High Prevalence of HPV 51 in an Unvaccinated Population and Implications for HPV Vaccines
Abstract
:1. Introduction
2. Materials and Methods
- (a)
- Repeat of original GP5+/6+ PCR EIA (gold standard): the GP5+/6+ HPV PCR-ELISA method [17] was performed on all specimens in a 96-well format with minor modifications (Text Supplement S2 in Supplementary Materials).
- (b)
- HPV 51 type-specific E7 Linear PCR followed by E7 Nested PCR (Text Supplements S3 and S4 in Supplementary Materials).
- (c)
- Repeat of Discordant Results: where there was discordance between GP5+/6+ PCR EIA and E7 PCR results, samples were retested using both methods. As we had previously observed increased specificity with Hotstar Taq (Supplement Figures S1 and S2), all samples were repeated with Hotstar Taq (Text Supplement S5 in Supplementary Materials).
- (d)
- HPV sequencing: to confirm the specificity of the E7 PCR method we sent a selection of positive samples for sequencing, analysis was undertaken using 4 peaks® software and a BLAST® search using megablast.
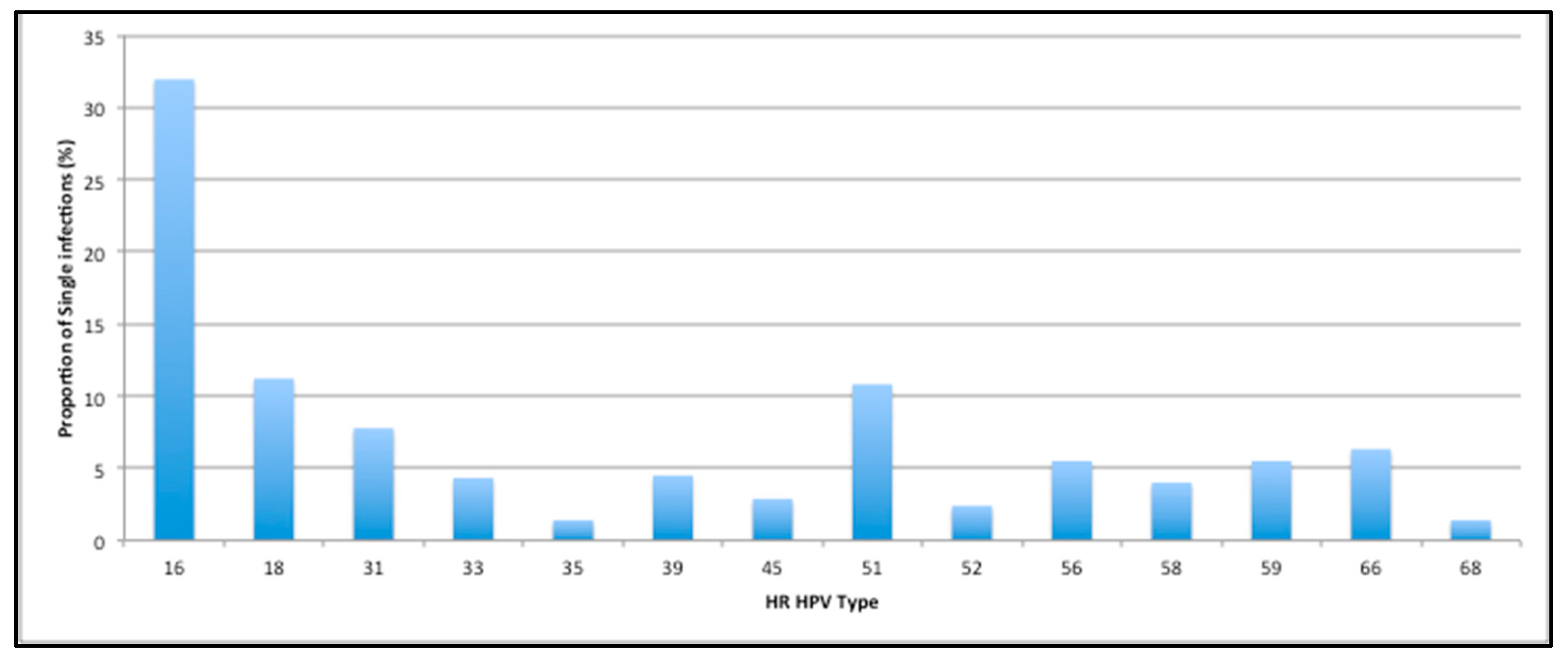
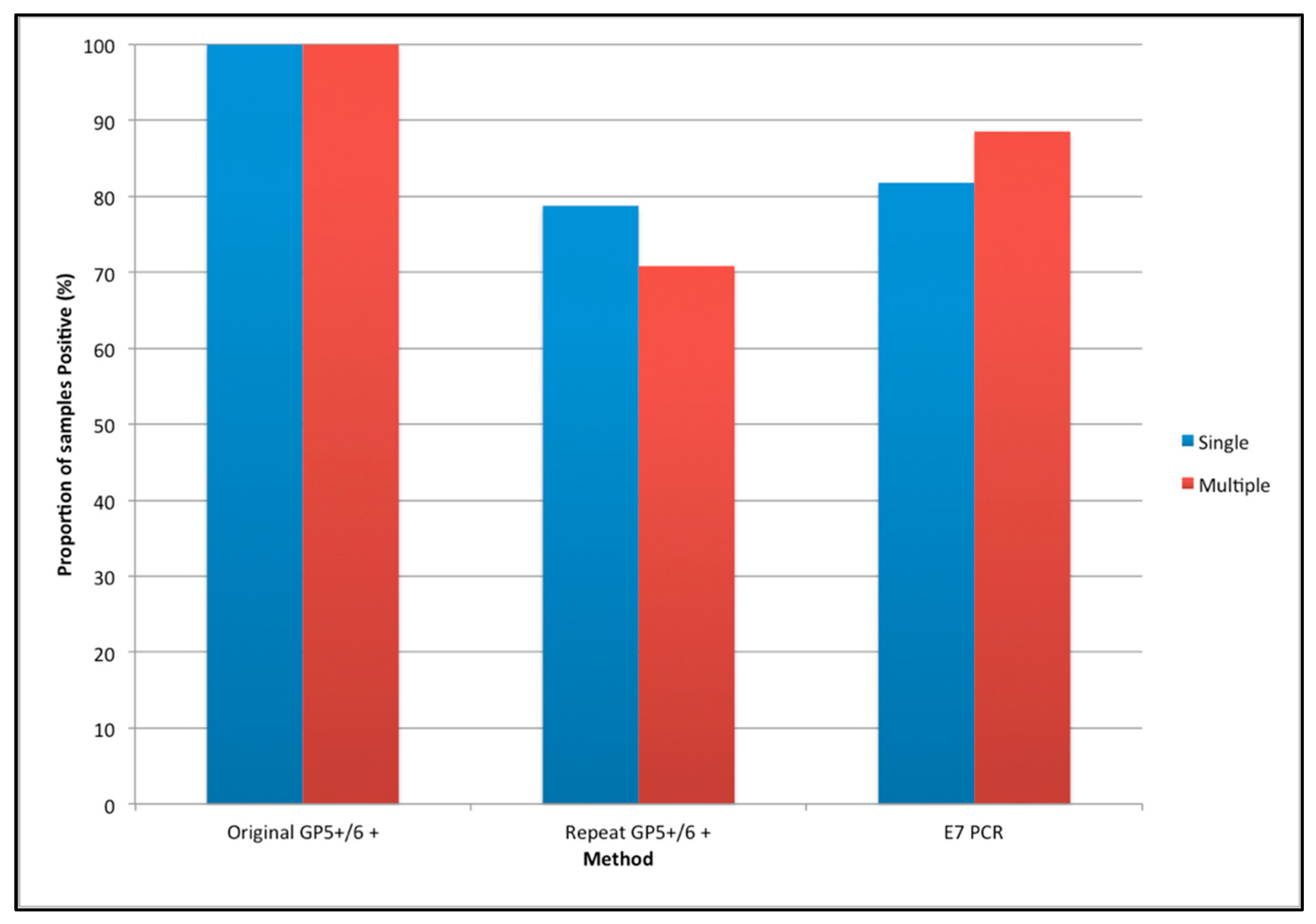
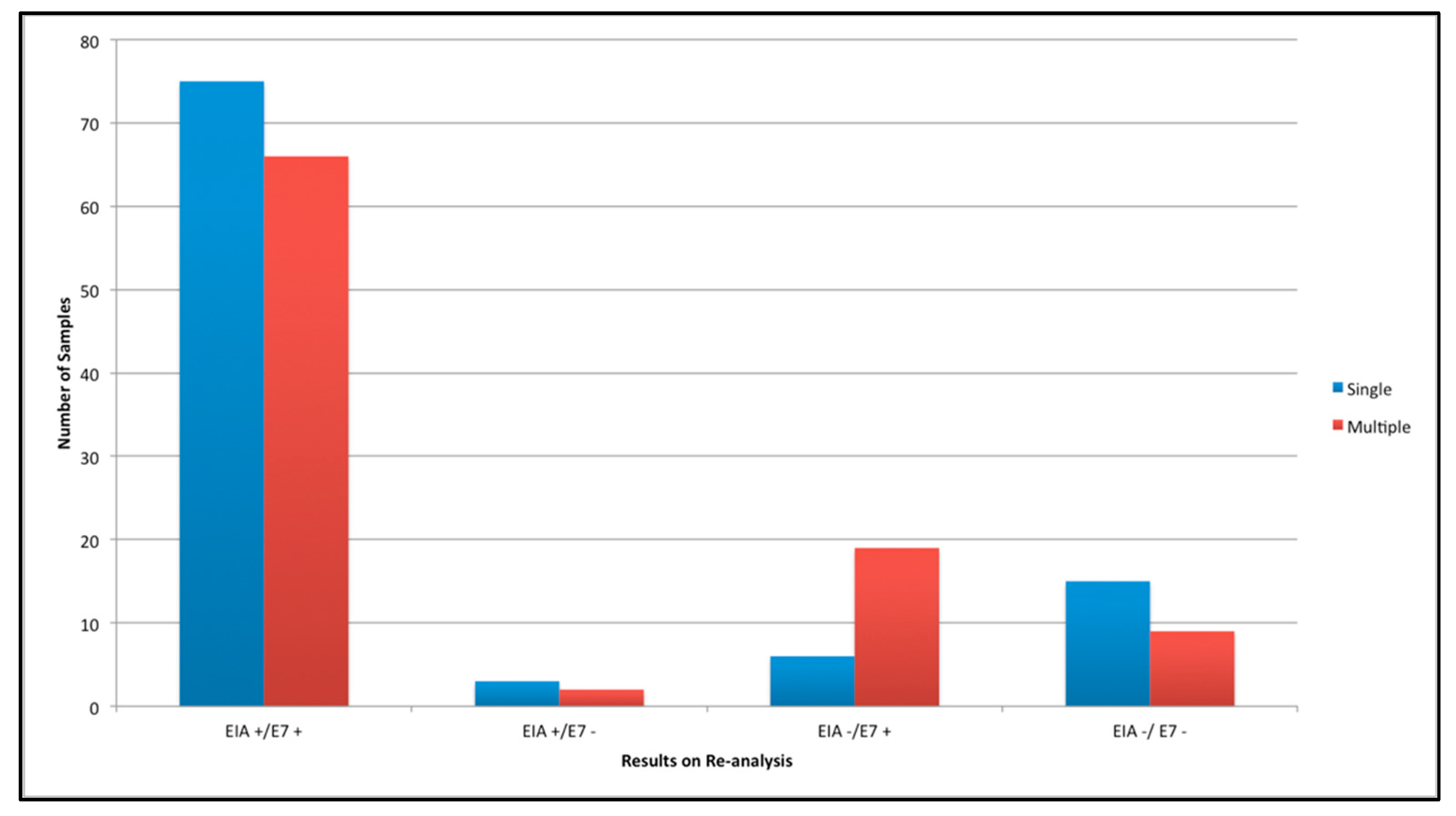
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Sanjosé, S.; Diaz, M.; Castellsagué, X.; Clifford, G.; Bruni, L.; Muñoz, N.; Bosch, F.X. Worldwide prevalence and genotype distribution of cervical human papillomavirus DNA in women with normal cytology: A meta-analysis. Lancet Infect. Dis. 2007, 7, 453–459. [Google Scholar] [CrossRef]
