Rapid Identity and Quantity CQA Test for Multivalent mRNA Drug Product Formulations
Abstract
:1. Introduction
2. Results
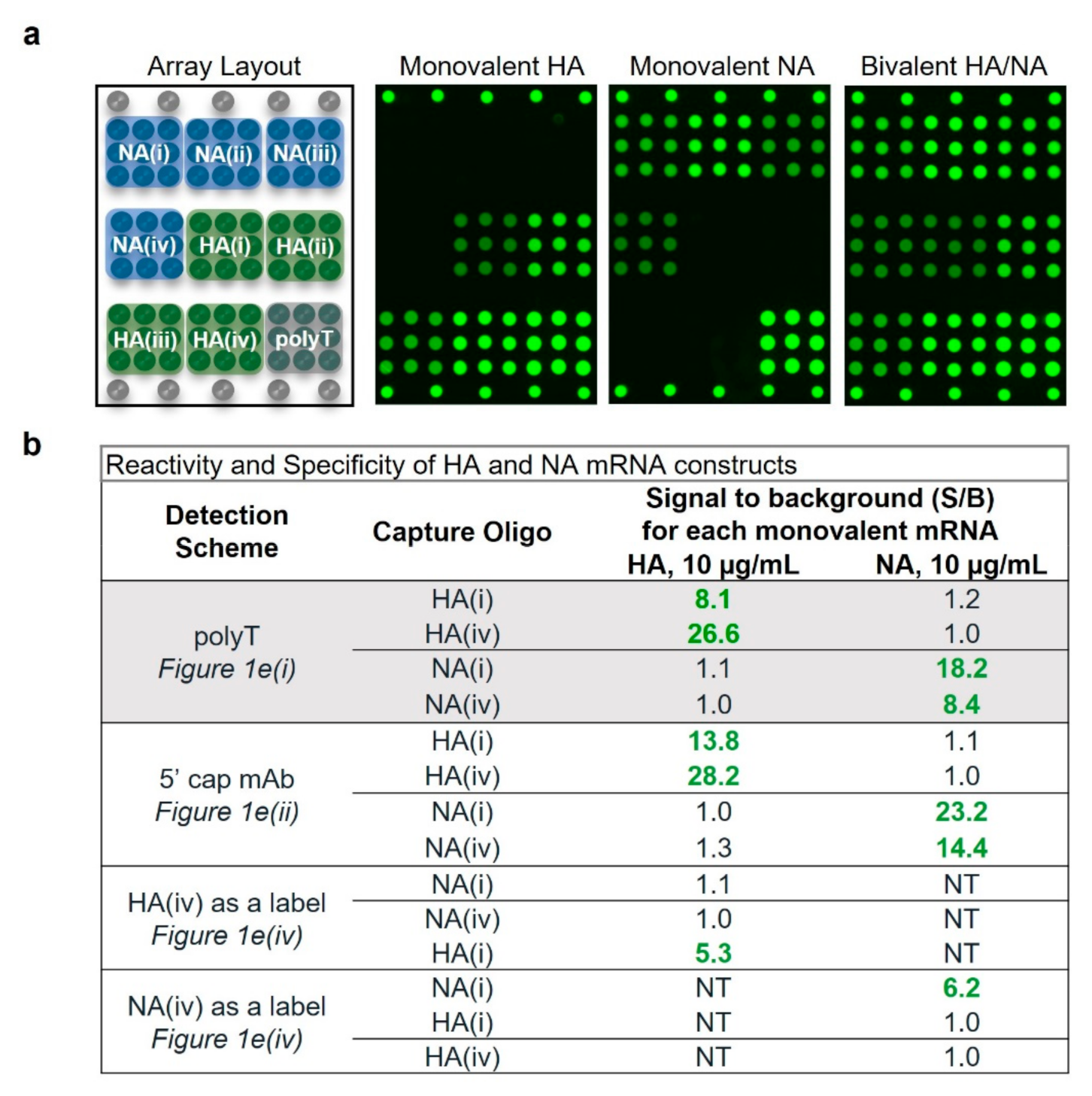
2.1. VaxArray mRNA Assay Design
2.2. Assay Shows High Specificity and Reactivity for HA and NA Targets
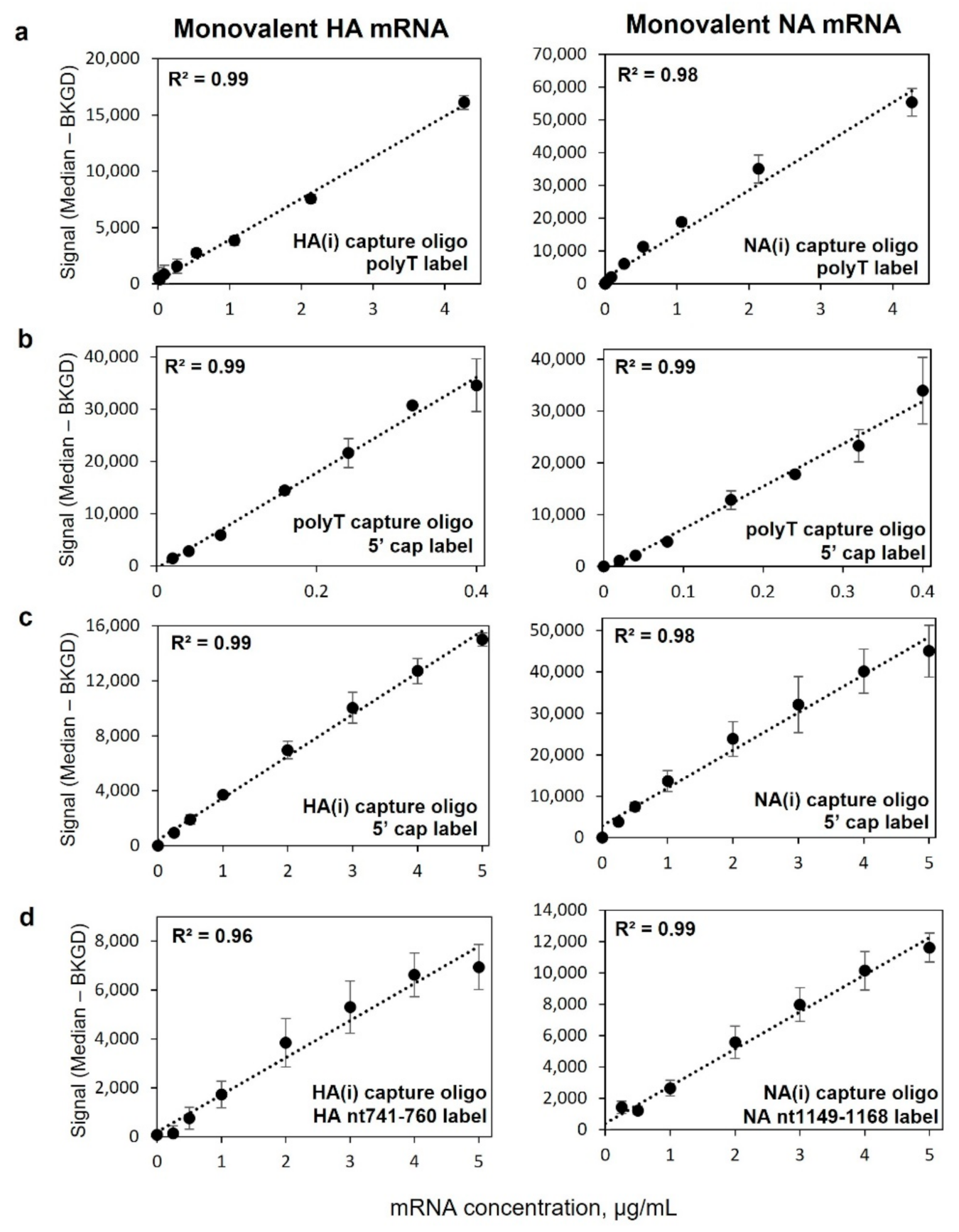
2.3. Monovalent mRNA Samples Show Excellent Linear Response, and Bivalent Response Matches That of Monovalent
2.4. Assay Has Vaccine-Relevant Sensitivity and At Least ~100× Working Range
2.5. Assay Demonstrates Average Accuracy of 104% Recovery, Average Precision of 9% RSD
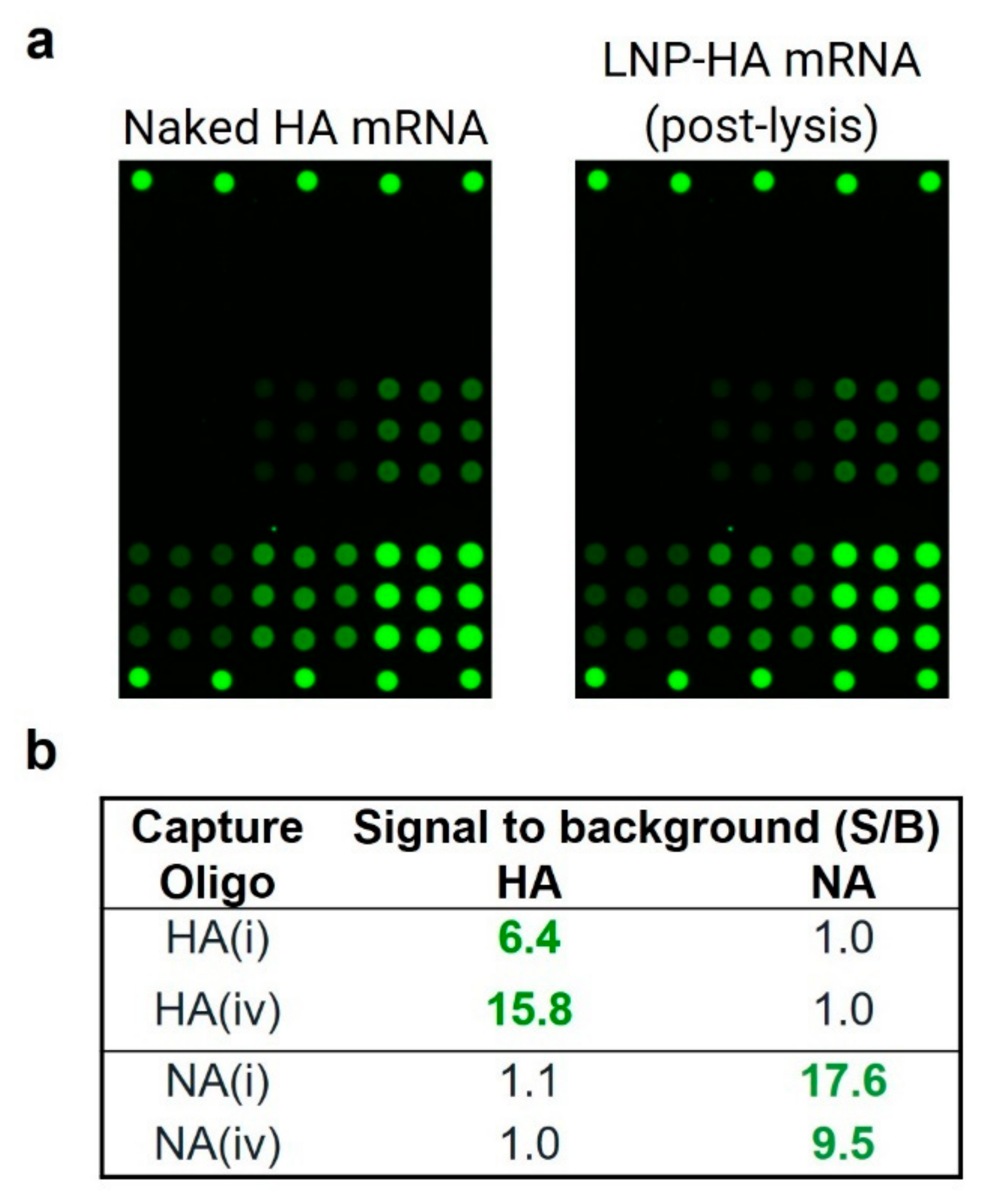
2.6. Assay Shows High Reactivity and Specificity for LNP-Encapsulated mRNA
2.7. LNP-Encapsulated mRNA Shows Equivalent Response in Mono- and Bivalent Samples, Equivalent Response to Naked mRNA, and Produces Good Accuracy and Precision
3. Discussion
4. Methods
4.1. mRNA Constructs
