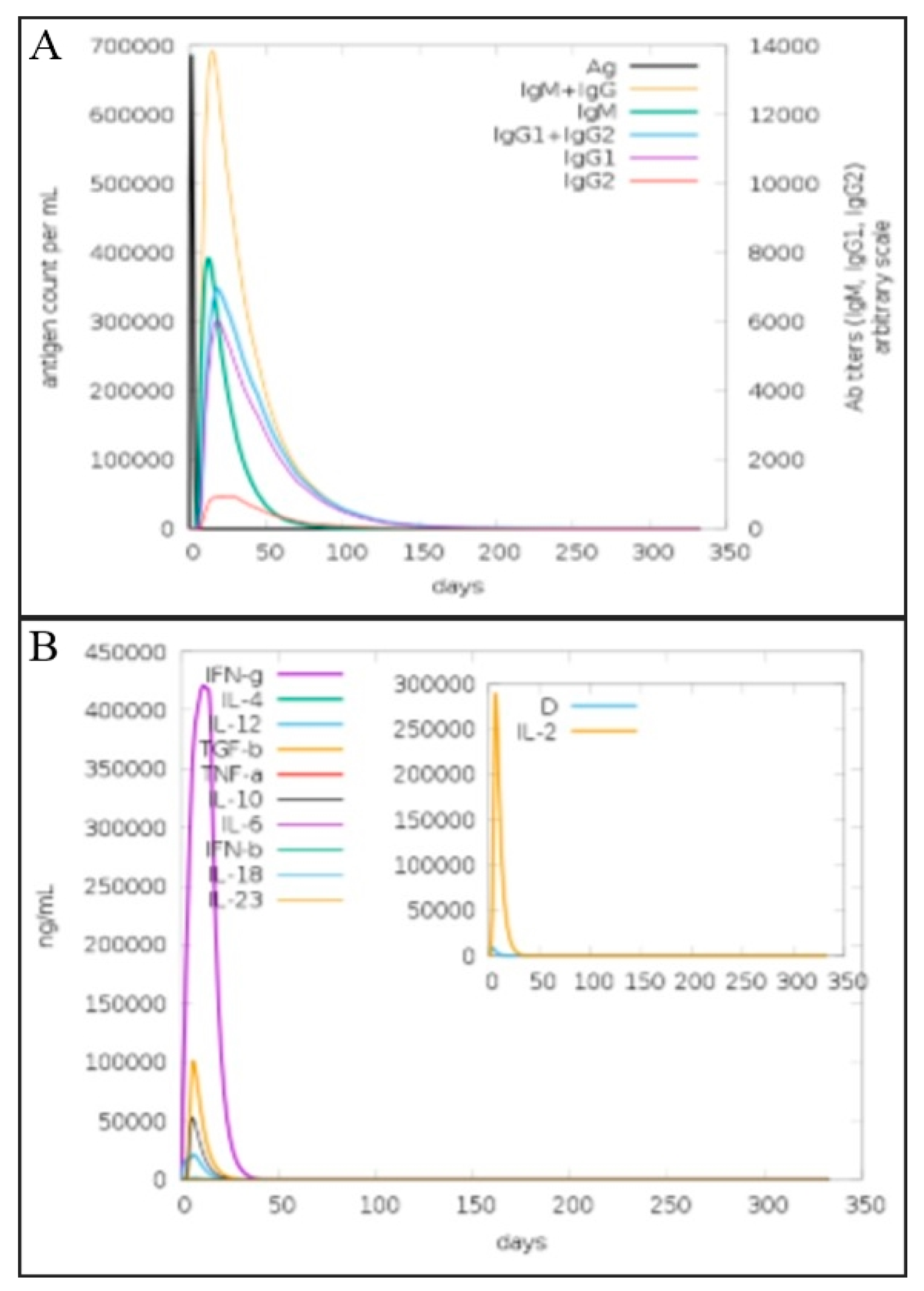
3.15. Vaccine Immune Simulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Caniça, M.; Manageiro, V.; Abriouel, H.; Moran-Gilad, J.; Franz, C.M.A.P. Antibiotic resistance in foodborne bacteria. Trends Food Sci. Technol. 2019, 84, 41–44. [Google Scholar] [CrossRef]
- MacLean, R.C.; San Millan, A. The evolution of antibiotic resistance. Science 2019, 365, 1082–1083. [Google Scholar] [CrossRef] [PubMed]
- Brooks, B.D.; Brooks, A.E. Therapeutic strategies to combat antibiotic resistance. Adv. Drug Deliv. Rev. 2014, 78, 14–27. [Google Scholar] [CrossRef] [PubMed]
- PCAST. National Action Plan for Combatting Antibiotic-Resistant Bacteria; White House: Washington, DC, USA, 2015. [Google Scholar]
- Reddick, L.E.; Alto, N.M. Bacteria fighting back: How pathogens target and subvert the host innate immune system. Mol. Cell 2014, 54, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Gagneux-Brunon, A.; Lucht, F.; Launay, O.; Berthelot, P.; Botelho-Nevers, E. Vaccines for healthcare-associated infections: Present, future, and expectations. Expert Rev. Vaccines 2018, 17, 421–433. [Google Scholar] [CrossRef]
- Vogel, U.; Taha, M.-K.; Vazquez, J.A.; Findlow, J.; Claus, H.; Stefanelli, P.; Caugant, D.A.; Kriz, P.; Abad, R.; Bambini, S. Predicted strain coverage of a meningococcal multicomponent vaccine (4CMenB) in Europe: A qualitative and quantitative assessment. Lancet Infect. Dis. 2013, 13, 416–425. [Google Scholar] [CrossRef]
- Bidmos, F.A.; Siris, S.; Gladstone, C.A.; Langford, P.R. Bacterial Vaccine Antigen Discovery in the Reverse Vaccinology 2.0 Era: Progress and Challenges. Front. Immunol. 2018, 9, 2315. [Google Scholar] [CrossRef]
- Dhiman, N.; Smith, D.I.; Poland, G.A. Next-generation sequencing: A transformative tool for vaccinology. Expert Rev. Vaccines 2009, 8, 963–967. [Google Scholar] [CrossRef]
- Dalsass, M.; Brozzi, A.; Medini, D.; Rappuoli, R. Comparison of open-source Reverse Vaccinology programs for bacterial vaccine antigen discovery. Front. Immunol. 2019, 10, 113. [Google Scholar] [CrossRef]
- Micoli, F.; Bagnoli, F.; Rappuoli, R.; Serruto, D. The role of vaccines in combatting antimicrobial resistance. Nat. Rev. Microbiol. 2021, 19, 287–302. [Google Scholar] [CrossRef]
- Moriel, D.G.; Scarselli, M.; Serino, L.; Mora, M.; Rappuoli, R.; Masignani, V. Genome-based vaccine development: A short cut for the future. Hum. Vaccin. 2008, 4, 184–188. [Google Scholar] [CrossRef][Green Version]
- Gul, H.; Ali, S.S.; Saleem, S.; Khan, S.; Khan, J.; Wadood, A.; Rehman, A.U.; Ullah, Z.; Ali, S.; Khan, H. Subtractive proteomics and immunoinformatics approaches to explore Bartonella bacilliformis proteome (virulence factors) to design B and T cell multi-epitope subunit vaccine. Infect. Genet. Evol. 2020, 85, 104551. [Google Scholar] [CrossRef]
