Seven Fatty Acid Metabolism-Related Genes as Potential Biomarkers for Predicting the Prognosis and Immunotherapy Responses in Patients with Esophageal Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Fatty-Acid-Metabolism-Related Genes
2.2. Construction and Evaluation of a Predictive Risk Score Model
2.3. The Association between Risk Score and Clinical Characteristics
2.4. Construction of a Nomogram for Patients with ESCA
2.5. Association between the Risk Model and Immune Parameters
2.6. GSCA Analysis
2.7. A Protein–Protein Interaction Network of DEGs in Groups of Different Risk Score Groups
2.8. Validation of the Expression and Prognostic Value of Seven FRGs
3. Results
3.1. Identifying Fatty-Acid-Metabolism-Related DEGs in ESCA Samples
3.2. Establishing and Validating a Prognostic FRG Signature
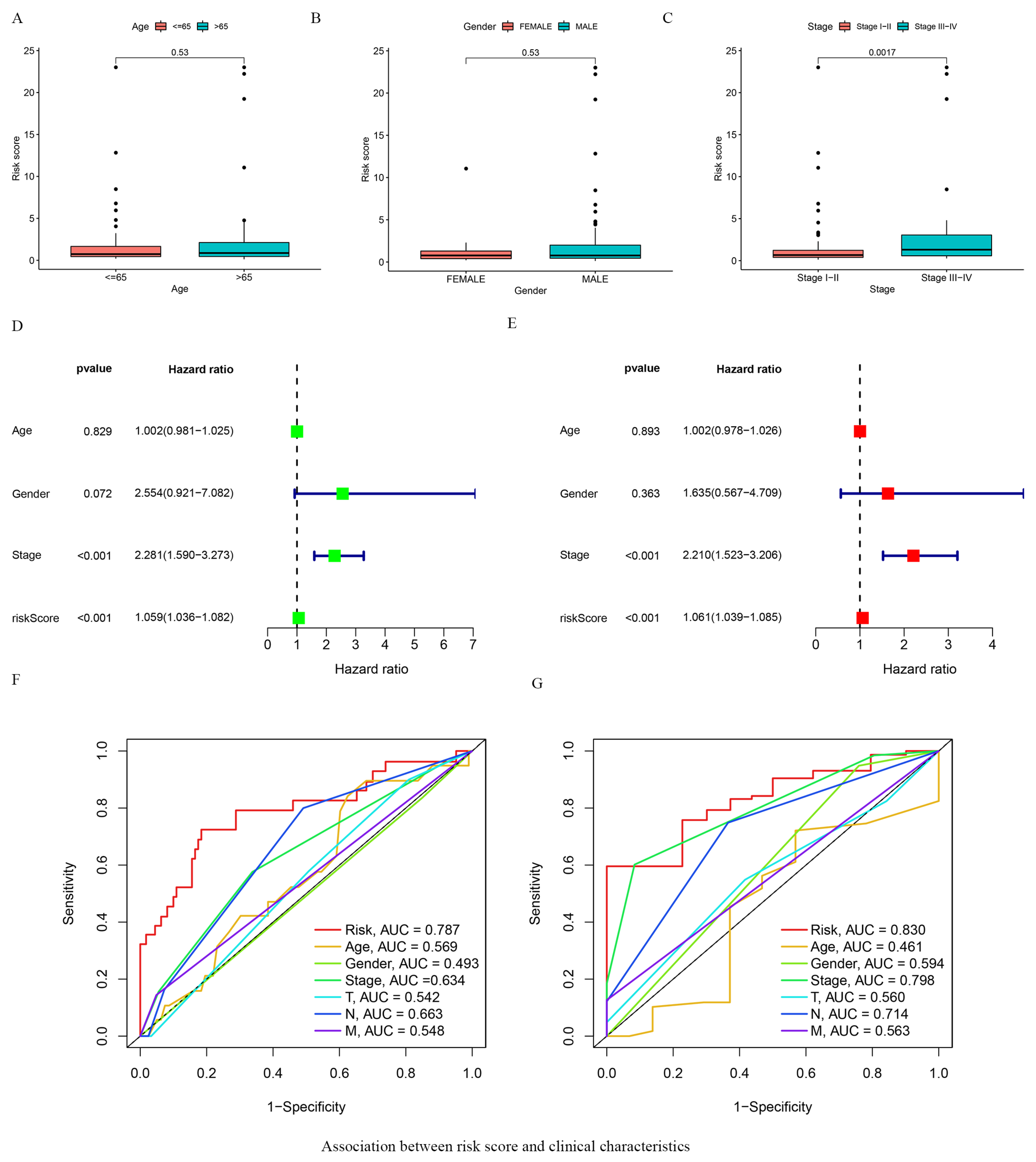
3.3. Association between Risk Score and Clinical Characteristics
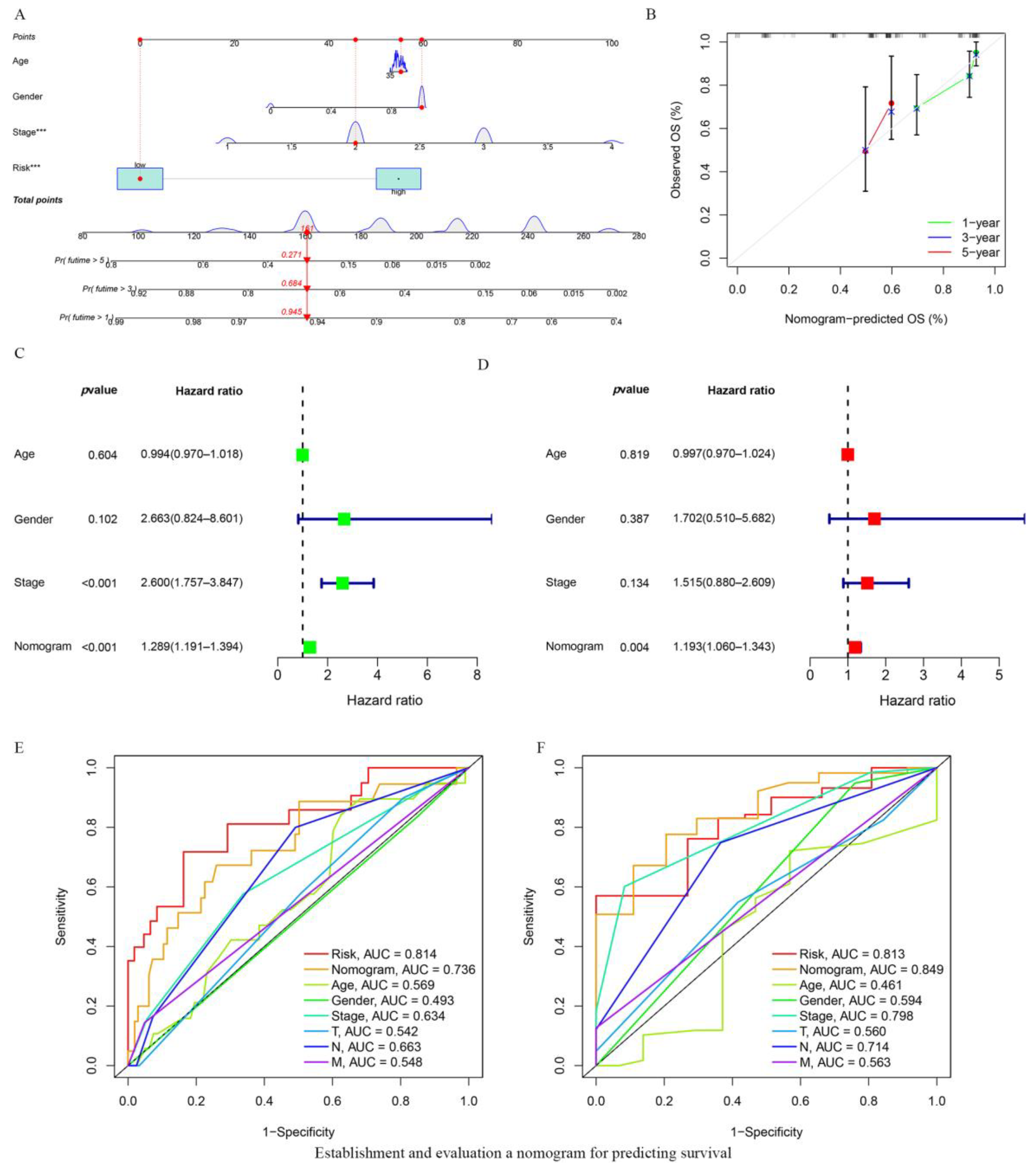
3.4. Establishing and Evaluating a Nomogram for Predicting Survival
3.5. Immunological Features of the Tumor and GSCA Analysis
3.6. DEGs in the Low- and High-Risk Score Groups
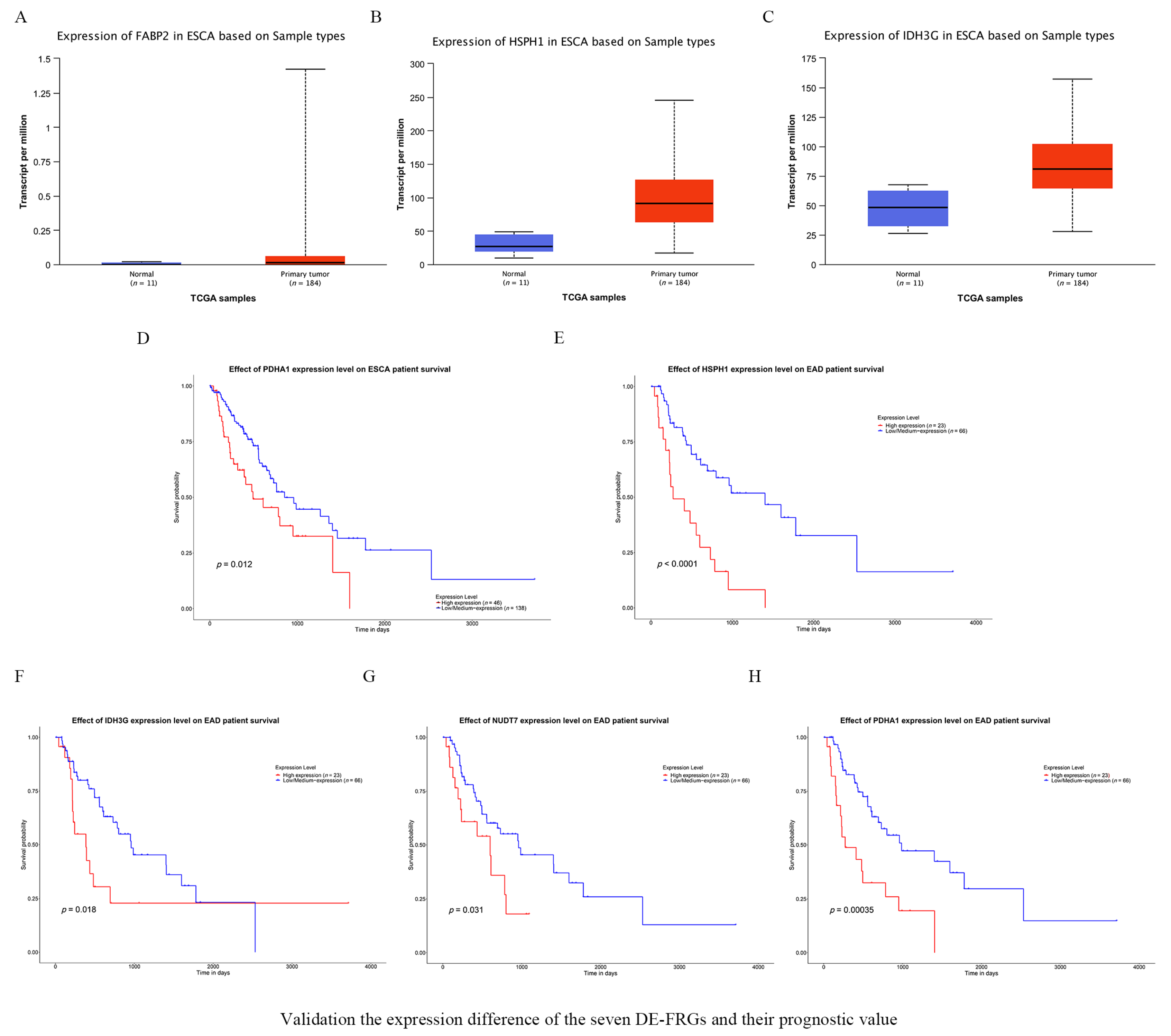
3.7. Validating the Expression of the Seven FRGs and Their Prognostic Value
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Li, Y.; Sebastian Zschaeck, Q.; Chen, S.; Chen, L.; Wu, H. Metabolic parameters of sequential 18F-FDG PET/CT predict overall survival of esophageal cancer patients treated with (chemo-) radiation. Radiat. Oncol. 2019, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Liu, X.-Y.; Cheng, Z.; Kai, W.; Song, Z. Comprehensive analysis of a new immune-related prognostic signature for esophageal cancer and its correlation with infiltrating immune cells and target genes. Ann. Transl. Med. 2021, 9, 1576. [Google Scholar] [CrossRef]
- Mao, Y.; Gao, S.; Wang, Q.; Shi, X.; Li, Y.; Gao, W.; Guan, F.; Li, X.; Han, Y.; Liu, Y.; et al. Analysis of a registry database for esophageal cancer from high-volume centers in China. Dis. Esophagus 2020, 33, doz091. [Google Scholar] [CrossRef]
- Ding, C.; Shan, C.; Li, M.; Chen, H.; Li, X.; Jin, Z. Characterization of the fatty acid metabolism in colorectal cancer to guide clinical therapy. Mol. Ther. Oncolytics 2021, 20, 532–544. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Harris, A. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 2012, 491, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhang, H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell. Mol. Life Sci. CMLS 2016, 73, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Arkaitz Carracedo, L.C.C.; Pandolfi, P.P. Cancer metabolism: Fatty acid oxidation in the limelight. Nat. Rev. Cancer 2013, 13, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Chen, D.; Lu, Q.; Yao, Y.; Ji, C. Bioinformatic Profiling Identifies a Fatty Acid Metabolism-Related Gene Risk Signature for Malignancy, Prognosis, and Immune Phenotype of Glioma. Dis. Markers 2019, 2019, 3917040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liao, Y.; Liu, P.; Du, Q.; Liang, Y.; Ooi, S.; Qin, S.; He, S.; Yao, S.; Wang, W. FABP5 promotes lymph node metastasis in cervical cancer by reprogramming fatty acid metabolism. Theranostics 2020, 10, 6561–6580. [Google Scholar] [CrossRef]
- Tabe, Y.; Konopleva, M.; Andreeff, M. Fatty Acid Metabolism, Bone Marrow Adipocytes, and AML. Front. Oncol. 2020, 10, 155. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Wei, R.; Zhang, X.; Jiang, N.; Fan, M.; Huang, J.; Xie, B.; Zhang, L.; Miao, W.; Butler, A.; et al. CPT1A/2-Mediated FAO Enhancement-A Metabolic Target in Radioresistant Breast Cancer. Front. Oncol. 2019, 9, 1201. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Fabritius, M.; Ip, C. Chemotherapeutic sensitization by endoplasmic reticulum stress: Increasing the efficacy of taxane against prostate cancer. Cancer Biol. Ther. 2009, 8, 146–152. [Google Scholar] [CrossRef]
- Corn, K.; Windham, M.; Rafat, M. Lipids in the tumor microenvironment: From cancer progression to treatment. Prog. Lipid Res. 2020, 80, 101055. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Cai, L.; Huang, W.; Weng, Q.; Lin, X.; You, M.; Liao, S. Prognostic value of fatty acid metabolism-related genes in patients with hepatocellular carcinoma. Aging 2021, 13, 17847–17863. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhang, P.; Li, F.; Shen, Y.; Chen, H.; Feng, Y.; He, A.; Wang, F. CD138 multiple myeloma cells express high level of CHK1 which correlated to overall survival in MM patient. Aging 2020, 12, 23067–23081. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Du, M.; Lu, L. A Novel 16-Genes Signature Scoring System as Prognostic Model to Evaluate Survival Risk in Patients with Glioblastoma. Biomedicines 2022, 10, 317. [Google Scholar] [CrossRef]
- Liu, Z.-K.; Wu, K.-F.; Zhang, R.-Y.; Kong, L.-M.; Shang, R.-Z.; Lv, J.-J.; Li, C.; Lu, M.; Yong, Y.-L.; Zhang, C.; et al. Pyroptosis-Related LncRNA Signature Predicts Prognosis and Is Associated With Immune Infiltration in Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 794034. [Google Scholar] [CrossRef]
- Huang, J.; Chen, W.; Chen, C.; Jie, Z.; Xiao, T. Comprehensive Analysis and Prognosis Prediction of N6-Methyladenosine-Related lncRNAs in Immune Microenvironment Infiltration of Gastric Cancer. Int. J. Gen. Med. 2022, 15, 2629–2643. [Google Scholar] [CrossRef] [PubMed]
