Identification of B-Cell Epitopes of HspA from Helicobacter pylori and Detection of Epitope Antibody Profiles in Naturally Infected Persons
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Construction, Expression, and Purification of H. pylori Recombinant HspA (rHspA) and GST Fusion Peptides
2.3. Synthesis of Peptides and Keyhole Limpet Hemocyanin (KLH)-Conjugated Peptides
2.4. Immunization of Mice and Sample Collection
2.5. Acquisition and Testing of Human Serum
2.6. ELISAs for Peptides and rHspA
2.7. SDS-PAGE and Western Blot Analysis
2.8. Lymphocyte Proliferation Responses
2.9. Subsection Statistical Analysis
3. Results
3.1. Subsection Preliminary Screening of Antigen Immunodominant Peptide Fragment in HspA
3.2. Fine Localization of the Epitope in HP1
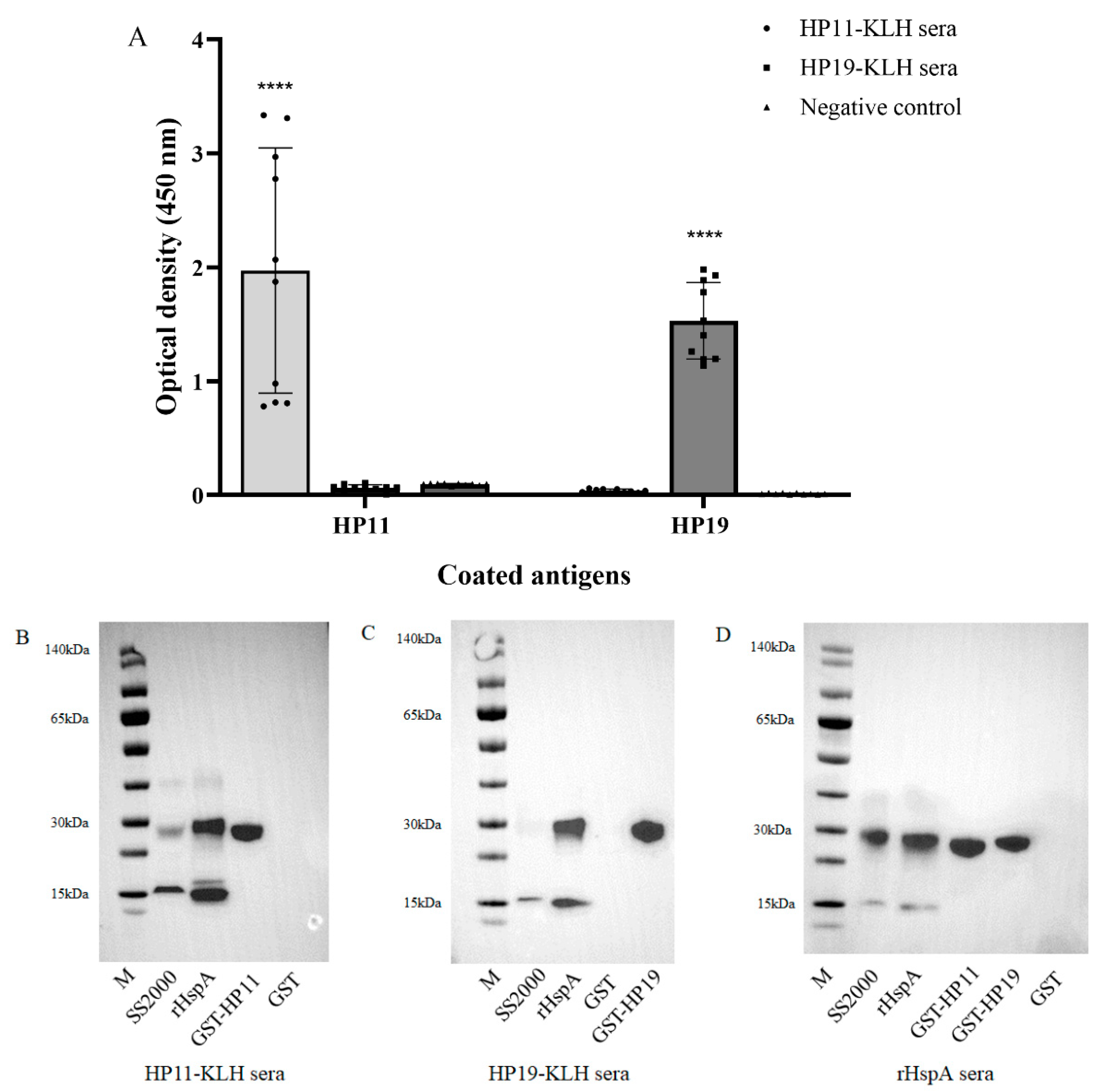
3.3. Detection of Immunogenicity and Immunoreactivity of Epitope HP11 and HP19
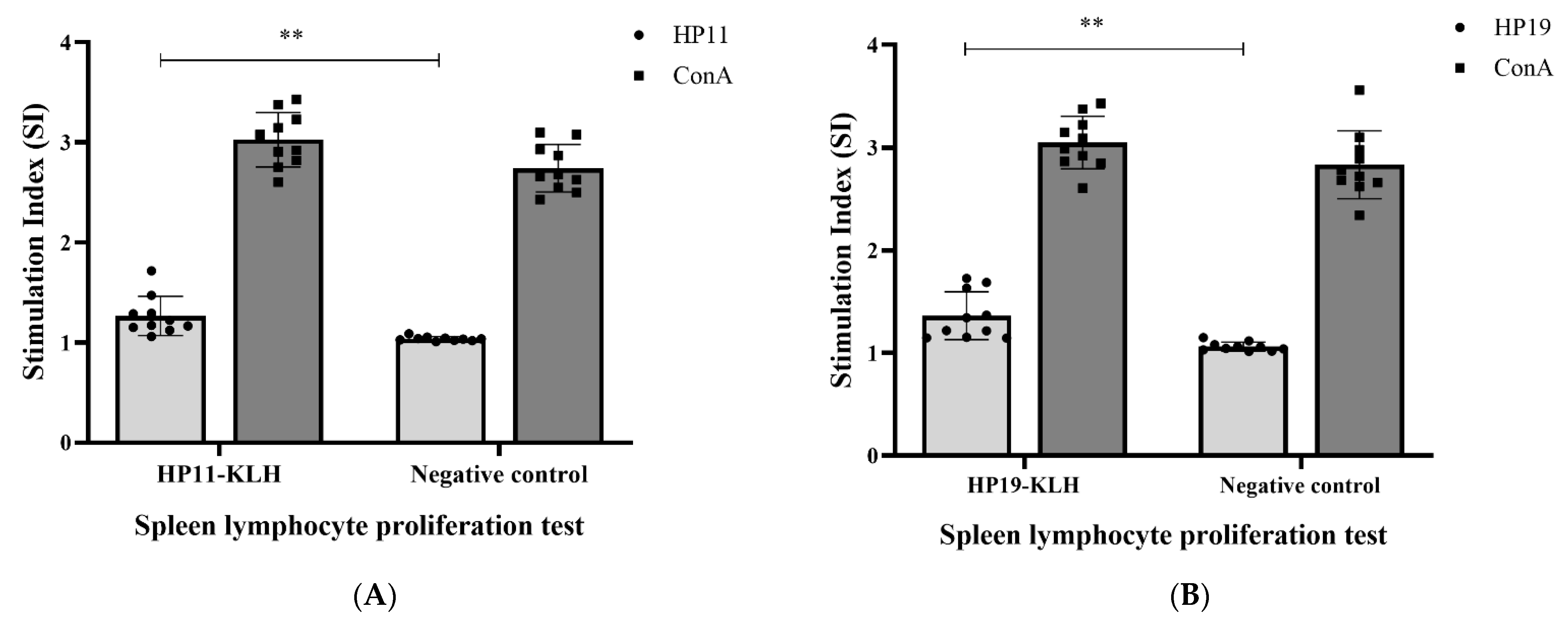
3.4. Lymphocyte Proliferation Responses Test
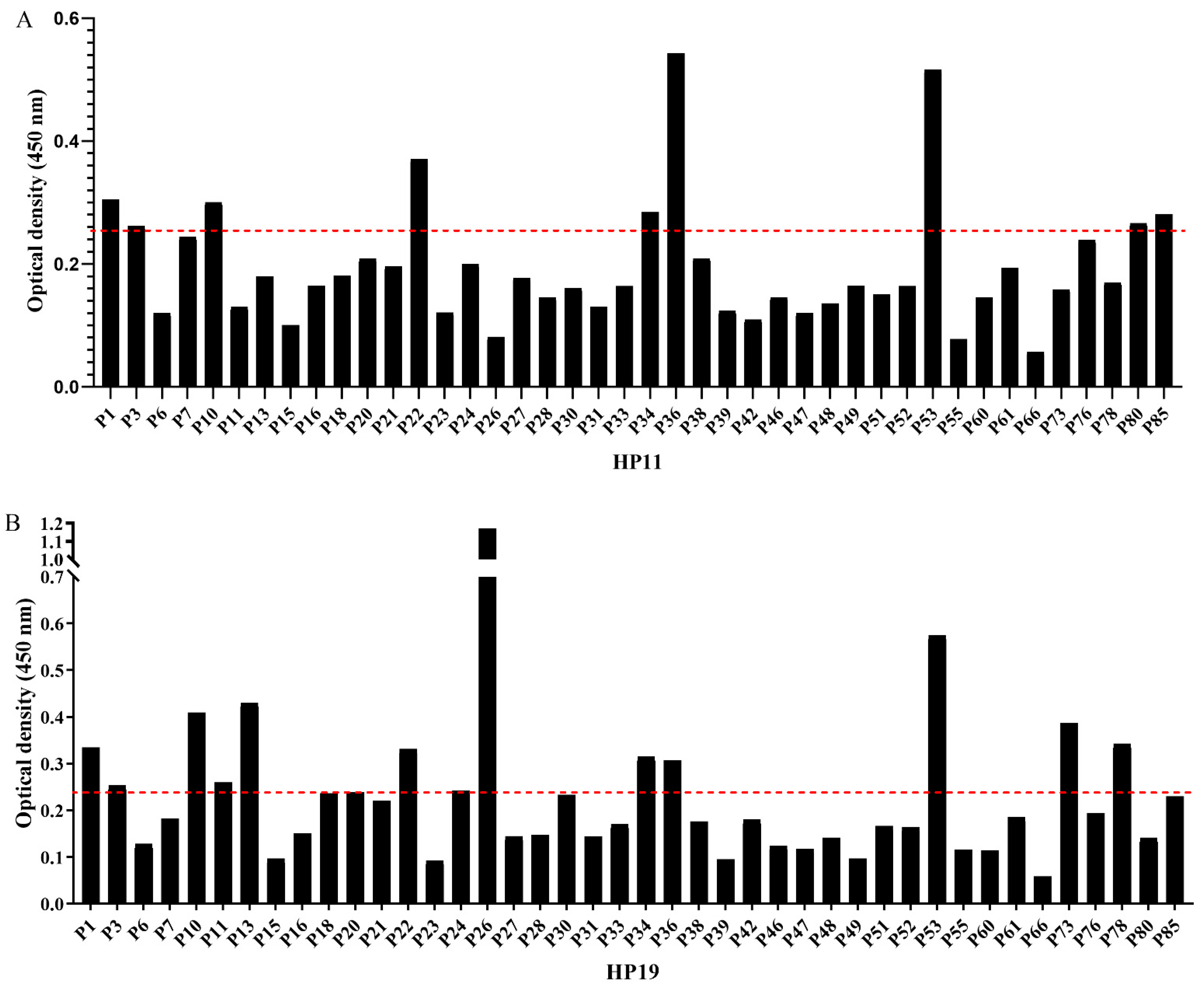
3.5. Amino Acid Homology and Human Serum Antibody Profile Analysis of Identified Epitopes
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Blaser, M.J.; Pérez-Pérez, G.I.; Lindenbaum, J.; Schneidman, D.; Van Deventer, G.; MarinSorensen, M.; Weinstein, W.M. Association of infection due to Helicobacter pylori with specific upper gastrointestinal pathology. Rev. Infect. Dis. 1991, 13 (Suppl. S8), S704–S708. [Google Scholar] [CrossRef]
- Forman, D.; Newell, D.G.; Fullerton, F.; Yarnell, J.W.; Stacey, A.R.; Wald, N.; Sitas, F. Association between infection with Helicobacter pylori and risk of gastric cancer: Evidence from a prospective investigation. BMJ 1991, 302, 1302–1305. [Google Scholar] [CrossRef]