- Wheeler, C.; Castellsagué, X.; Garland, S.; Szarewski, A.; Paavonen, J.; Naud, P.; Salmerón, J.; Chow, S.-N.; Apter, D.; Kitchener, H. Cross-protective efficacy of HPV-16/18 AS04-adjuvanted vaccine against cervical infection and precancer caused by non-vaccine oncogenic HPV types: 4-year end-of-study analysis of the randomised, double-blind PATRICIA trial. Lancet Oncol. 2012, 13, 100–110. [Google Scholar] [CrossRef]
- Hibbitts, S.; Jones, J.; Powell, N.; Dallimore, N.; Mcrea, J.; Beer, H.; Tristram, A.; Fielder, H.; Fiander, A.N. Human papillomavirus prevalence in women attending routine cervical screening in South Wales, UK: A cross-sectional study. Br. J. Cancer 2008, 99, 1929–1933. [Google Scholar] [CrossRef] [Green Version]
- Moody, C.A.; Laimins, L.A. Human papillomavirus oncoproteins: Pathways to transformation. Nat. Rev. Cancer 2010, 10, 550–560. [Google Scholar] [CrossRef]
- Doorbar, J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 2006, 110, 525–541. [Google Scholar] [CrossRef] [Green Version]
- Tumban, E.; Peabody, J.; Peabody, D.S.; Chackerian, B. A Pan-HPV Vaccine Based on Bacteriophage PP7 VLPs Displaying Broadly Cross-Neutralizing Epitopes from the HPV Minor Capsid Protein, L2. PLoS ONE 2011, 6, e23310. [Google Scholar] [CrossRef]
- Powell, N.; Boyde, A.; Tristram, A.; Hibbitts, S.; Fiander, A. The potential impact of human papillomavirus vaccination in contemporary cytologically screened populations may be underestimated: An observational retrospective analysis of invasive cervical cancers. Int. J. Cancer 2009, 125, 2425–2427. [Google Scholar] [CrossRef]