- Xiang, L.; Lan, C. Significance of a Tumor Mutation Burden Gene Signature with Prognosis and Immune Feature of Gastric Cancer Patients. Int. J. Genom. 2022, 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Cheng, Q.; Chang, K.; Bao, L.; Yi, X. Integrated Analysis of Ferroptosis-Related Biomarker Signatures to Improve the Diagnosis and Prognosis Prediction of Ovarian Cancer. Front. Cell Dev. Biol. 2021, 9, 807862. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Hu, F.; Xia, M.; Han, L.; Zhang, Q.; Guo, A. GSCALite: A web server for gene set cancer analysis. Bioinformatics 2018, 34, 3771–3772. [Google Scholar] [CrossRef]
- Xia, Q.-D.; Sun, J.-X.; Liu, C.-Q.; Xu, J.-Z.; An, Y.; Xu, M.-Y.; Liu, Z.; Hu, J.; Wang, S.-G. Ferroptosis Patterns and Tumor Microenvironment Infiltration Characterization in Bladder Cancer. Front. Cell Dev. Biol. 2022, 10, 832892. [Google Scholar] [CrossRef]
- Niu, Z.; Wang, X.; Xu, Y.; Li, Y.; Gong, X.; Zeng, Q.; Zhang, B.; Xi, J.; Pei, X.; Yue, W.; et al. Development and Validation of a Novel Survival Model for Cutaneous Melanoma Based on Necroptosis-Related Genes. Front. Oncol. 2022, 12, 852803. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Zhang, M.; Guan, B.; Chen, Q.; Dong, Z.; Wang, C. Identification of hub genes associated with prognosis, diagnosis, immune infiltration and therapeutic drug in liver cancer by integrated analysis. Hum. Genom. 2021, 15, 39. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.; Bashel, B.; Balasubramanya, S.; Creighton, C.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Wu, J.; Zhao, H.; Nie, X.C. RPP25 as a Prognostic-Related Biomarker That Correlates With Tumor Metabolism in Glioblastoma. Front. Oncol. 2021, 11, 714904. [Google Scholar] [CrossRef]
- Wei, X.; Mao, T.; Li, S.; He, J.; Hou, X.; Li, H.; Zhan, M.; Yang, X.; Li, R.; Xiao, J.; et al. DT-13 inhibited the proliferation of colorectal cancer via glycolytic metabolism and AMPK/mTOR signaling pathway. Phytomedicine Int. J. Phytother. Phytopharm. 2019, 54, 120–131. [Google Scholar] [CrossRef]
- Vincent, Z.; Urakami, K.; Maruyama, K.; Yamaguchi, K.; Kusuhara, M. CD133-positive cancer stem cells from Colo205 human colon adenocarcinoma cell line show resistance to chemotherapy and display a specific metabolomic profile. Genes Cancer 2014, 5, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Zeki, S.S.; Graham, T.A.; Wright, N.A. Stem cells and their implications for colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Zhou, B.; Yang, Q.; Pan, Y.; Yang, W.; Freedland, S.; Ding, L.; Freeman, M.; Breunig, J.; Bhowmick, N.; et al. A Transcriptional Regulatory Loop of Master Regulator Transcription Factors, PPARG, and Fatty Acid Synthesis Promotes Esophageal Adenocarcinoma. Cancer Res. 2021, 81, 1216–1229. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Hua, R.; Li, B.; Gu, H.; Sun, Y.; Li, Z. Loss of FBP1 promotes proliferation, migration, and invasion by regulating fatty acid metabolism in esophageal squamous cell carcinoma. Aging 2020, 13, 4986–4998. [Google Scholar] [CrossRef]