4.2. Oligonucleotide Sequence Design and Microarray Printing
4.3. Detection Labels
4.4. VaxArray mRNA Assay
4.5. Reactivity/Specificity
4.6. Analytical Sensitivity and Dynamic Range
4.7. Accuracy and Precision of Naked mRNA
4.8. LNP Encapsulation of mRNA
4.9. Response Curves, Accuracy and Precision of Encapsulated mRNA
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ball, P. The lightning-fast quest for COVID vaccines—And what it means for other diseases. Nature 2020, 589, 3. [Google Scholar] [CrossRef] [PubMed]
- Graham, B. Rapid COVID-19 vaccine development. Science 2020, 368, 2. [Google Scholar] [CrossRef] [PubMed]
- Developing COVID-19 Vaccines. Available online: https://www.cdc.gov/coronavirus/2019-ncov/vaccines/distributing/steps-ensure-safety.html (accessed on 29 August 2022).
- Kashte, S.; Gulbake, A.; El-Amin Iii, S.F.; Gupta, A. COVID-19 vaccines: Rapid development, implications, challenges and future prospects. Hum. Cell. 2021, 34, 711–733. [Google Scholar] [CrossRef] [PubMed]
- Mayo Clinic. COVID-19 and Related Vaccine Development and Research. Available online: https://www.mayoclinic.org/coronavirus-covid-19/history-disease-outbreaks-vaccine-timeline/covid-19 (accessed on 29 August 2022).
- Jackson, N.A.C.; Kester, K.E.; Casimiro, D.; Gurunathan, S.; DeRosa, F. The promise of mRNA vaccines: A biotech and industrial perspective. NPJ Vaccines 2020, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Pardi, N. mRNA Innovates the Vaccine Field. Vaccines 2021, 9, 486. [Google Scholar] [CrossRef]
- Bok, K.; Sitar, S.; Graham, B.S.; Mascola, J.R. Accelerated COVID-19 vaccine development: Milestones, lessons, and prospects. Immunity 2021, 54, 1636–1651. [Google Scholar] [CrossRef]
- Yu, A.M.; Tu, M.J. Deliver the promise: RNAs as a new class of molecular entities for therapy and vaccination. Pharmacol. Ther. 2022, 230, 107967. [Google Scholar] [CrossRef]
- Webb, C.; Ip, S.; Bathula, N.V.; Popova, P.; Soriano, S.K.; Ly, H.H.; Eryilmaz, B.; Nguyen Huu, V.A.; Broadhead, R.; Rabel, M.; et al. Current Status and Future Perspectives on MRNA Drug Manufacturing. Mol. Pharm. 2022, 19, 1047–1058. [Google Scholar] [CrossRef]
- Barbier, A.J.; Jiang, A.Y.; Zhang, P.; Wooster, R.; Anderson, D.G. The clinical progress of mRNA vaccines and immunotherapies. Nat. Biotechnol. 2022, 40, 840–854. [Google Scholar] [CrossRef]
- How Have Covid-19 Vaccines Been Made Quickly and Safely? Available online: https://wellcome.org/news/quick-safe-covid-vaccine-development (accessed on 30 August 2022).
- FDA. Emergency Use Authorization for Coronavirus Disease 2019 (COVID-19) EUA Information. Available online: https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization (accessed on 18 August 2022).