- Hussell, T.; Isaacson, P.G.; Crabtree, J.E.; Spencer, J. The response of cells from low-grade B-cell gastric lymphomas of mucosa-associated lymphoid tissue to Helicobacter pylori. Lancet 1993, 342, 571–574. [Google Scholar] [CrossRef]
- Cellini, L. Helicobacter pylori: A chameleon-like approach to life. World J. Gastroenterol. 2014, 20, 5575–5582. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhu, Y.; Lu, N.H. Recent progress in Helicobacter pylori treatment. Chin. Med. J. 2020, 133, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Thung, I.; Aramin, H.; Vavinskaya, V.; Gupta, S.; Park, J.Y.; Crowe, S.E.; Valasek, M.A. Review article, the global emergence of Helicobacter pylori antibiotic resistance. Aliment. Pharmacol. Ther. 2016, 43, 514–533. [Google Scholar] [CrossRef]
- Ferrero, R.L.; Thiberge, J.M.; Kansau, I.; Wuscher, N.; Huerre, M.; Labigne, A. The GroES homolog of Helicobacter pylori confers protective immunity against mucosal infection in mice. Proc. Natl. Acad. Sci. USA 1995, 92, 6499–6503. [Google Scholar] [CrossRef]
- Suerbaum, S.; Thiberge, J.M.; Kansau, I.; Ferrero, R.L.; Labigne, A. Helicobacter pylori hspA-hspB heat-shock gene cluster, nucleotide sequence.; expression.; putative function and immunogenicity. Mol. Microbiol. 1994, 14, 959–974. [Google Scholar] [CrossRef]
- Cun, S.; Li, H.; Ge, R.; Lin, M.C.; Sun, H. A histidine-rich and cysteinerich metal-binding domain at the C terminus of heat shock protein A from Helicobacter pylori: Implication for nickel homeostasis and bismuth susceptibility. J. Biol. Chem. 2008, 283, 15142–15151. [Google Scholar] [CrossRef]
