Oxidative Stress-Generating Antimicrobials, a Novel Strategy to Overcome Antibacterial Resistance
Abstract
1. Introduction
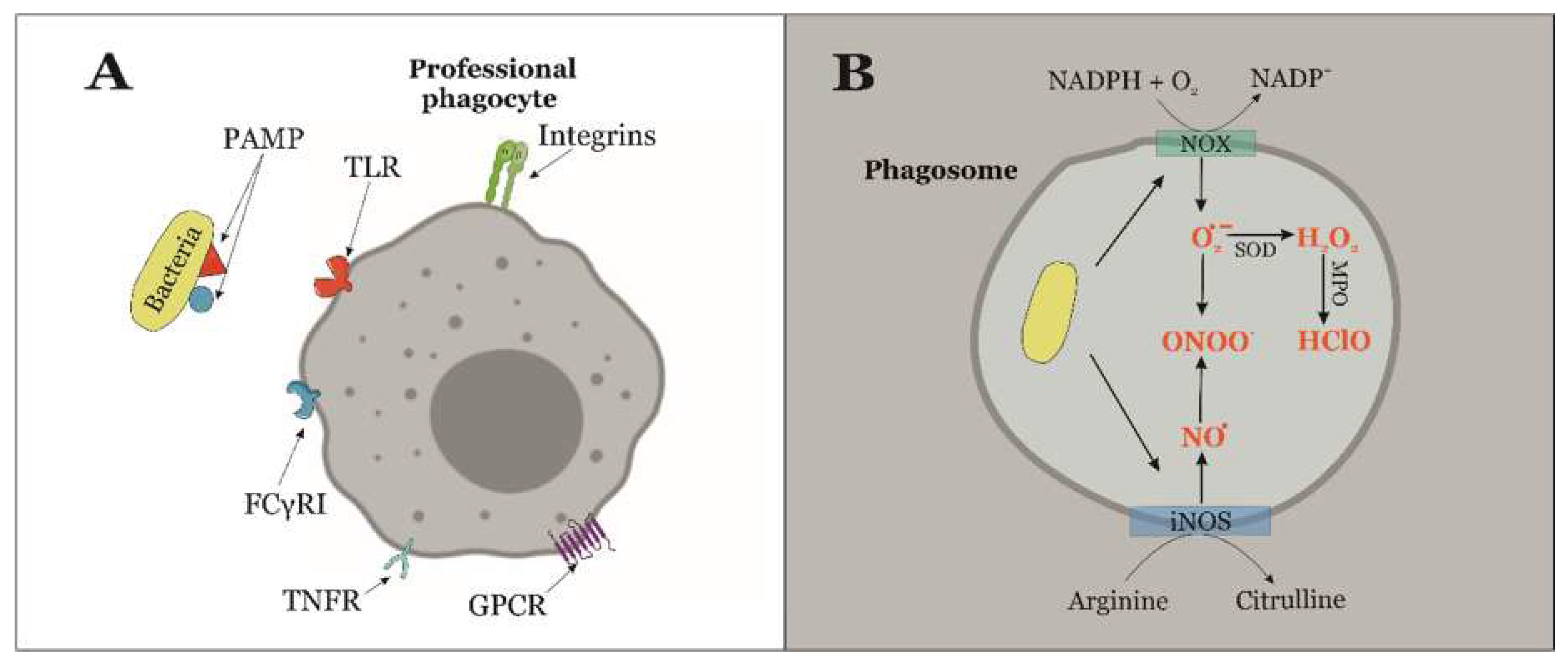
2. Oxidative Stress Response in Intracellular Bacterial Pathogens
3. Molecular Pathways of RONS-Biosynthesis in Immune Cells
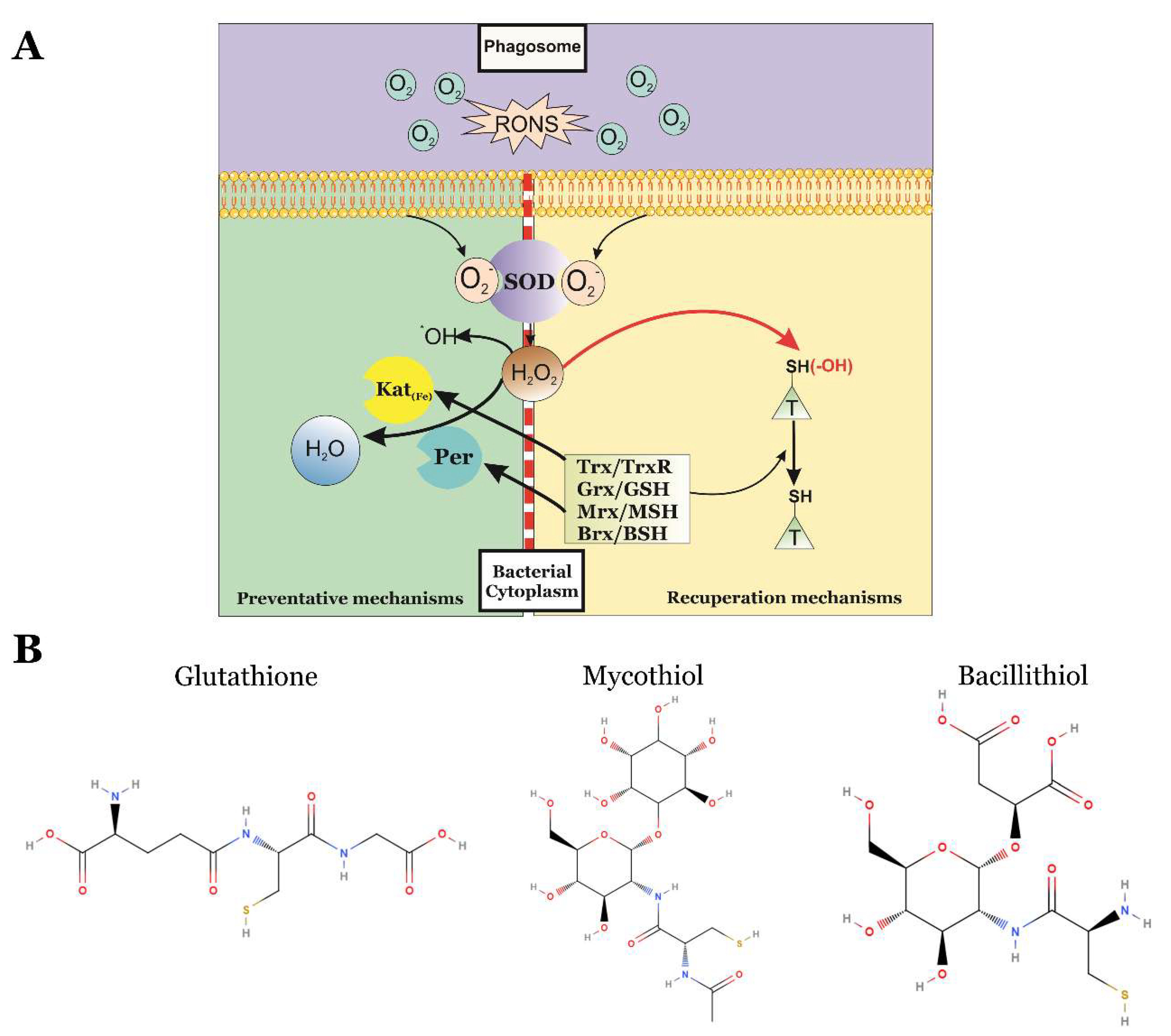
4. Antioxidant Systems of Intracellular Bacterial Pathogens
4.1. Enzymatic Preventative Mechanisms of Oxidative Stress
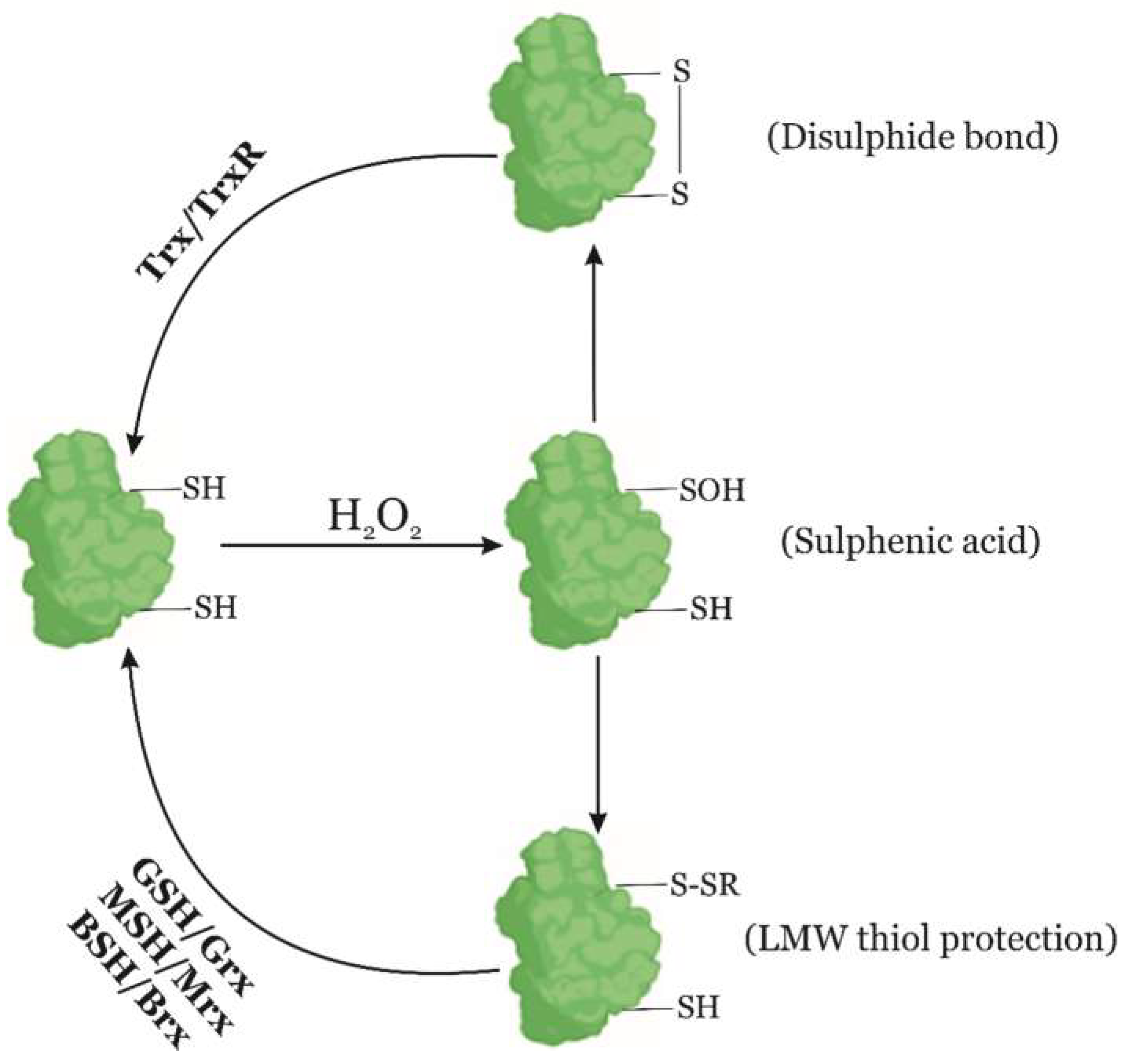
4.2. Enzymatic Reparation Mechanisms of Protein Oxidation
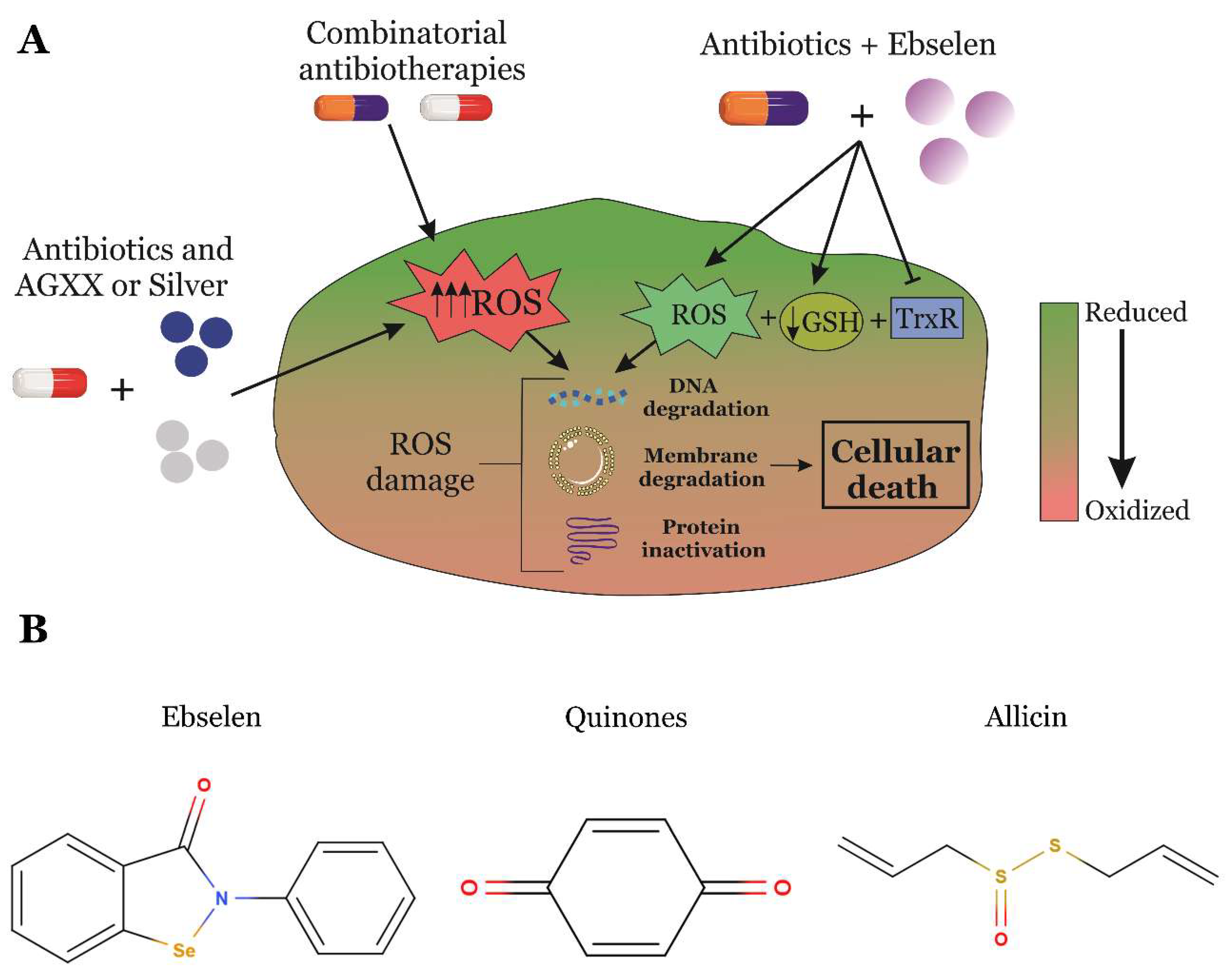
5. RONS-Producing Anti-infectives Are an Attractive Strategy to Overcome Antimicrobial Resistance
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sies, H. Oxidative Stress: Introductory Remarks; Academic Press: London, UK, 1985; pp. 1–8. [Google Scholar]
- Jones, D.P. Redefining Oxidative Stress. Antioxid. Redox Signal. 2006, 8, 1865–1879. [Google Scholar] [CrossRef] [PubMed]
- Sies, H.; Berndt, C.; Jones, D.P. Oxidative Stress. Annu. Rev. Biochem. 2017, 86, 15–48. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Sun, Q.; Chen, S. Oxidative stress: A major pathogenesis and potential therapeutic target of antioxidative agents in Parkinson’s disease and Alzheimer’s disease. Prog. Neurobiol. 2016, 147, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bonnefont-Rousselot, D. The Role of Antioxidant Micronutrients in the Prevention of Diabetic Complications. Treat. Endocrinol. 2004, 3, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Cantin, A.M. Potential for antioxidant therapy of cystic fibrosis. Curr. Opin. Pulm. Med. 2004, 10, 531–536. [Google Scholar] [CrossRef]
- Gill, J.G.; Piskounova, E.; Morrison, S.J. Cancer, Oxidative Stress, and Metastasis. Cold Spring Harb. Symp. Quant. Biol. 2016, 81, 163–175. [Google Scholar] [CrossRef]
- Slauch, J.M. How does the oxidative burst of macrophages kill bacteria? still an open question. Mol. Microbiol. 2011, 80, 580–583. [Google Scholar] [CrossRef]