- Lehtinen, M.; Paavonen, J.; Wheeler, C.M.; Jaisamrarn, U.; Garland, S.M.; Castellsagué, X.; Skinner, S.R.; Apter, D.; Naud, P.; Salmerón, J.; et al. Overall efficacy of HPV-16/18 AS04-adjuvanted vaccine against grade 3 or greater cervical intraepithelial neoplasia: 4-year end-of-study analysis of the randomised, double-blind PATRICIA trial. Lancet Oncol. 2012, 13, 89–99. [Google Scholar] [CrossRef]
- Lehtinen, M.; Paavonen, J. Vaccination against human papillomaviruses shows great promise. Lancet 2004, 364, 1731–1732. [Google Scholar] [CrossRef]
- Tota, J.E.; Ramanakumar, A.V.; Jiang, M.; Dillner, J.; Walter, S.D.; Kaufman, J.S.; Coutlée, F.; Villa, L.L.; Franco, E.L. Epidemiologic approaches to evaluating the potential for human papillomavirus type replacement postvaccination. Am. J. Epidemiol. 2013, 178, 625–634. [Google Scholar] [CrossRef]
- Hibbitts, S.; Tristram, A.; Beer, H.; McRea, J.; Rose, B.; Hauke, A.; Nuttall, D.; Dallimore, N.; Newcombe, R.G.; Fiander, A. UK population based study to predict impact of HPV vaccination. J. Clin. Virol. 2014, 59, 109–114. [Google Scholar] [CrossRef]