- U.S. Food & Drug Administration. Vaccines Licensed for Use in the United States. Available online: https://www.fda.gov/vaccines-blood-biologics/vaccines/vaccines-licensed-use-united-states (accessed on 23 August 2022).
- Hogan, M.J.; Pardi, N. mRNA Vaccines in the COVID-19 Pandemic and Beyond. Ann. Rev. Med. 2022, 73, 17–39. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Clinical Trials. Available online: www.clinicaltrials.gov (accessed on 18 August 2022).
- Ladak, R.J.; He, A.J.; Huang, Y.H.; Ding, Y. The Current Landscape of mRNA Vaccines Against Viruses and Cancer-A Mini Review. Front. Immunol. 2022, 13, 885371. [Google Scholar] [CrossRef]
- Rosa, S.S.; Prazeres, D.M.F.; Azevedo, A.M.; Marques, M.P.C. mRNA vaccines manufacturing: Challenges and bottlenecks. Vaccine 2021, 39, 2190–2200. [Google Scholar] [CrossRef]
- Whitley, J.; Zwolinski, C.; Denis, C.; Maughan, M.; Hayles, L.; Clarke, D.; Snare, M.; Liao, H.; Chiou, S.; Marmura, T.; et al. Development of mRNA manufacturing for vaccines and therapeutics: mRNA platform requirements and development of a scalable production process to support early phase clinical trials. Transl. Res. 2022, 242, 38–55. [Google Scholar] [CrossRef]
- Sandbrink, J.B.; Shattock, R.J. RNA Vaccines: A Suitable Platform for Tackling Emerging Pandemics? Front. Immunol. 2020, 11, 608460. [Google Scholar] [CrossRef]
- FDA Approves First COVID-19 Vaccine. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-covid-19-vaccine (accessed on 8 August 2022).
- Lurie, N.; Saville, M.; Hatchett, R.; Halton, J. Developing Covid-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 2020, 382, 1969–1973. [Google Scholar] [CrossRef]
- Krammer, F. SARS-CoV-2 vaccines in development. Nature 2020, 586, 516–527. [Google Scholar] [CrossRef]
- Coronavirus (COVID-19) Update: FDA Authorizes Moderna, Pfizer-BioNTech Bivalent COVID-19 Vaccines for Use as a Booster Dose. Available online: https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-authorizes-moderna-pfizer-biontech-bivalent-covid-19-vaccines-use (accessed on 9 September 2022).
- Moderna Receives FDA Authorization for Emergency Use of Omicron-Targeting Bivalent COVID-19 Booster Vaccine for Adults 18 Years and Older. Available online: https://investors.modernatx.com/news/news-details/2022/Moderna-Receives-FDA-Authorization-for-Emergency-Use-of-Omicron-Targeting-Bivalent-COVID-19-Booster-Vaccine-for-Adults-18-Years-and-Older/default.aspx (accessed on 9 September 2022).
- Pfizer and BioNTech Granted FDA Emergency Use Authorization of Omicron BA.4/BA.5-Adapted Bivalent COVID-19 Vaccine Booster for Ages 12 Years and Older|Pfizer. Available online: https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-granted-fda-emergency-use-authorization (accessed on 9 September 2022).
- Abbasi, J. Moderna’s mRNA Vaccine for Seasonal Flu Enters Clinical Trials. JAMA 2021, 326, 1365. [Google Scholar] [CrossRef]
- A Study of mRNA-1010 Seasonal Influenza Vaccine in Healthy Adults. Moderna. Available online: https://clinicaltrials.gov/ct2/show/NCT04956575 (accessed on 22 August 2022).
- Abbasi, J. Pfizer Launches Phase 1 mRNA Flu Vaccine Trial. JAMA 2021, 326, 1784. [Google Scholar] [CrossRef]
- Pfizer. A Study to Evaluate the Safety, Tolerability, and Immunogenicity of a Modified RNA Vaccine Against Influenza. Available online: https://clinicaltrials.gov/ct2/show/NCT05052697 (accessed on 22 August 2022).