- Omoniyi, A.A.; Adebisi, S.S.; Musa, S.A.; Nzalak, J.O.; Bauchi, Z.M.; Bako, K.W.; Olatomide, O.D.; Zachariah, R.; Nyengaard, J.R. In silico design and analyses of a multi-epitope vaccine against Crimean-Congo hemorrhagic fever virus through reverse vaccinology and immunoinformatics approaches. Sci. Rep. 2022, 12, 8736. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L.; McIver, C.J. Plesiomonas shigelloides revisited. Clin. Microbiol. Rev. 2016, 29, 349–374. [Google Scholar] [CrossRef]
- Kelly, D.F.; Rappuoli, R. Reverse vaccinology and vaccines for serogroup B Neisseria meningitidis. In Hot Topics in Infection and Immunity in Children II; Springer: Berlin/Heidelberg, Germany, 2005; pp. 217–223. [Google Scholar]
- Selim, S.; Almuhayawi, M.S.; Zakai, S.A.; Salama, A.A.; Warrad, M. Distinction between Antimicrobial Resistance and Putative Virulence Genes Characterization in Plesiomonas shigelloides Isolated from Different Sources. Antibiotics 2022, 11, 85. [Google Scholar] [CrossRef]
- Taylor, D.N.; Trofa, A.C.; Sadoff, J.; Chu, C.; Bryla, D.; Shiloach, J.; Cohen, D.; Ashkenazi, S.; Lerman, Y.; Egan, W. Synthesis, characterization, and clinical evaluation of conjugate vaccines composed of the O-specific polysaccharides of Shigella dysenteriae type 1, Shigella flexneri type 2a, and Shigella sonnei (Plesiomonas shigelloides) bound to bacterial toxoids. Infect. Immun. 1993, 61, 3678–3687. [Google Scholar] [CrossRef]
- Barel, L.-A.; Mulard, L.A. Classical and novel strategies to develop a Shigella glycoconjugate vaccine: From concept to efficacy in human. Hum. Vaccines Immunother. 2019, 15, 1338–1356. [Google Scholar] [CrossRef]
- Herrera, C.M.; Schmitt, J.S.; Chowdhry, E.I.; Riddle, M.S. From Kiyoshi Shiga to Present-Day Shigella Vaccines: A Historical Narrative Review. Vaccines 2022, 10, 645. [Google Scholar] [CrossRef]
- Qin, C.; Schumann, B.; Zou, X.; Pereira, C.L.; Tian, G.; Hu, J.; Seeberger, P.H.; Yin, J. Total synthesis of a densely functionalized Plesiomonas shigelloides serotype 51 aminoglycoside trisaccharide antigen. J. Am. Chem. Soc. 2018, 140, 3120–3127. [Google Scholar] [CrossRef]
- Rashid, M.I.; Naz, A.; Ali, A.; Andleeb, S. Prediction of vaccine candidates against Pseudomonas aeruginosa: An integrated genomics and proteomics approach. Genomics 2017, 109, 274–283. [Google Scholar] [CrossRef]
- Hassan, A.; Naz, A.; Obaid, A.; Paracha, R.Z.; Naz, K.; Awan, F.M.; Muhmmad, S.A.; Janjua, H.A.; Ahmad, J.; Ali, A. Pangenome and immuno-proteomics analysis of Acinetobacter baumannii strains revealed the core peptide vaccine targets. BMC Genom. 2016, 17, 732. [Google Scholar] [CrossRef] [PubMed]