- Hibbitts, S.; Rieck, G.C.; Hart, K.; Powell, N.G.; Beukenholdt, R.; Dallimore, N.; Mcrea, J.; Hauke, A.; Tristram, A.; Fiander, A.N. Human papillomavirus infection: An Anonymous Prevalence Study in South Wales, UK. Br. J. Cancer 2006, 95, 226–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arbyn, M.; Simon, M.; Peeters, E.; Xu, L.; Meijer, C.J.L.M.; Berkhof, J.; Cuschieri, K.; Bonde, J.; Ostrbenk Vanlencak, A.; Zhao, F.-H.; et al. 2020 list of human papillomavirus assays suitable for primary cervical cancer screening. Clin. Microbiol. Infect. 2021, 27, 1083–1095. [Google Scholar] [CrossRef] [PubMed]
- Castle, P.E.; Solomon, D.; Wheeler, C.M.; Gravitt, P.E.; Wacholder, S.; Schiffman, M. Human Papillomavirus Genotype Specificity of Hybrid Capture 2. J. Clin. Microbiol. 2008, 46, 2595–2604. [Google Scholar] [CrossRef] [Green Version]
- Cytology BSfC. Recommended Code of Practice for Laboratories. Participating in the UK Cervical Screening Programmes 2010. Available online: http://www.britishcytology.org.uk/resources/BSCC_COP_2010.pdf (accessed on 1 August 2022).
- Desjardins, P.; Conklin, D. NanoDrop microvolume quantitation of nucleic acids. J. Vis. Exp. 2010, 2465. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, M.; Snijders, P.; van den Brule, A.; Helmerhorst, T.; Meijer, C.; Walboomers, J. A general primer GP5+/GP6+ -mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 1997, 791–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cymru, L.; Government, W. Welsh Index of Multiple Deprivation (WIMD) Cardiff: National Statistics Local Government Data Unit 2019. Available online: https://wimd.gov.wales/ (accessed on 23 August 2022).