- Chen, T.; Hsieh, Y.; Huang, J.; Liu, C.; Chuang, L.; Huang, P.; Kuo, T.; Chia, H.; Chou, C.; Chang, C.; et al. Determination of Pyruvate Metabolic Fates Modulates Head and Neck Tumorigenesis. Neoplasia 2019, 21, 641–652. [Google Scholar] [CrossRef]
- Peng, M.; Han, J.; Li, L.; Ma, H. Suppression of fat deposition in broiler chickens by (-)-hydroxycitric acid supplementation: A proteomics perspective. Sci. Rep. 2016, 6, 32580. [Google Scholar] [CrossRef]
- Zhong, Y.; Li, X.; Ji, Y.; Li, X.; Li, Y.; Yu, D.; Yuan, Y.; Liu, J.; Li, H.; Zhang, M.; et al. Pyruvate dehydrogenase expression is negatively associated with cell stemness and worse clinical outcome in prostate cancers. Oncotarget 2017, 8, 13344–13356. [Google Scholar] [CrossRef]
- Zhong, Y.; Huang, R.; Li, X.; Xu, R.; Zhou, F.; Wang, J.; Fan, H.; Goscinski, M.; Zhang, M.; Wen, J.-G.; et al. Decreased Expression of PDHE1α Predicts Worse Clinical Outcome in Esophageal Squamous Cell Carcinoma. Anticancer. Res. 2015, 35, 5533–5538. [Google Scholar]
- Liu, L.; Cao, J.; Zhao, J.; Li, X.; Suo, Z.; Li, H. PDHA1 Gene Knockout In Human Esophageal Squamous Cancer Cells Resulted In Greater Warburg Effect And Aggressive Features In Vitro And In Vivo. OncoTargets Ther. 2019, 12, 9899–9913. [Google Scholar] [CrossRef]
- Li, S.; Liu, D.; Zhang, X.; Zhu, X.; Lu, X.; Huang, J.; Yang, L.; Wu, Y. Low expression of PDHA1 predicts poor prognosis in gastric cancer. Pathol. Res. Pract. 2019, 215, 478–482. [Google Scholar] [CrossRef]
- Fan, G.; Tu, Y.; Wu, N.; Xiao, H. The expression profiles and prognostic values of HSPs family members in Head and neck cancer. Cancer Cell Int. 2020, 20, 220. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, N.; Zhang, L.; Song, Y.; Liu, J.; Yu, J.; Yang, M. Oncogene HSPH1 modulated by the rs2280059 genetic variant diminishes EGFR-TKIs efficiency in advanced lung adenocarcinoma. Carcinogenesis 2020, 41, 1195–1202. [Google Scholar] [CrossRef]
- Berthenet, K.; Christophe, B.; Collura, A.; Svrcek, M.; Richaud, S.; Hammann, A.; Yousfi, S.C.N.; Wanherdrick, K.; Duplomb, L. Extracellular HSP110 skews macrophage polarization in colorectal cancer. Oncoimmunology 2016, 5, e1170264. [Google Scholar] [CrossRef]