- Winterbourn, C.C.; Kettle, A.J. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid. Redox Signal. 2013, 18, 642–660. [Google Scholar] [CrossRef]
- Winterbourn, C.C.; Kettle, A.J.; Hampton, M.B. Reactive oxygen species and neutrophil function. Annu. Rev. Biochem. 2016, 85, 765–792. [Google Scholar] [CrossRef]
- Fang, F.C. Antimicrobial reactive oxygen and nitrogen species: Concepts and controversies. Nat. Rev. Microbiol. 2004, 2, 820–832. [Google Scholar] [CrossRef]
- Mitchell, G.; Chen, C.; Portnoy, D.A. Strategies used by bacteria to grow in macrophages. Microbiol. Spectr. 2016, 4, 701–725. [Google Scholar] [CrossRef] [PubMed]
- Fauci, A.S.; Morens, D.M. The perpetual challenge of infectious diseases. N. Engl. J. Med. 2012, 366, 454–461. [Google Scholar] [CrossRef]
- Casadevall, A.; Fang, F.C. The intracellular pathogen concept. Mol. Microbiol. 2019, 113, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Flannagan, R.S.; Cosío, G.; Grinstein, S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 2009, 7, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.T.; Green, E.R.; Mecsas, J. Neutrophils to the ROScue: Mechanisms of NADPH oxidase activation and bacterial resistance. Front. Cell. Infect. Microbiol. 2017, 7, 373. [Google Scholar] [CrossRef]
- Rugemalira, E.; Roine, I.; Kuligowski, J.; Sánchez-Illana, Á.; Piñeiro-Ramos, J.D.; Andersson, S.; Peltola, H.; Cruzeiro, M.L.; Pelkonen, T.; Vento, M. Protein oxidation biomarkers and myeloperoxidase activation in cerebrospinal fluid in childhood bacterial meningitis. Antioxidants 2019, 8, 441. [Google Scholar] [CrossRef]
- Heyworth, P.G.; Cross, A.R.; Curnutte, J.T. Chronic granulomatous disease. Curr. Opin. Immunol. 2003, 15, 578–584. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Toxicity of iron and hydrogen peroxide: The Fenton reaction. Toxicol. Lett. 1995, 82–83, 969–974. [Google Scholar] [CrossRef]
- Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef]