- Li, N.; Franceschi, S.; Howell-Jones, R.; Snijders, P.J.F.; Clifford, G.M. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: Variation by geographical region, histological type and year of publication. Int. J. Cancer 2011, 128, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Bruni, L.; Diaz, M.; Castellsagué, X.; Ferrer, E.; Bosch, X.F.; De Sanjosé, S. Cervical Human Papillomavirus Prevalence in 5 Continents: Meta-Analysis of 1 Million Women with Normal Cytological Findings. J. Infect. Dis. 2010, 202, 1789–1799. [Google Scholar] [CrossRef] [Green Version]
- Brotherton, J.M.L.; Tabrizi, S.N.; Phillips, S.; Pyman, J.; Cornall, A.M.; Lambie, N.; Anderson, L.; Cummings, M.; Payton, D.; Scurry, J.P.; et al. Looking beyond human papillomavirus (HPV) genotype 16 and 18: Defining HPV genotype distribution in cervical cancers in Australia prior to vaccination. Int. J. Cancer 2017, 141, 1576–1584. [Google Scholar] [CrossRef] [Green Version]
- Cuschieri, K.S. Multiple high risk HPV infections are common in cervical neoplasia and young women in a cervical screening population. J. Clin. Pathol. 2004, 57, 68–72. [Google Scholar] [CrossRef]
- O’Leary, M.C.; Sinka, K.; Robertson, C.; Cuschieri, K.; Lyman, R.; Lacey, M.; Potts, A.; Cubie, H.A.; Donaghy, M. HPV type-specific prevalence using a urine assay in unvaccinated male and female 11- to 18-year olds in Scotland. Br. J. Cancer 2011, 104, 1221–1226. [Google Scholar] [CrossRef] [PubMed]
- Piana, A.; Sotgiu, G.; Castiglia, P.; Pischedda, S.; Cocuzza, C.; Capobianco, G.; Marras, V.; Dessole, S.; Muresu, E. Prevalence and type distribution of human papillomavirus infection in women from North Sardinia, Italy. BMC Public Health 2011, 11, 785. [Google Scholar] [CrossRef] [Green Version]
- Levi, J.E.; Kleter, B.; Quint, W.G.V.; Fink, M.C.S.; Canto, C.L.M.; Matsubara, R.; Linhares, I.; Segurado, A.S.; Vanderborght, B.; Neto, J.E.; et al. High Prevalence of Human Papillomavirus (HPV) Infections and High Frequency of Multiple HPV Genotypes in Human Immunodeficiency Virus-Infected Women in Brazil. J. Clin. Microbiol. 2002, 40, 3341–3345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levi, J.E.; Fernandes, S.; Tateno, A.F.; Motta, E.; Lima, L.P.; Eluf-Neto, J.; Pannuti, C.S. Presence of multiple human papillomavirus types in cervical samples from HIV-infected women. Gynecol. Oncol. 2004, 92, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Wang, C.; Liu, S.; He, J.; Jiang, Y. HPV genotypic spectrum in Jilin province, China, where non-vaccine-covered HPV53 and 51 are prevalent, exhibits a bimodal age-specific pattern. PLoS ONE 2020, 15, e0230640. [Google Scholar] [CrossRef]
- Yan, X.; Shen, L.; Xiao, Y.; Wang, Q.; Li, F.; Qian, Y. Prevalence, characteristics, and distribution of HPV genotypes in women from Zhejiang Province, 2016–2020. Virol. J. 2021, 18, 208. [Google Scholar] [CrossRef]
- Sabet, F.; Mosavat, A.; Ahmadi Ghezeldasht, S.; Basharkhah, S.; Shamsian, S.A.A.; Abbasnia, S.; Shamsian, K.; Rezaee, S.A. Prevalence, genotypes and phylogenetic analysis of human papillomaviruses (HPV) in northeast Iran. Int. J. Infect. Dis. 2021, 103, 480–488. [Google Scholar] [CrossRef]
- Rideg, O.; Dergez, T.; Farkas, K.; Kovács, K.; Kálmán, E.; Tornóczky, T.; Oszter, A. High Prevalence of Non-Vaccinated Oncogenic Human Papillomavirus Genotypes in High-Grade Squamous Intraepithelial Lesions of the Cervix: Thought-Provoking Results of a Detailed HPV Genotype Analysis. Vaccines 2022, 10, 748. [Google Scholar] [CrossRef]
- Falcaro, M.; Castañon, A.; Ndlela, B.; Checchi, M.; Soldan, K.; Lopez-Bernal, J.; Elliss-Brookes, L.; Sasieni, P. The effects of the national HPV vaccination programme in England, UK, on cervical cancer and grade 3 cervical intraepithelial neoplasia incidence: A register-based observational study. Lancet 2021, 398, 2084–2092. [Google Scholar] [CrossRef]