- Zappasodi, R.; Ruggiero, G.; Tortoreto, M.; Tringali, C.; Cavanè, A.; Cabras, A.D.; Castagnoli, L.; Venerando, B.; Zaffaroni, N.; Gianni, A.M.; et al. HSPH1 inhibition downregulates Bcl-6 and c-Myc and hampers the growth of human aggressive B-cell non-Hodgkin lymphoma. Blood 2015, 125, 1768–1771. [Google Scholar] [CrossRef] [PubMed]
- Shumar, S.; Kerr, E.; Fagone, P.; Infante, A.; Leonardi, R. Overexpression of Nudt7 decreases bile acid levels and peroxisomal fatty acid oxidation in the liver. J. Lipid Res. 2019, 60, 1005–1019. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zheng, W.; Chen, J.; Lin, S.; Zou, Z.; Li, X.; Tan, Z. Systematic analysis of survival-associated alternative splicing signatures in clear cell renal cell carcinoma. J. Cell. Biochem. 2020, 121, 4074–4084. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhou, Y.; Sun, Y.; Wang, Q. Intestinal fatty acid binding protein: A rising therapeutic target in lipid metabolism. Prog. Lipid Res. 2022, 87, 101178. [Google Scholar] [CrossRef]
- Duan, J.; Xie, Y.; Qu, L.; Wang, L.; Zhou, S.; Wang, Y.; Fan, Z.; Yang, S.; Jiao, S. A nomogram-based immunoprofile predicts overall survival for previously untreated patients with esophageal squamous cell carcinoma after esophagectomy. J. Immunother. Cancer 2018, 6, 100. [Google Scholar] [CrossRef]
- Gao, Y.; Guo, W.; Geng, X.; Zhang, Y.; Zhang, G.; Qiu, B.; Tan, F.; Xue, Q.; Gao, S.; He, J. Prognostic value of tumor-infiltrating lymphocytes in esophageal cancer: An updated meta-analysis of 30 studies with 5122 patients. Ann. Transl. Med. 2020, 8, 822. [Google Scholar] [CrossRef] [PubMed]
- Murakami, T.; Tanaka, N.; Takamatsu, K.; Hakozaki, K.; Fukumoto, K.; Masuda, T.; Mikami, S.; Kakimi, T.S.K.; Tsunoda, T.; Sawada, K.; et al. Multiplexed single-cell pathology reveals the association of CD8 T-cell heterogeneity with prognostic outcomes in renal cell carcinoma. Cancer Immunol. Immunother. CII 2021, 70, 3001–3013. [Google Scholar] [CrossRef]
- Taniguchi, H.; Caeser, R.; Chavan, S.; Zhan, Y.; Chow, A.; Manoj, P.; Uddin, F.; Kitai, H.; Qu, R.; Hayatt, O.; et al. WEE1 inhibition enhances the antitumor immune response to PD-L1 blockade by the concomitant activation of STING and STAT1 pathways in SCLC. Cell Rep. 2022, 39, 110814. [Google Scholar] [CrossRef]
- Hänze, J.; Wegner, M.; Noessner, E.; Hofmann, R.; Hegele, A. Co-Regulation of Immune Checkpoint PD-L1 with Interferon-Gamma Signaling is Associated with a Survival Benefit in Renal Cell Cancer. Target. Oncol. 2020, 15, 377–390. [Google Scholar] [CrossRef]
- Ayers, M.; Lunceford, J.; Nebozhyn, M.; Murphy, E.; Loboda, A.; Kaufman, D.; Albright, A.; Cheng, J.; Kang, S.; Shankaran, V.; et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017, 127, 2930–2940. [Google Scholar] [CrossRef]