- Rodrigues, M.R.; Rodriguez, D.; Russo, M.; Campa, A. Macrophage activation includes high intracellular myeloperoxidase activity. Biochem. Biophys. Res. Commun. 2002, 292, 869–873. [Google Scholar] [CrossRef]
- Rosen, H.; Klebanoff, S.J.; Wang, Y.; Brot, N.; Heinecke, J.W.; Fu, X. Methionine oxidation contributes to bacterial killing by the myeloperoxidase system of neutrophils. Proc. Natl. Acad. Sci. USA 2009, 106, 18686–18691. [Google Scholar] [CrossRef] [PubMed]
- Dukan, S.; Touati, D. Hypochlorous acid stress in Escherichia coli: Resistance, DNA damage, and comparison with hydrogen peroxide stress. J. Bacteriol. 1996, 178, 6145–6150. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C.; Hampton, M.B. Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 2008, 45, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Boronat, S.; Domènech, A.; Paulo, E.; Calvo, I.A.; García-Santamarina, S.; García, P.; Encinar del Dedo, J.; Barcons, A.; Serrano, E.; Carmona, M.; et al. Thiol-based H2O2 signalling in microbial systems. Redox Biol. 2014, 2, 395–399. [Google Scholar] [CrossRef]
- Pedre, B.; Young, D.; Charlier, D.; Mourenza, Á.; Rosado, L.A.; Marcos-Pascual, L.; Wahni, K.; Martens, E.G.; de la Rubia, A.; Belousov, V.V.; et al. Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2 sensing. Proc. Natl. Acad. Sci. USA 2018, 115, E11623–E11632. [Google Scholar] [CrossRef]
- Teramoto, H.; Inui, M.; Yukawa, H. OxyR acts as a transcriptional repressor of hydrogen peroxide-inducible antioxidant genes in Corynebacterium glutamicum R. FEBS J. 2013, 280, 3298–3312. [Google Scholar] [CrossRef]
- Aslund, F.; Zheng, M.; Beckwith, J.; Stortz, G. Regulation of the OxyR transcription factor by hydrogen peroxide and the cellular thiol-disulfide status. Proc. Natl. Acad. Sci. USA 1999, 96, 6161–6165. [Google Scholar] [CrossRef]