- Sanober, G.; Ahmad, S.; Azam, S.S. Identification of plausible drug targets by investigating the druggable genome of MDR Staphylococcus epidermidis. Gene Rep. 2017, 7, 147–153. [Google Scholar] [CrossRef]
- Vilela Rodrigues, T.C.; Jaiswal, A.K.; de Sarom, A.; de Castro Oliveira, L.; Freire Oliveira, C.J.; Ghosh, P.; Tiwari, S.; Miranda, F.M.; de Jesus Benevides, L.; de Carvalho Azevedo, V.; et al. Reverse vaccinology and subtractive genomics reveal new therapeutic targets against Mycoplasma pneumoniae: A causative agent of pneumonia. R. Soc. Open Sci. 2019, 6, 190907. [Google Scholar] [CrossRef] [PubMed]
- Baseer, S.; Ahmad, S.; Ranaghan, K.E.; Azam, S.S. Towards a peptide-based vaccine against Shigella sonnei: A subtractive reverse vaccinology based approach. Biologicals 2017, 50, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Butt, A.M.; Tahir, S.; Nasrullah, I.; Idrees, M.; Lu, J.; Tong, Y. Mycoplasma genitalium: A comparative genomics study of metabolic pathways for the identification of drug and vaccine targets. Infect. Genet. Evol. 2012, 12, 53–62. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Yuan, C.; Wei, Y.; Zhang, S.; Cheng, J.; Cheng, X.; Qian, C.; Wang, Y.; Zhang, Y.; Yin, Z.; Chen, H. Comparative Genomic Analysis Reveals Genetic Mechanisms of the Variety of Pathogenicity, Antibiotic Resistance, and Environmental Adaptation of Providencia Genus. Front. Microbiol. 2020, 11, 572642. [Google Scholar] [CrossRef]
- Naz, A.; Awan, F.M.; Obaid, A.; Muhammad, S.A.; Paracha, R.Z.; Ahmad, J.; Ali, A. Identification of putative vaccine candidates against Helicobacter pylori exploiting exoproteome and secretome: A reverse vaccinology based approach. Infect. Genet. Evol. 2015, 32, 280–291. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, P.; Luo, J.; Jiang, Y. Secreted protein prediction system combining CJ-SPHMM, TMHMM, and PSORT. Mamm. Genome 2003, 14, 859–865. [Google Scholar] [CrossRef]
- Asad, Y.; Ahmad, S.; Rungrotmongkol, T.; Ranaghan, K.E.; Azam, S.S. Immuno-informatics driven proteome-wide investigation revealed novel peptide-based vaccine targets against emerging multiple drug resistant Providencia stuartii. J. Mol. Graph. Model. 2018, 80, 238–250. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Springer: Berlin/Heidelberg, Germany, 2005; pp. 571–607. [Google Scholar]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Tusnady, G.E.; Simon, I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001, 17, 849–850. [Google Scholar] [CrossRef]
- Doytchinova, I.A.; Flower, D.R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007, 8, 4. [Google Scholar] [CrossRef]
- Vita, R.; Overton, J.A.; Greenbaum, J.A.; Ponomarenko, J.; Clark, J.D.; Cantrell, J.R.; Wheeler, D.K.; Gabbard, J.L.; Hix, D.; Sette, A.; et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2014, 43, D405–D412. [Google Scholar] [CrossRef]