- CureVac AG. A Study to Evaluate the Safety, Reactogenicity and Immunogenicity of Vaccine CVSQIV in Healthy Adults. Available online: https://clinicaltrials.gov/ct2/show/NCT05252338 (accessed on 22 August 2022).
- Moderna. A Safety, Reactogenicity, and Immunogenicity Study of mRNA- 1073 (COVID-19/Influenza) Vaccine in Adults 18 to 75 Years Old. Available online: https://clinicaltrials.gov/ct2/show/NCT05375838 (accessed on 22 August 2022).
- Analytical Procedures for mRNA Vaccine Quality. Available online: https://www.uspnf.com/notices/analytical-procedures-mrna-vaccines-20220210 (accessed on 10 February 2022).
- Baldwin, J.; Piplani, S.; Sakala, I.G.; Honda-Okubo, Y.; Li, L.; Petrovsky, N. Rapid development of analytical methods for evaluating pandemic vaccines: A COVID-19 perspective. Bioanalysis 2021, 13, 1805–1826. [Google Scholar] [CrossRef]
- Sanyal, G.; Särnefält, A.; Kumar, A. Considerations for bioanalytical characterization and batch release of COVID-19 vaccines. NPJ Vaccines 2021, 6, 53. [Google Scholar] [CrossRef]
- Freyn, A.W.; da Silva, J.R.; Rosado, V.C.; Bliss, C.M.; Pine, M.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; de Souza Ferreira, L.C.; Weissman, D.; et al. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020, 28, 1569–1584. [Google Scholar] [CrossRef]
- Dawson, E.D.; Taylor, A.W.; Johnson, J.E.; Hu, T.; McCormick, C.; Thomas, K.N.; Gao, R.Y.; Wahid, R.; Mahmood, K.; Rowlen, K.L. VaxArray immunoassay for the multiplexed quantification of poliovirus D-antigen. J. Immunol. Methods 2022, 504, 113259. [Google Scholar] [CrossRef]
- Gillis, J.H.; Thomas, K.N.; Manoharan, S.; Panchakshari, M.; Taylor, A.W.; Miller, D.F.; Byrne-Nash, R.T.; Riley, C.; Rowlen, K.L.; Dawson, E. Multiplexed VaxArray immunoassay for rapid antigen quantification in measles and rubella vaccine manufacturing. Vaccine X. 2021, 9, 100113. [Google Scholar] [CrossRef]
- Kuck, L.R.; Byrne-Nash, R.; Gillis, J.; Bueter, K.; Couzens, L.K.; Eichelberger, M.C.; Rowlen, K.L. VaxArray for hemagglutinin and neuraminidase potency testing of influenza vaccines. Vaccine 2018, 36, 2937–2945. [Google Scholar] [CrossRef]
- Toth, E.; Dawson, E.D.; Taylor, A.W.; Stoughton, R.S.; Blair, R.H.; Johnson, J.E., Jr.; Slinskey, A.; Fessler, R.; Smith, C.B.; Talbot, S.; et al. FluChip-8G Insight: HA and NA subtyping of potentially pandemic influenza A viruses in a single assay. Influenza Other Respir. Viruses 2020, 14, 55–60. [Google Scholar] [CrossRef] [Green Version]
- Blair, R.H.; Dawson, E.D.; Taylor, A.W.; Johnson, J.E., Jr.; Slinskey, A.H.; O’Neil, K.; Smolak, A.W.; Toth, E.; Liikanen, K.; Stoughton, R.S.; et al. Clinical validation of the FluChip-8G Influenza A+B Assay for influenza type and subtype identification. J. Clin. Virol. 2019, 118, 20–27. [Google Scholar] [CrossRef]