- Milse, J.; Petri, K.; Rückert, C.; Kalinowski, J. Transcriptional response of Corynebacterium glutamicum ATCC 13032 to hydrogen peroxide stress and characterization of the OxyR regulon. J. Biotechnol. 2014, 190, 40–54. [Google Scholar] [CrossRef]
- Imlay, J.A. Transcription factors that defend bacteria against reactive oxygen species. Annu. Rev. Microbiol. 2015, 69, 93–108. [Google Scholar] [CrossRef]
- Beggs, G.A.; Brennan, R.G.; Arshad, M. MarR family proteins are important regulators of clinically relevant antibiotic resistance. Protein Sci. 2019, 29, 647–653. [Google Scholar] [CrossRef]
- Antelmann, H.; Helmann, J.D. Thiol-based redox switches and gene regulation. Antioxid. Redox Signal. 2011, 14, 1049–1063. [Google Scholar] [CrossRef] [PubMed]
- Ngoc, Q.; Busche, T.; Loi, V.V.; Kalinowski, J.; Antelmann, H. The redox-sensing MarR-type repressor HypS controls hypochlorite and antimicrobial resistance in Mycobacterium smegmatis. Free Radic. Biol. Med. 2020, 147, 252–261. [Google Scholar]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Bollela, V.R.; Namburete, E.I.; Feliciano, C.S.; Macheque, D.; Harrison, L.H.; Caminero, J.A. Detection of katG and inhA mutations to guide isoniazid and ethionamide use for drug-resistant tuberculosis. Int. J. Tuberc. Lung Dis. 2016, 20, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Shastri, M.D.; Shukla, S.D.; Chong, W.C.; Dua, K.; Peterson, G.M.; Patel, R.P.; Hansbro, P.M.; Eri, R.; O’Toole, R.F. Role of oxidative stress in the pathology and management of human tuberculosis. Oxid. Med. Cell. Longev. 2018, 7695364. [Google Scholar] [CrossRef]
- Ma, Z.; Strickland, K.T.; Cherne, M.D.; Sehanobish, E.; Rohde, K.H.; Self, W.T.; Davidson, V.L. The Rv2633c protein of Mycobacterium tuberculosis is a non-heme di-iron catalase with a possible role in defenses against oxidative stress. J. Biol. Chem. 2018, 293, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Bidaud, P.; He, L.; Sanguinetti, M.; Laugier, C.; Petry, S. Rhodococcus equi’s extreme resistance to hydrogen peroxide is mainly conferred by one of its four catalase genes. PLoS ONE 2012, 7, e42396. [Google Scholar] [CrossRef]