- Otsubo, T.; Hagiwara, T.; Tamura-Nakano, M.; Sezaki, T.; Miyake, O.; Hinohara, C.; Shimizu, T.; Yamada, K.; Dohi, T.; Kawamura, Y.I. Aberrant DNA hypermethylation reduces the expression of the desmosome-related molecule periplakin in esophageal squamous cell carcinoma. Cancer Med. 2015, 4, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Tonoike, Y.; Matsushita, K.; Tomonaga, T.; Katada, K.; Tanaka, N.; Shimada, H.; Nakatani, Y.; Okamoto, Y.; Nomura, F. Adhesion molecule periplakin is involved in cellular movement and attachment in pharyngeal squamous cancer cells. BMC Cell Biol. 2011, 12, 41. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.L.; Zhang, L.; Zhang, H.; Zhang, Y.; Liu, D. Mechanism of periplakin on ovarian cancer cell phenotype and its influence on prognosis. Transl. Cancer Res. 2022, 11, 1372–1385. [Google Scholar] [CrossRef]
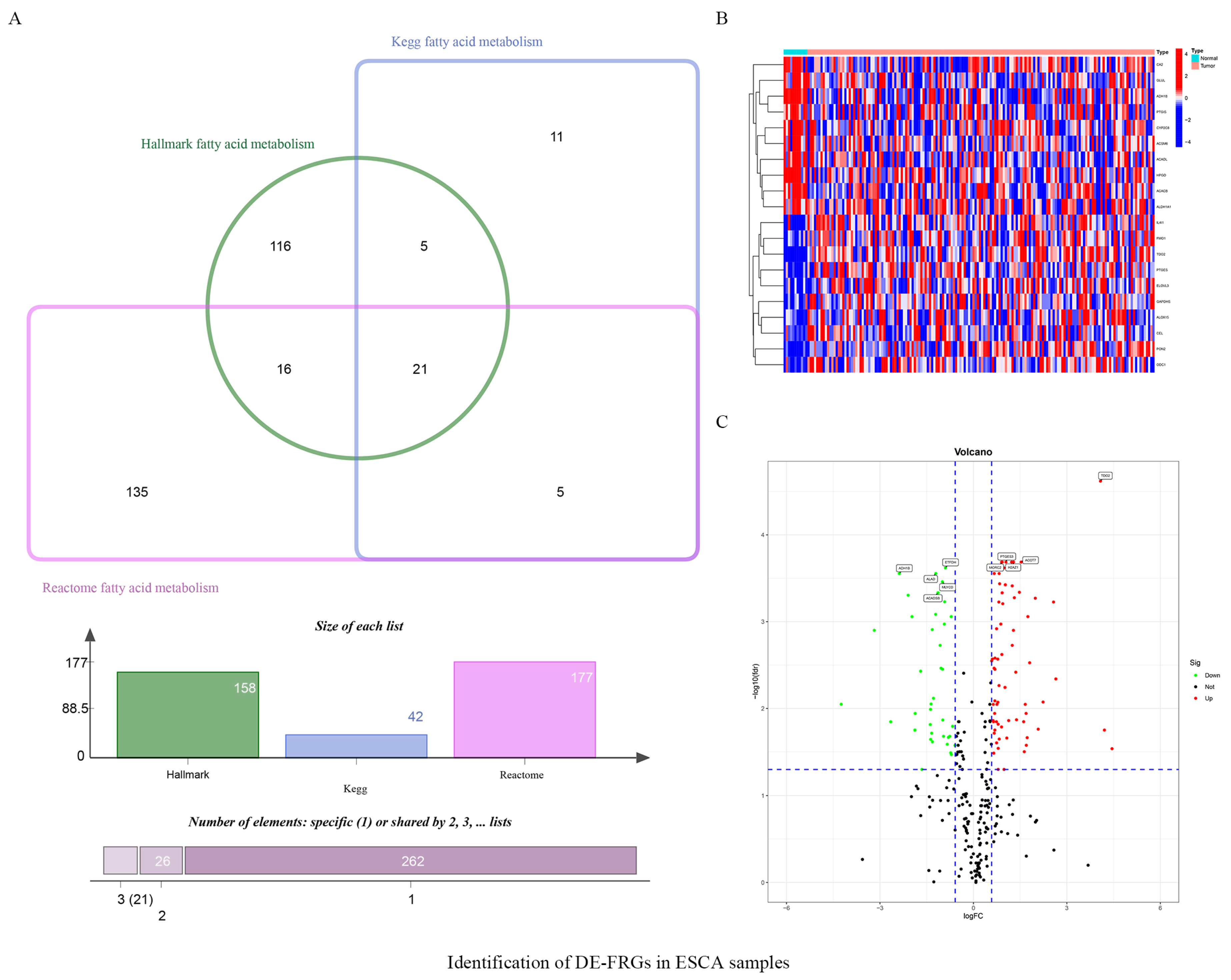



| Gene | logFC | p-Value | FDR |
|---|---|---|---|
| FADS2 | 1.6647 | 0.0022 | 0.0090 |
| HACD1 | −0.8386 | 0.0109 | 0.0258 |
| PTGES2 | 0.8793 | 0.0001 | 0.0011 |
| HPGD | −1.9718 | 0.0001 | 0.0009 |
| PTGES | 2.5728 | 0.0001 | 0.0006 |
| MAOA | −0.9333 | 0.0001 | 0.0011 |
| CD36 | −0.6685 | 0.0053 | 0.0161 |
| ELOVL2 | 1.7260 | 0.0084 | 0.0217 |
| GPX1 | 0.6824 | 0.0031 | 0.0114 |
| ENO3 | 0.7959 | 0.0125 | 0.0288 |
| CEL | 4.4503 | 0.0128 | 0.0291 |
| ACBD6 | 0.8421 | 0.0000 | 0.0004 |
| MORC2 | 0.9267 | 0.0000 | 0.0002 |
| UBE2L6 | 1.4763 | 0.0000 | 0.0005 |
| CYP2C9 | −1.3856 | 0.0050 | 0.0153 |
| SLC25A1 | 0.6648 | 0.0006 | 0.0034 |
| ALDH1A1 | −1.6541 | 0.0256 | 0.0499 |
| PRKAA2 | −1.3722 | 0.0091 | 0.0227 |
| ACOT7 | 1.5324 | 0.0000 | 0.0002 |
| ABCD1 | 1.0133 | 0.0012 | 0.0057 |
| GLUL | −1.6938 | 0.0007 | 0.0037 |
| ACOT9 | 0.6684 | 0.0071 | 0.0193 |
| NCAPH2 | 0.8264 | 0.0000 | 0.0003 |
| HACD2 | 0.7813 | 0.0004 | 0.0027 |
| OSTC | 0.5871 | 0.000 | 0.0028 |
| FADS1 | 1.6133 | 0.0046 | 0.0142 |
| HMGCS2 | −0.9822 | 0.0038 | 0.0136 |
| ALOX15 | 4.2060 | 0.0063 | 0.0177 |
| CPOX | 0.9419 | 0.0001 | 0.0006 |
| ACSM6 | −2.6558 | 0.0043 | 0.0142 |
| NTHL1 | 0.9232 | 0.0000 | 0.0005 |
| MIX23 | 1.0243 | 0.0000 | 0.0004 |
| HSPH1 | 1.3146 | 0.0000 | 0.0005 |
| PTGDS | −0.7081 | 0.0160 | 0.0339 |
| CYP2C8 | −4.2436 | 0.0021 | 0.0089 |
| PTGES3 | 1.0439 | 0.0000 | 0.0002 |
| CPT1B | 0.9034 | 0.0055 | 0.0163 |
| ADH1C | −1.3389 | 0.0071 | 0.0193 |
| BLVRA | 0.6532 | 0.0152 | 0.0327 |
| SUCLG2 | −1.3197 | 0.0002 | 0.0012 |
| THRSP | −1.3148 | 0.0099 | 0.0242 |
| ACOX3 | −0.5875 | 0.0115 | 0.0266 |
| ELOVL3 | 2.6467 | 0.0009 | 0.0046 |
| TDO2 | 4.0796 | 0.0000 | 0.0000 |
| ACADS | −0.9901 | 0.0007 | 0.0035 |
| YWHAH | 0.7439 | 0.0002 | 0.0012 |