- Schauer, K.; Muller, C.; Carrière, M.; Labigne, A.; Cavazza, C.; De Reuse, H. The Helicobacter pylori GroES Cochaperonin HspA Functions as a Specialized Nickel Chaperone and Sequestration Protein through Its Unique C-Terminal Extension. J. Bacteriol. 2010, 192, 1231–1237. [Google Scholar] [CrossRef]
- Ha, N.C.; Oh, S.T.; Sung, J.Y.; Cha, K.A.; Lee, M.H.; Oh, B.H. Supramolecular assembly and acid resistance of Helicobacter pylori urease. Nat. Struct. Biol. 2001, 8, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Maier, R.J.; Fu, C.; Gilbert, J.; Moshiri, F.; Olson, J.; Plaut, A.G. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol. Lett. 1996, 141, 71–76. [Google Scholar] [CrossRef]
- Eamranond, P.P.; Torres, J.; Muñoz, O.; Pérez-Pérez, G.I. Age-specific immune response to HspA in Helicobacter pylori-positive persons in Mexico. Clin. Diagn. Lab. Immunol. 2004, 11, 983–985. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rowinska-Zyrek, M.; Witkowska, D.; Bielinska, S.; Kamysz, W.; Kozlowski, H. The -Cys-Cys- motif in Helicobacter pylori’s Hpn and HspA proteins is an essential anchoring site for metal ions. Dalton Trans. 2011, 40, 5604–5610. [Google Scholar] [CrossRef] [PubMed]
- Iankov, I.D.; Kurokawa, C.; Viker, K.; Robinson, S.I.; Ammayappan, A.; Panagioti, E.; Federspiel, M.J.; Galanis, E. Live Attenuated Measles Virus Vaccine Expressing Helicobacter pylori Heat Shock Protein A. Mol. Ther. Oncolytics 2020, 19, 136–148. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, J.; Yang, F.; Wu, W.; Sun, H.; Xie, Q.; Si, W.; Zou, Q.; Yang, Z. Immunization with Heat Shock Protein A and γ-Glutamyl Transpeptidase Induces Reduction on the Helicobacter pylori Colonization in Mice. PLoS ONE 2015, 10, e0130391. [Google Scholar] [CrossRef]