- Roden, R.; Wu, T.-C. How will HPV vaccines affect cervical cancer? Nat. Rev. Cancer 2006, 6, 753–763. [Google Scholar] [CrossRef]
- Debrah, O.; Agyemang-Yeboah, F.; Donkoh, E.T.; Asmah, R.H. Prevalence of vaccine and non-vaccine human papillomavirus types among women in Accra and Kumasi, Ghana: A cross-sectional study. BMC Women’s Health 2021, 21, 372. [Google Scholar] [CrossRef] [PubMed]
- Rahmat, F.; Kuan, J.Y.; Hajiman, Z.; Mohamed Shakrin, N.N.S.; Che Roos, N.A.; Mustapa, M.; Ahmad Zaidi, N.A.; Ahmad, A. Human Papillomavirus (HPV) Prevalence and Type Distribution in Urban Areas of Malaysia. Asian Pac. J. Cancer Prev. 2021, 22, 2969–2976. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Peng, M.; Wei, X.; Pan, D.; Xue, H.; Xu, Y.; Dong, B. Prevalence of Human Papillomavirus and Genotype Distribution in Pregnant and Non-Pregnant Women in China. Risk Manag. Healthc. Policy 2021, 14, 3147–3157. [Google Scholar] [CrossRef] [PubMed]
- Gray, P.; Palmroth, J.; Luostarinen, T.; Apter, D.; Dubin, G.; Garnett, G.; Eriksson, T.; Natunen, K.; Merikukka, M.; Pimenoff, V.; et al. Evaluation of HPV type-replacement in unvaccinated and vaccinated adolescent females-Post-hoc analysis of a community-randomized clinical trial (II). Int. J. Cancer 2018, 142, 2491–2500. [Google Scholar] [CrossRef] [Green Version]
- Molina-Pineda, A.; López-Cardona, M.G.; Limón-Toledo, L.P.; Cantón-Romero, J.C.; Martínez-Silva, M.G.; Ramos-Sánchez, H.V.; Flores-Miramontes, M.G.; De La Mata-González, P.; Jave-Suárez, L.F.; Aguilar-Lemarroy, A. High frequency of HPV genotypes 59, 66, 52, 51, 39 and 56 in women from Western Mexico. BMC Infect. Dis. 2020, 20, 889. [Google Scholar] [CrossRef]
- Wheeler, M.C.; Kjaer, K.S.; Sigurdsson, K.; Iversen, O.E.; Hernandez-Avila, M.; Perez, G.; Brown, D.R.; Koutsky, L.A.; Tay, E.H.; García, P.; et al. The Impact of Quadrivalent Human Papillomavirus (HPV; Types 6, 11, 16, and 18) L1 Virus-Like Particle Vaccine on Infection and Disease Due to Oncogenic Nonvaccine HPV Types in Sexually Active Women Aged 16–26 Years. J. Infect. Dis. 2009, 199, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.R.; Kjaer, S.K.; Sigurdsson, K.; Iversen, O.-E.; Hernandez-Avila, M.; Wheeler, C.M.; Perez, G.; Koutsky, L.A.; Tay, E.H.; Garcia, P.; et al. The Impact of Quadrivalent Human Papillomavirus (HPV; Types 6, 11, 16, and 18) L1 Virus-like Particle Vaccine on Infection and Disease Due to Oncogenic Nonvaccine HPV Types in Generally HPV-Naive Women Aged 16–26 Years. J. Infect. Dis. 2009, 199, 926–935. [Google Scholar] [CrossRef]
- Herrero, R. Human Papillomavirus (HPV) Vaccines: Limited Cross-Protection against Additional HPV Types. J. Infect. Dis. 2009, 199, 919–922. [Google Scholar] [CrossRef]
- Garnett, G.P.; Waddell, H.C. Public health paradoxes and the epidemiological impact of an HPV vaccine. J. Clin. Virol. 2000, 19, 101–111. [Google Scholar] [CrossRef]
- Durham, D.P.; Poolman, E.M.; Ibuka, Y.; Townsend, J.P.; Galvani, A.P. Reevaluation of epidemiological data demonstrates that it is consistent with cross-immunity among human papillomavirus types. J. Infect. Dis. 2012, 206, 1291–1298. [Google Scholar] [CrossRef]
- Merikukka, M.; Kaasila, M.; Namujju, P.B.; Palmroth, J.; Kirnbauer, R.; Paavonen, J.; Surcel, H.M.; Lehtinen, M. Differences in incidence and co-occurrence of vaccine and nonvaccine human papillomavirus types in Finnish population before human papillomavirus mass vaccination suggest competitive advantage for HPV33. Int. J. Cancer 2011, 128, 1114–1119. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.K.; Madeleine, M.M.; Biggar, R.J.; Engels, E.A. Risk of Human Papillomavirus–Associated Cancers Among Persons With AIDS. JNCI J. Natl. Cancer Inst. 2009, 101, 1120–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elbasha, E.H.; Galvani, A.P. Vaccination against multiple HPV types. Math. Biosci. 2005, 197, 88–117. [Google Scholar] [CrossRef]