- Garg, V.K.; Avashthi, H.; Tiwari, A.; Jain, P.A.; Ramkete, P.W.; Kayastha, A.M.; Singh, V.K. MFPPI–multi FASTA ProtParam interface. Bioinformation 2016, 12, 74. [Google Scholar] [CrossRef]
- Saadi, M.; Karkhah, A.; Nouri, H.R. Development of a multi-epitope peptide vaccine inducing robust T cell responses against brucellosis using immunoinformatics based approaches. Infect. Genet. Evol. 2017, 51, 227–234. [Google Scholar] [CrossRef]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef]
- Goodman, D.B.; Church, G.M.; Kosuri, S. Causes and effects of N-terminal codon bias in bacterial genes. Science 2013, 342, 475–479. [Google Scholar] [CrossRef]
- Sussman, J.L.; Lin, D.; Jiang, J.; Manning, N.O.; Prilusky, J.; Ritter, O.; Abola, E.E. Protein Data Bank (PDB): Database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998, 54, 1078–1084. [Google Scholar] [CrossRef]
- Schneidman-Duhovny, D.; Inbar, Y.; Nussinov, R.; Wolfson, H.J. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res. 2005, 33, W363–W367. [Google Scholar] [CrossRef]
- Andrusier, N.; Nussinov, R.; Wolfson, H.J. FireDock: Fast interaction refinement in molecular docking. Proteins Struct. Funct. Bioinform. 2007, 69, 139–159. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Mbelle, N.; Osei Sekyere, J.; Feldman, C.; Maningi, N.E.; Modipane, L.; Essack, S.Y. Genomic analysis of two drug-resistant clinical Morganella morganii strains isolated from UTI patients in Pretoria, South Africa. Lett. Appl. Microbiol. 2020, 70, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Ren, Y.; Zhou, Z.; Guo, X.; Li, Y.; Feng, L.; Wang, L. Complete genome sequence of Enterobacter cloacae subsp. cloacae type strain ATCC 13047. J. Bacteriol. 2010, 192, 2463–2464. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Lee, T.-S.; Allen, B.K.; Giese, T.J.; Guo, Z.; Li, P.; Lin, C.; McGee Jr, T.D.; Pearlman, D.A.; Radak, B.K.; Tao, Y. Alchemical binding free energy calculations in AMBER20: Advances and best practices for drug discovery. J. Chem. Inf. Model. 2020, 60, 5595–5623. [Google Scholar] [CrossRef]
- Rapin, N.; Lund, O.; Bernaschi, M.; Castiglione, F. Computational immunology meets bioinformatics: The use of prediction tools for molecular binding in the simulation of the immune system. PLoS ONE 2010, 5, 9862. [Google Scholar] [CrossRef]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Ekundayo, T.C.; Okoh, A.I. Pathogenomics of virulence traits of Plesiomonas shigelloides that were deemed inconclusive by traditional experimental approaches. Front. Microbiol. 2018, 9, 3077. [Google Scholar] [CrossRef]
- Andreano, E.; D’Oro, U.; Rappuoli, R.; Finco, O. Vaccine evolution and its application to fight modern threats. Front. Immunol. 2019, 10, 1722. [Google Scholar] [CrossRef]