- Staerck, C.; Gastebois, A.; Vandeputte, P.; Calenda, A.; Larcher, G.; Gillmann, L.; Papon, N.; Bouchara, J.P.; Fleury, M.J.J. Microbial antioxidant defense enzymes. Microb. Pathog. 2017, 110, 56–65. [Google Scholar] [CrossRef]
- Hall, A.; Parsonage, D.; Poole, L.B.; Karplus, P.A. Structural evidence that peroxiredoxin catalytic power is based on transition-state stabilization. J. Mol. Biol. 2010, 402, 194–209. [Google Scholar] [CrossRef]
- Fomenko, D.E.; Koc, A.; Agisheva, N.; Jacobsen, M.; Kaya, A.; Malinouski, M.; Rutherford, J.C.; Siu, K.L.; Jin, D.Y.; Winge, D.R.; et al. Thiol peroxidases mediate specific genome-wide regulation of gene expression in response to hydrogen peroxide. Proc. Natl. Acad. Sci. USA 2011, 108, 2729–2734. [Google Scholar] [CrossRef]
- Hanschmann, E.M.; Godoy, J.R.; Berndt, C.; Hudemann, C.; Lillig, C.H. Thioredoxins, glutaredoxins, and peroxiredoxins—molecular mechanisms and health significance: From cofactors to antioxidants to redox signaling. Antioxid. Redox Signal. 2013, 19, 1539–1605. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.; Nelson, K.; Poole, L.B.; Karplus, P.A. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid. Redox Signal. 2011, 15, 795–815. [Google Scholar] [CrossRef] [PubMed]
- Tosatto, S.C.E.; Bosello, V.; Fogolari, F.; Mauri, P.; Roveri, A.; Toppo, S.; Flohé, L.; Ursini, F.; Maiorino, M. The catalytic site of glutathione peroxidases. Antioxid. Redox Signal. 2008, 10, 1515–1525. [Google Scholar] [CrossRef]
- Koh, C.S.; Didierjean, C.; Navrot, N.; Panjikar, S.; Mulliert, G.; Rouhier, N.; Jacquot, J.P.; Aubry, A.; Shawkataly, O.; Corbier, C. Crystal structures of a poplar thioredoxin peroxidase that exhibits the structure of glutathione peroxidases: Insights into redox-driven conformational changes. J. Mol. Biol. 2007, 370, 512–529. [Google Scholar] [CrossRef] [PubMed]