- Fappani, C.; Bianchi, S.; Panatto, D.; Petrelli, F.; Colzani, D.; Scuri, S.; Gori, M.; Amendola, A.; Grappasonni, I.; Tanzi, E.; et al. HPV Type-Specific Prevalence a Decade after the Implementation of the Vaccination Program: Results from a Pilot Study. Vaccines 2021, 9, 336. [Google Scholar] [CrossRef] [PubMed]
- Carozzi, F.; Puliti, D.; Ocello, C.; Anastasio, P.S.; Moliterni, E.A.; Perinetti, E.; Serradell, L.; Burroni, E.; Confortini, M.; Mantellini, P.; et al. Monitoring vaccine and non-vaccine HPV type prevalence in the post-vaccination era in women living in the Basilicata region, Italy. BMC Infect. Dis. 2018, 18, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saccucci, M.; Franco, E.L.; Ding, L.; Bernstein, D.I.; Brown, D.; Kahn, J.A. Non-Vaccine-Type Human Papillomavirus Prevalence After Vaccine Introduction: No Evidence for Type Replacement but Evidence for Cross-Protection. Sex. Transm. Dis. 2018, 45, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Covert, C.; Ding, L.; Brown, D.; Franco, E.L.; Bernstein, D.I.; Kahn, J.A. Evidence for cross-protection but not type-replacement over the 11 years after human papillomavirus vaccine introduction. Hum. Vaccines Immunother. 2019, 15, 1962–1969. [Google Scholar] [CrossRef] [Green Version]
- Gray, P.; Kann, H.; Pimenoff, V.N.; Adhikari, I.; Eriksson, T.; Surcel, H.M.; Vänskä, S.; Dillner, J.; Faust, H.; Lehtinen, M. Long-term follow-up of human papillomavirus type replacement among young pregnant Finnish females before and after a community-randomised HPV vaccination trial with moderate coverage. Int. J. Cancer 2020, 147, 3511–3522. [Google Scholar] [CrossRef]
- Tota, J.E.; Struyf, F.; Merikukka, M.; Gonzalez, P.; Kreimer, A.R.; Bi, D.; Castellsagué, X.; de Carvalho, N.S.; Garland, S.M.; Harper, D.M.; et al. Evaluation of Type Replacement Following HPV16/18 Vaccination: Pooled Analysis of Two Randomized Trials. J. Natl. Cancer Inst. 2017, 109, djw300. [Google Scholar] [CrossRef]
- De Vincenzo, R.; Ricci, C.; Conte, C.; Scambia, G. HPV vaccine cross-protection: Highlights on additional clinical benefit. Gynecol. Oncol. 2013, 130, P642–P651. [Google Scholar] [CrossRef]
- Stanley, M.; Joura, E.; Yen, G.P.; Kothari, S.; Luxembourg, A.; Saah, A.; Walia, A.; Perez, G.; Khoury, H.; Badgley, D.; et al. Systematic literature review of neutralizing antibody immune responses to non-vaccine targeted high-risk HPV types induced by the bivalent and the quadrivalent vaccines. Vaccine 2021, 39, 2214–2223. [Google Scholar] [CrossRef] [PubMed]
- Schiffman, M.; Clifford, G.; Buonaguro, F.M. Classification of weakly carcinogenic human papillomavirus types: Addressing the limits of epidemiology at the borderline. Infect. Agents Cancer 2009, 4, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancer WHOIAfRo. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans: Human Papillomavirus; Group IW: Lyon, France, 2007. [Google Scholar]
- de Sanjose, S.; Quint, W.G.V.; Alemany, L.; Geraets, D.T.; Klaustermeier, J.E.; Lloveras, B.; Tous, S.; Felix, A.; Bravo, L.E.; Shin, H.-R.; et al. Human papillomavirus genotype attribution in invasive cervical cancer: A retrospective cross-sectional worldwide study. Lancet Oncol. 2010, 11, 1048–1056. [Google Scholar] [CrossRef]
- England, P.H. Cervical Screening: Programme and Colposcopy Management 2021. Guidelines for Commissioners, Screening Providers and Programme Managers for NHS Cervical Screening. Available online: https://www.gov.uk/government/publications/cervical-screening-programme-and-colposcopy-management (accessed on 18 July 2022).