| AMACR | −0.7589 | 0.0080 | 0.0209 |
| PCCA | −0.9226 | 0.0001 | 0.0006 |
| ODC1 | 1.8059 | 0.0005 | 0.0030 |
| ALAD | −1.2068 | 0.0000 | 0.0003 |
| CA2 | −1.8684 | 0.0031 | 0.0114 |
| RDH11 | 0.6085 | 0.0004 | 0.0027 |
| LDHA | 1.2471 | 0.0003 | 0.0019 |
| ACADSB | −1.1380 | 0.0000 | 0.0005 |
| ACAT1 | −1.2122 | 0.0001 | 0.0008 |
| GABARAPL1 | −0.9593 | 0.0080 | 0.0209 |
| ALOX5AP | 1.0724 | 0.0086 | 0.0219 |
| NSDHL | 0.7857 | 0.0020 | 0.0085 |
| FMO1 | 2.2368 | 0.0019 | 0.0084 |
| ACAT2 | 0.7943 | 0.0049 | 0.0152 |
| GPX4 | 0.6893 | 0.0018 | 0.0082 |
| ACSS1 | −0.7269 | 0.0150 | 0.0324 |
| ETFDH | −0.8984 | 0.0000 | 0.0002 |
| ACSM3 | −1.3758 | 0.0026 | 0.0102 |
| ACBD7 | 1.6988 | 0.0113 | 0.0266 |
| AUH | −0.8033 | 0.0083 | 0.0215 |
| H2AZ1 | 1.2866 | 0.0000 | 0.0002 |
| SMS | 0.9141 | 0.0004 | 0.0024 |
| ELOVL5 | 1.3837 | 0.0037 | 0.0135 |
| ALDOA | 0.7531 | 0.0023 | 0.0090 |
| CYP4B1 | −1.2830 | 0.0017 | 0.0076 |
| ACO2 | −0.7125 | 0.0001 | 0.0009 |
| MDH2 | 0.6924 | 0.0004 | 0.0026 |
| PPT1 | 1.4317 | 0.0000 | 0.0002 |
| PON2 | 1.7812 | 0.0000 | 0.0002 |
| PSME1 | 0.9799 | 0.0000 | 0.0002 |
| PTGS2 | 1.6912 | 0.0031 | 0.0114 |
| HSD17B10 | 0.6922 | 0.0007 | 0.0035 |
| ACLY | 1.2460 | 0.0000 | 0.0004 |
| HMGCS1 | 0.8293 | 0.0089 | 0.0225 |
| ECI2 | −1.3581 | 0.0021 | 0.0089 |
| METAP1 | 0.6304 | 0.0040 | 0.0138 |
| APEX1 | 0.8167 | 0.0001 | 0.0006 |
| MIF | 1.2857 | 0.0002 | 0.0013 |
| ADSL | 0.9045 | 0.0000 | 0.0002 |
| SCD | 1.3604 | 0.0007 | 0.0038 |
| RDH16 | 1.6334 | 0.0139 | 0.0315 |
| PRXL2B | 0.7951 | 0.0256 | 0.0499 |
| IL4I1 | 1.9874 | 0.0000 | 0.0005 |
| ACADL | −3.1819 | 0.0002 | 0.0013 |
| PTGIS | −1.8784 | 0.0062 | 0.0177 |
| ACAD11 | 0.7438 | 0.0103 | 0.0249 |
| ACACB | −2.0952 | 0.0000 | 0.0005 |
| ADH1B | −2.3779 | 0.0000 | 0.0003 |
| ENO2 | 1.1322 | 0.0040 | 0.0138 |
| NBN | 0.6371 | 0.0022 | 0.0090 |
| LGALS1 | 1.7481 | 0.0001 | 0.0009 |
| GAPDHS | 2.0802 | 0.0059 | 0.0173 |
| SLC27A2 | 0.9832 | 0.0256 | 0.0499 |
| HSP90AA1 | 1.2248 | 0.0000 | 0.0002 |
| MLYCD | −0.9958 | 0.0000 | 0.0003 |
| SLC25A17 | 0.6682 | 0.0000 | 0.0003 |
| BCKDHB | −1.0404 | 0.0006 | 0.0034 |
| DBI | 0.6881 | 0.0062 | 0.0177 |
| GPD2 | 0.6431 | 0.0045 | 0.0142 |
| S100A10 | 0.7390 | 0.0046 | 0.0142 |
| HSD17B7 | 0.8224 | 0.0011 | 0.0054 |
| CYP4A11 | −1.0684 | 0.0003 | 0.0019 |
| Gene | logFC | p-Value | FDR |
|---|---|---|---|
| KRT16P2 | −1.1383 | 0.0004 | 0.0175 |
| RNF225 | −1.5708 | 0.0038 | 0.0488 |
| FOXE1 | −1.1111 | 0.0035 | 0.0477 |
| USH1G | −1.1662 | 0.0024 | 0.0386 |
| BCAT1 | −1.1360 | 0.0015 | 0.0301 |
| MIEN1 | 1.1579 | 0.0012 | 0.0275 |
| MLXIPL | 1.2968 | 0.0012 | 0.0270 |
| PPL | −1.3393 | 0.0006 | 0.0204 |