- Mermelstein, D.J.; Lin, C.; Nelson, G.; Kretsch, R.; McCammon, J.A.; Walker, R.C. Fast and flexible gpu accelerated binding free energy calculations within the amber molecular dynamics package. Comput. Chem. 2018, 39, 1354–1358. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Mahajan, S.; Paul, S.; Yan, Z.; Kim, H.; Jespersen, M.C.; Jurtz, V.; Andreatta, M.; Greenbaum, J.A.; Marcatili, P. IEDB-AR: Immune epitope database—Analysis resource in 2019. Nucleic Acids Res. 2019, 47, W502–W506. [Google Scholar] [CrossRef]
- Witkowski, P.T.; Drexler, J.F.; Kallies, R.; Ličková, M.; Bokorova, S.; Mananga, G.D.; Szemes, T.; Leroy, E.M.; Krueger, D.H.; Drosten, C. Phylogenetic analysis of a newfound bat-borne hantavirus supports a laurasiatherian host association for ancestral mammalian hantaviruses. Infect. Genet. Evol. 2016, 41, 113–119. [Google Scholar] [CrossRef]
- Yero, D.; Conchillo-Solé, O.; Daura, X. Antigen Discovery in Bacterial Panproteomes. In Vaccine Delivery Technology; Springer: Berlin/Heidelberg, Germany, 2021; pp. 43–62. [Google Scholar]
- Taiwo, A.A.; Falilat, A.J.; Ezemuel, Y.S. Computational design of peptide vaccine against Acinetobacter baumannii infection using comparative genomic approach. Comput. Biol. Bioinform. 2014, 2, 13–18. [Google Scholar] [CrossRef]
- Ismail, M.; Sajid, Z.; Ali, A.; Wu, X.; Muhammad, S.A.; Shaikh, R.S. Prediction of Prophylactic Peptide Vaccine Candidates for Human Papillomavirus (HPV): Immunoinformatics and Reverse Vaccinology Approaches. Curr. Proteom. 2021, 18, 178–192. [Google Scholar] [CrossRef]


| Strain | Size (Mb) | GC% |
|---|---|---|
| MS-17-188 | 3.97036 | 51.4817 |
| NCTC10360 | 3.40598 | 52 |
| Protein | Gene Ontology | Human | Lactobacillus rhamnosus | Lactobacillus casei | Lactobacillus johnsonii | Antigenicity |
|---|---|---|---|---|---|---|
| Flagellar hook protein FlgE | Bacterial-type flagellum basal body | No-Similarity | No Similarity | 0.82 | ||
| Hypothetical protein | Membrane protein | 0.70 | ||||
| Hemoglobin/transferrin/ lactoferrin family receptor | Integral component of membrane | 0.70 | ||||
| MHcPred | Antigenicity | Allergenicity | Solubility | ToxinPred |
|---|---|---|---|---|
| GFKESRAEF | 0.52 | Non-Allergen | Soluble | Non-Toxin |
| VQVPTEAGQ | 0.50 | |||
| KINENGVVV | 0.77 | |||
| ENKALSQET | 0.70 | |||
| QGYASANDE | 0.70 | |||
| RLNPTDSRW | 1.28 | |||
| TLDYRLNPT | 2.23 | |||
| RVTKKQSDK | 1.49 | |||
| GEREGKNRP | 2.24 | |||
| RDKKTNQPL | 1.19 |
| S.N | A.A | S.N | A.A | Chi3 | Energy | Sum B-Factors |
|---|---|---|---|---|---|---|
| 11 | Thr | 29 | Leu | 90.87 | 5.4 | 0 |
| 19 | Ala | 22 | Thr | 68.11 | 4.56 | 0 |
| 19 | Thr | 41 | Leu | 89.79 | 2.9 | 0 |
| 38 | Pro | 77 | Gln | 115.92 | 3.87 | 0 |