- Brenot, A.; King, K.Y.; Janowiak, B.; Griffith, O.; Caparon, M.G. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect. Immun. 2004, 72, 408–413. [Google Scholar] [CrossRef]
- Gebicka, L.; Didik, J. Catalytic scavenging of peroxynitrite by catalase. J. Inorg. Biochem. 2009, 103, 1375–1379. [Google Scholar] [CrossRef]
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef]
- Horst, C.; Berndt, C.; Holmgren, A. Glutaredoxin systems. Biochim. Biophys. Acta Gen. Subj. 2008, 1780, 1304–1317. [Google Scholar]
- Laurent, T.C.; Moore, E.C.; Reichard, P. Enzymatic synthesis of deoxyribonicleotides. J. Biol. Chem. 1964, 239, 3436–3444. [Google Scholar]
- Comtois, S.L.; Gidley, M.D.; Kelly, D.J. Role of the thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology 2003, 149, 121–129. [Google Scholar] [CrossRef]
- Li, K.; Härtig, E.; Klug, G. Thioredoxin 2 is involved in oxidative stress defence and redox-dependent expression of photosynthesis genes in Rhodobacter capsulatus. Microbiology 2003, 149, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, C.; Assemat, K.; Clément-Métral, J.D.; Klug, G. Thioredoxin is essential for Rhodobacter sphaeroides growth by aerobic and anaerobic respiration. Microbiology 1997, 143, 83–91. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scharf, C.; Riethdorf, S.; Ernst, H.; Engelmann, S.; Völker, U.; Hecker, M. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 1998, 180, 1869–1877. [Google Scholar] [CrossRef] [PubMed]
- Small, J.L.; Park, S.W.; Kana, B.D.; Ioerger, T.R.; Sacchettini, J.C.; Ehrt, S. Perturbation of cytochrome c maturation reveals adaptability of the respiratory chain in Mycobacterium tuberculosis. MBio 2013, 4, e00475-13. [Google Scholar] [CrossRef]
- Mourenza, Á.; Collado, C.; Bravo-Santano, N.; Gil, J.A.; Mateos, L.M.; Letek, M. The extracellular thioredoxin Etrx3 is required for host cell infection in Rhodococcus equi. Vet. Res. 2020, 51, 1–7. [Google Scholar] [CrossRef]