- Taylor, A.W.; Dawson, E.D.; Blair, R.H.; Johnson, J.E., Jr.; Slinskey, A.H.; Smolak, A.W.; Toth, E.; Liikanen, K.; Stoughton, R.S.; Smith, C.; et al. Analytical evaluation of the microarray-based FluChip-8G Influenza A+B Assay. J. Virol. Methods 2019, 273, 113686. [Google Scholar] [CrossRef]
- Byrne-Nash, R.T.; Gillis, J.H.; Miller, D.F.; Bueter, K.M.; Kuck, L.R.; Rowlen, K.L. A neuraminidase potency assay for quantitative assessment of neuraminidase in influenza vaccines. NPJ Vaccines 2019, 4, 3. [Google Scholar] [CrossRef]




| Capture Oligo | LLOQ (µg/mL) | ULOQ (µg/mL) | Linear Dynamic Range (ULOQ/LLOQ) |
|---|---|---|---|
| HA(i) | 0.20 | 18 | 90 |
| HA(iv) | 0.08 | 17 | 213 |
| NA(i) | 0.08 | 14 | 175 |
| NA(iv) | 0.20 | 18 | 90 |
| Capture Oligo | Accuracy (% Expected) | Precision (% RSD) | ||||||
|---|---|---|---|---|---|---|---|---|
| Operator 1 | Operator 2 | Operator 3 | By Capture | Operator 1 | Operator 2 | Operator 3 | By Capture | |
| HA(i) | 102% | 101% | 105% | 102 ± 2% | 12% | 6% | 2% | 8 ± 5% |
| HA(iv) | 107% | 105% | 97% | 103 ± 5% | 10% | 7% | 3% | 6 ± 4% |
| NA(i) | 110% | 106% | 100% | 105 ± 5% | 13% | 12% | 7% | 11 ± 3% |
| NA(iv) | 105% | 107% | 108% | 107 ± 2% | 10% | 8% | 8% | 9 ± 1% |
| by Operator | 106 ± 3% | 105% ± 3% | 103% ± 5% | 11 ± 2% | 8% ± 3% | 5% ± 3% | ||
| Over all capture oligos, all operators (n = 96) | 104 ± 2% | 9 ± 2% | ||||||
| Capture Oligo | Accuracy (% Recovery) | Precision (% of RSD of Measured Conc.) |
|---|---|---|
| HA(i) | 104% | 10% |
| HA(iv) | 98% | 7% |
| NA(i) | 125% | 7% |
| NA(iv) | 102% | 9% |
| Overall capture oligos (n = 32) | 108% ± 12% | 8% ± 1% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, R.Y.; Riley, C.M.; Toth, E.; Blair, R.H.; Gerold, M.N.; McCormick, C.; Taylor, A.W.; Hu, T.; Rowlen, K.L.; Dawson, E.D. Rapid Identity and Quantity CQA Test for Multivalent mRNA Drug Product Formulations. Vaccines 2022, 10, 1704. https://doi.org/10.3390/vaccines10101704
Gao RY, Riley CM, Toth E, Blair RH, Gerold MN, McCormick C, Taylor AW, Hu T, Rowlen KL, Dawson ED. Rapid Identity and Quantity CQA Test for Multivalent mRNA Drug Product Formulations. Vaccines. 2022; 10(10):1704. https://doi.org/10.3390/vaccines10101704
Chicago/Turabian StyleGao, Rachel Y., Christine M. Riley, Evan Toth, Rebecca H. Blair, Megan N. Gerold, Caitlin McCormick, Amber W. Taylor, Tianjing Hu, Kathy L. Rowlen, and Erica D. Dawson. 2022. "Rapid Identity and Quantity CQA Test for Multivalent mRNA Drug Product Formulations" Vaccines 10, no. 10: 1704. https://doi.org/10.3390/vaccines10101704
APA StyleGao, R. Y., Riley, C. M., Toth, E., Blair, R. H., Gerold, M. N., McCormick, C., Taylor, A. W., Hu, T., Rowlen, K. L., & Dawson, E. D. (2022). Rapid Identity and Quantity CQA Test for Multivalent mRNA Drug Product Formulations. Vaccines, 10(10), 1704. https://doi.org/10.3390/vaccines10101704