| 74 | Glu | 116 | Ala | 80.19 | 3.86 | 0 |
| 32 | Trp | 112 | Lys | 79.07 | 1.07 | 0 |
| 104 | Lys | 135 | Arg | 103.02 | 0.99 | 0 |
| Vaccine Complex | Interactive Residues |
|---|---|
| MHC-I | Ala128,Asn24,His145,Phe131,Ala149,Asp106,Ile52,Pro20,Ala136,Asp223,Leu 272,Ser132,Ala150,Glu148,Leu201,Thr80,Arg157,Gly 104,Lys 19,Trp 167,Arg 75,Gln 141,Met 4,Trp 51,Arg 169,Glu 16,Met 99 |
| MHC-II | Arg256,Gln77,Leu219,Ser191,Asn10,Gln197,Lys84,Thr4,Asn124,Gly25,Lys232,Thr230,Asn192,Gly197,Met122,Trp109,Ala19,His20,Phe09,Trp208,Ala191, His74,Pro18,Tyr188,Asp 43,His77,Pro224, Val 108,Gln 02,Ile45,Pro 238,Val 164 |
| TLR-4 | Arg355,Gln 145,Ile38,Met 1,Ser312,Ala128,Gln152,Ile454,Met58,Ser569,Asn 65,Glu50,Lys3,Phe 06,Thr27,Asn544,Glu137,Val524,Phe09,Thr 260,Asp 100,Glu 161,Lys 44,Phe 396,Val 73,Asp194,Gly 153,Lys55,Phe500,Val122,Cys 40,His115,Lys64,Pro140,Val165,Gln70,Ile36,Lys247,Pro23,Val 146 |
| Energy Parameter | TLR-4-Vaccine Complex | MHC-I-Vaccine Complex | MHC-II-Vaccine Complex |
|---|---|---|---|
| MM-GBSA | |||
| VDWAALS | −75.06 | −69.84 | −76.32 |
| EEL | −66.75 | −56.06 | −45.25 |
| Delta G gas | −141.81 | −125.9 | −121.57 |
| Delta G solv | 29.67 | 33.64 | 32.47 |
| Delta Total | −112.14 | −92.26 | −89.1 |
| MM-PBSA | |||
| VDWAALS | −75.06 | −69.84 | −76.32 |
| EEL | −66.75 | −56.06 | −45.25 |
| Delta G gas | −141.81 | −125.9 | −121.57 |
| Delta G solv | 28.99 | 30.34 | 28.14 |
| Delta Total | −112.82 | −95.56 | −93.43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mushtaq, M.; Khan, S.; Hassan, M.; Al-Harbi, A.I.; Hameed, A.R.; Khan, K.; Ismail, S.; Irfan, M.; Ahmad, S. Computational Design of a Chimeric Vaccine against Plesiomonas shigelloides Using Pan-Genome and Reverse Vaccinology. Vaccines 2022, 10, 1886. https://doi.org/10.3390/vaccines10111886
Mushtaq M, Khan S, Hassan M, Al-Harbi AI, Hameed AR, Khan K, Ismail S, Irfan M, Ahmad S. Computational Design of a Chimeric Vaccine against Plesiomonas shigelloides Using Pan-Genome and Reverse Vaccinology. Vaccines. 2022; 10(11):1886. https://doi.org/10.3390/vaccines10111886
Chicago/Turabian StyleMushtaq, Mahnoor, Saifullah Khan, Muhammad Hassan, Alhanouf I. Al-Harbi, Alaa R. Hameed, Khadeeja Khan, Saba Ismail, Muhammad Irfan, and Sajjad Ahmad. 2022. "Computational Design of a Chimeric Vaccine against Plesiomonas shigelloides Using Pan-Genome and Reverse Vaccinology" Vaccines 10, no. 11: 1886. https://doi.org/10.3390/vaccines10111886
APA StyleMushtaq, M., Khan, S., Hassan, M., Al-Harbi, A. I., Hameed, A. R., Khan, K., Ismail, S., Irfan, M., & Ahmad, S. (2022). Computational Design of a Chimeric Vaccine against Plesiomonas shigelloides Using Pan-Genome and Reverse Vaccinology. Vaccines, 10(11), 1886. https://doi.org/10.3390/vaccines10111886