- Saleh, M.; Bartual, S.G.; Abdullah, M.R.; Jensch, I.; Asmat, T.M.; Petruschka, L.; Pribyl, T.; Gellert, M.; Lillig, C.H.; Antelmann, H.; et al. Molecular architecture of Streptococcus pneumoniae surface thioredoxin-fold lipoproteins crucial for extracellular oxidative stress resistance and maintenance of virulence. EMBO Mol. Med. 2013, 5, 1852–1870. [Google Scholar] [CrossRef]
- Ribes, S.; Abdullah, M.R.; Saleh, M.; Hanisch, U.K.; Nau, R.; Hammerschmidt, S. Thioredoxins and methionine sulfoxide reductases in the pathophysiology of pneumococcal meningitis. J. Infect. Dis. 2016, 214, 953–961. [Google Scholar] [CrossRef][Green Version]
- Achard, M.E.S.; Hamilton, A.J.; Dankowski, T.; Heras, B.; Schembri, M.S.; Edwards, J.L.; Jennings, M.P.; McEwan, A.G. A periplasmic thioredoxin-like protein plays a role in defense against oxidative stress in Neisseria gonorrhoeae. Infect. Immun. 2009, 77, 4934–4939. [Google Scholar] [CrossRef]
- Tanboon, W.; Chuchue, T.; Vattanaviboon, P.; Mongkolsuk, S. Inactivation of thioredoxin-like gene alters oxidative stress resistance and reduces cytochrome c oxidase activity in Agrobacterium tumefaciens. FEMS Microbiol. Lett. 2009, 295, 110–116. [Google Scholar] [CrossRef]
- Abicht, H.K.; Schärer, M.A.; Quade, N.; Ledermann, R.; Mohorko, E.; Capitani, G.; Hennecke, H.; Glockshuber, R. How periplasmic thioredoxin TlpA reduces bacterial copper chaperone ScoI and cytochrome oxidase subunit II (CoxB) prior to metallation. J. Biol. Chem. 2014, 289, 32431–32444. [Google Scholar] [CrossRef]
- Ordóñez, E.; Van Belle, K.; Roos, G.; De Galan, S.; Letek, M.; Gil, J.A.; Wyns, L.; Mateos, L.M.; Messens, J. Arsenate reductase, mycothiol, and mycoredoxin concert thiol/disulfide exchange. J. Biol. Chem. 2009, 284, 15107–15116. [Google Scholar] [CrossRef] [PubMed]
- Helmann, J.D. Bacillithiol, a new player in bacterial redox homeostasis. Antioxid. Redox Signal. 2011, 15, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Loi, V.V.; Rossius, M.; Antelmann, H. Redox regulation by reversible protein S-thiolation in bacteria. Front. Microbiol. 2015, 6, 187. [Google Scholar] [CrossRef] [PubMed]