- Kansau, I.; Guillain, F.; Thiberge, J.M.; Labigne, A. Nickel binding and immunological properties of the C-terminal domain of the Helicobacter pylori GroES homologue (HspA). Mol. Microbiol. 1996, 22, 1013–1023. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.K.; Thompson, S.A.; Pérez-Pérez, G.I.; Kansau, I.; Van der Ende, A.; Labigne, A.; Sung, J.J.; Chung, S.C.; Blaser, M.J. Helicobacter pylori heat shock protein A, serologic responses and genetic diversity. Clin. Diagn. Lab. Immunol. 1999, 6, 377–382. [Google Scholar] [CrossRef]
- Pérez-Pérez, G.I.; Thiberge, J.M.; Labigne, A.; Blaser, M.J. Relationship of immune response to heat-shock protein and characteristics of Helicobacter pylori-infected patients. J. Infect. Dis. 1996, 174, 1046–1050. [Google Scholar] [CrossRef][Green Version]
- Widmer, M.; de Korwin, J.D.; Aucher, P.; Thiberge, J.M.; Suerbaum, S.; Labigne, A.; Fauchère, J.L. Performance of native and recombinant antigens for diagnosis of Helicobacter pylori infection. Eur. J. Clin. Microbiol. Infect. Dis. 1999, 18, 823–826. [Google Scholar] [CrossRef]
- Vanet, A.; Labigne, A. Evidence for specific secretion rather than autolysis in the release of some Helicobacter pylori proteins. Infect. Immun. 1998, 66, 1023–1027. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Zhao, W.; Zou, Z.; Kong, L.; Yang, L. Oral multivalent epitope vaccine; based on UreB, HpaA, CAT, and LTB, for prevention and treatment of Helicobacter pylori infection in C57BL/6 mice. Helicobacter 2021, 26, e12807. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Hong, D.; Wang, S.; Zhang, F.; Tang, F.; Wu, T.; Chu, Y.; Liu, H.; He, M.; Yang, H.; et al. Therapeutic Protection Against, H. pylori Infection in Mongolian Gerbils by Oral Immunization With a Tetravalent Epitope-Based Vaccine With Polysaccharide Adjuvant. Front. Immunol. 2019, 10, 1185. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Sang, S.; Guan, Q.; Tao, H.; Wang, Y.; Liu, C. Identification of B-Cell Epitopes of HspA from Helicobacter pylori and Detection of Epitope Antibody Profiles in Naturally Infected Persons. Vaccines 2022, 10, 65. https://doi.org/10.3390/vaccines10010065
Zhang X, Sang S, Guan Q, Tao H, Wang Y, Liu C. Identification of B-Cell Epitopes of HspA from Helicobacter pylori and Detection of Epitope Antibody Profiles in Naturally Infected Persons. Vaccines. 2022; 10(1):65. https://doi.org/10.3390/vaccines10010065
Chicago/Turabian StyleZhang, Xin, Shuli Sang, Qing Guan, Haoxia Tao, Yanchun Wang, and Chunjie Liu. 2022. "Identification of B-Cell Epitopes of HspA from Helicobacter pylori and Detection of Epitope Antibody Profiles in Naturally Infected Persons" Vaccines 10, no. 1: 65. https://doi.org/10.3390/vaccines10010065
APA StyleZhang, X., Sang, S., Guan, Q., Tao, H., Wang, Y., & Liu, C. (2022). Identification of B-Cell Epitopes of HspA from Helicobacter pylori and Detection of Epitope Antibody Profiles in Naturally Infected Persons. Vaccines, 10(1), 65. https://doi.org/10.3390/vaccines10010065