| TMEM74B | 1.0136 | 0.0003 | 0.0142 |
| NEFM | −2.1322 | 0.0017 | 0.0325 |
| GBP6 | −1.4412 | 0.0018 | 0.0334 |
| KRT16P6 | −1.1958 | 0.0025 | 0.0398 |
| ALOX15B | −1.1045 | 0.0008 | 0.0223 |
| AHNAK2 | −1.0332 | 0.0002 | 0.0111 |
| AMBP | 2.2574 | 0.0031 | 0.0450 |
| MIR559 | 1.1306 | 0.0013 | 0.0275 |
| ALOX12 | −1.2123 | 0.0007 | 0.0212 |
| PCK1 | 2.4876 | 0.0000 | 0.0039 |
| PDX1 | 1.2546 | 0.0005 | 0.0187 |
| PINLYP | −1.1081 | 0.0003 | 0.0151 |
| FAM83C | −1.0035 | 0.0034 | 0.0471 |
| ANXA1 | −1.1213 | 0.0006 | 0.0199 |
| YBX2 | 1.1975 | 0.0000 | 0.0047 |
| MIR3189 | 1.1694 | 0.0028 | 0.0425 |
| NOX1 | 1.1536 | 0.0002 | 0.0125 |
| MMP9 | −1.3834 | 0.0012 | 0.0272 |
| EMP1 | −1.1129 | 0.0009 | 0.0241 |
| SBSN | −1.0531 | 0.0030 | 0.0439 |
| CCL15 | 1.6283 | 0.0036 | 0.0480 |
| ACY3 | 1.2213 | 0.0003 | 0.0153 |
| ASCL2 | 1.4926 | 0.0034 | 0.0467 |
| IL1RN | −1.2627 | 0.0002 | 0.0138 |
| A2ML1 | −1.6120 | 0.0008 | 0.0233 |
| TGM1 | −1.8265 | 0.0014 | 0.0295 |
| NEFL | −1.1713 | 0.0006 | 0.0200 |
| CLRN3 | 1.2620 | 0.0028 | 0.0427 |
| QPRT | 1.2541 | 0.0013 | 0.0275 |
| VIL1 | 1.4509 | 0.0032 | 0.0454 |
| WNK4 | 1.2133 | 0.0034 | 0.0469 |
| GOLT1A | 1.2505 | 0.0006 | 0.0206 |
| ANKRD33B | −1.0516 | 0.0016 | 0.0314 |
| CLDN3 | 1.4046 | 0.0004 | 0.0170 |
| PRAP1 | 1.5993 | 0.0020 | 0.0354 |
| ZBED2 | −1.3404 | 0.0001 | 0.0100 |
| ECM1 | −1.2862 | 0.0022 | 0.0368 |
| GPA33 | 1.5683 | 0.0023 | 0.0380 |
| BCAN-AS1 | 1.1560 | 0.0004 | 0.0175 |
| RNF157 | 1.2480 | 0.0011 | 0.0262 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Pan, S.; Ke, Y.; Pan, J.; Li, Y.; Ma, H. Seven Fatty Acid Metabolism-Related Genes as Potential Biomarkers for Predicting the Prognosis and Immunotherapy Responses in Patients with Esophageal Cancer. Vaccines 2022, 10, 1721. https://doi.org/10.3390/vaccines10101721
Guo Y, Pan S, Ke Y, Pan J, Li Y, Ma H. Seven Fatty Acid Metabolism-Related Genes as Potential Biomarkers for Predicting the Prognosis and Immunotherapy Responses in Patients with Esophageal Cancer. Vaccines. 2022; 10(10):1721. https://doi.org/10.3390/vaccines10101721
Chicago/Turabian StyleGuo, Ya, Shupei Pan, Yue Ke, Jiyuan Pan, Yuxing Li, and Hongbing Ma. 2022. "Seven Fatty Acid Metabolism-Related Genes as Potential Biomarkers for Predicting the Prognosis and Immunotherapy Responses in Patients with Esophageal Cancer" Vaccines 10, no. 10: 1721. https://doi.org/10.3390/vaccines10101721
APA StyleGuo, Y., Pan, S., Ke, Y., Pan, J., Li, Y., & Ma, H. (2022). Seven Fatty Acid Metabolism-Related Genes as Potential Biomarkers for Predicting the Prognosis and Immunotherapy Responses in Patients with Esophageal Cancer. Vaccines, 10(10), 1721. https://doi.org/10.3390/vaccines10101721