- Jones, J.; Powell, N.G.; Tristram, A.; Fiander, A.N.; Hibbitts, S. Comparison of the PapilloCheck® DNA micro-array Human Papillomavirus detection assay with Hybrid Capture II and PCR-enzyme immunoassay using the GP5/6+ primer set. J. Clin. Virol. 2009, 45, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Kino, N.; Sata, T.; Sato, Y.; Sugase, M.; Matsukura, T. Molecular Cloning and Nucleotide Sequence Analysis of a Novel Human Papillomavirus (Type 82) Associated with Vaginal Intraepithelial Neoplasia. Clin. Diagn. Lab. Immunol. 2000, 7, 91–95. [Google Scholar] [CrossRef] [Green Version]
- de Villiers, E.M.; Fauquet, C.; Broker, T.R.; Bernard, H.U.; zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Viviano, M.; DeBeaudrap, P.; Tebeu, P.-M.; Fouogue, J.T.; Vassilakos, P.; Petignat, P. A review of screening strategies for cervical cancer in human immunodeficiency virus-positive women in sub-Saharan Africa. Int. J. Women’s Health 2017, 9, 69–79. [Google Scholar] [CrossRef] [Green Version]
- Rezhake, R.; Hu, S.Y.; Zhao, S.; Xu, X.Q.; Zhao, X.L.; Zhang, L.; Wang, Y.; Zhang, X.; Pan, Q.J.; Qiao, Y.L.; et al. Eight-type human papillomavirus E6/E7 oncoprotein detection as a novel and promising triage strategy for managing HPV-positive women. Int. J. Cancer 2019, 144, 34–42. [Google Scholar] [CrossRef]
- Agorastos, T.; Chatzistamatiou, K.; Moysiadis, T.; Kaufmann, A.M.; Skenderi, A.; Lekka, I.; Koch, I.; Soutschek, E.; Boecher, O.; Kilintzis, V.; et al. Human papillomavirus E7 protein detection as a method of triage to colposcopy of HPV positive women, in comparison to genotyping and cytology. Final results of the PIPAVIR study. Int. J. Cancer 2017, 141, 519–530. [Google Scholar] [CrossRef]
| Single (n) | Multiple (n) | |
|---|---|---|
| Age (years) | ||
| 20 | 62 | 65 |
| 21 | 23 | 18 |
| 22 | 15 | 17 |
| SDS | ||
| NA | 4 | 3 |
| Q1 | 17 | 20 |
| Q2 | 18 | 16 |
| Q3 | 23 | 17 |
| Q4 | 20 | 23 |
| Q5 | 18 | 21 |
| Cytology | ||
| Negative | 53 | 47 |
| Borderline | 22 | 26 |
| Mild | 23 | 22 |
| Moderate | 2 | 2 |
| Severe | 0 | 3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bowden, S.J.; Ellis, L.B.; Kyrgiou, M.; Fiander, A.N.; Hibbitts, S. High Prevalence of HPV 51 in an Unvaccinated Population and Implications for HPV Vaccines. Vaccines 2022, 10, 1754. https://doi.org/10.3390/vaccines10101754
Bowden SJ, Ellis LB, Kyrgiou M, Fiander AN, Hibbitts S. High Prevalence of HPV 51 in an Unvaccinated Population and Implications for HPV Vaccines. Vaccines. 2022; 10(10):1754. https://doi.org/10.3390/vaccines10101754
Chicago/Turabian StyleBowden, Sarah J., Laura Burney Ellis, Maria Kyrgiou, Alison N. Fiander, and Samantha Hibbitts. 2022. "High Prevalence of HPV 51 in an Unvaccinated Population and Implications for HPV Vaccines" Vaccines 10, no. 10: 1754. https://doi.org/10.3390/vaccines10101754
APA StyleBowden, S. J., Ellis, L. B., Kyrgiou, M., Fiander, A. N., & Hibbitts, S. (2022). High Prevalence of HPV 51 in an Unvaccinated Population and Implications for HPV Vaccines. Vaccines, 10(10), 1754. https://doi.org/10.3390/vaccines10101754