- Moldogazieva, N.T.; Mokhosoev, I.M.; Feldman, N.B.; Lutsenko, S.V. ROS and RNS signalling: Adaptive redox switches through oxidative/nitrosative protein modifications. Free Radic. Res. 2018, 52, 507–543. [Google Scholar] [CrossRef] [PubMed]
- Chi, B.K.; Gronau, K.; Mäder, U.; Hessling, B.; Becher, D.; Antelmann, H. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol. Cell. Proteom. 2011, 10, 35. [Google Scholar] [CrossRef] [PubMed]
- Linzner, N.; Van Loi, V.; Fritsch, V.N.; Tung, Q.N.; Stenzel, S.; Wirtz, M.; Hell, R.; Hamilton, C.J.; Tedin, K.; Fulde, M.; et al. Staphylococcus aureus uses the bacilliredoxin (BrxAB)/bacillithiol disulfide reductase (YpdA) redox pathway to defend against oxidative stress under infections. Front. Microbiol. 2019, 10, 1355. [Google Scholar] [CrossRef]
- Hugo, M.; Van Laer, K.; Reyes, A.M.; Vertommen, D.; Messens, J.; Radi, R.; Trujillo, M. Mycothiol/mycoredoxin 1-dependent reduction of the peroxiredoxin AhpE from Mycobacterium tuberculosis. J. Biol. Chem. 2014, 289, 5228–5239. [Google Scholar] [CrossRef]
- Van Laer, K.; Buts, L.; Foloppe, N.; Vertommen, D.; Van Belle, K.; Wahni, K.; Roos, G.; Nilsson, L.; Mateos, L.M.; Rawat, M.; et al. Mycoredoxin-1 is one of the missing links in the oxidative stress defence mechanism of Mycobacteria. Mol. Microbiol. 2012, 86, 787–804. [Google Scholar] [CrossRef]
- Mourenza, Á.; Bravo-santano, N.; Gil, J.A.; Mateos, L.M.; Letek, M. Mycoredoxins are required for redox homeostasis and intracellular survival in the actinobacterial pathogen Rhodococcus equi. Antioxidants 2019, 8, 558. [Google Scholar] [CrossRef]
- Imber, M.; Pietrzyk-Brzezinska, A.J.; Antelmann, H. Redox regulation by reversible protein S-thiolation in Gram-positive bacteria. Redox Biol. 2019, 20, 130–145. [Google Scholar] [CrossRef]
- Hopkins, B.L.; Neumann, C.A. Redoxins as gatekeepers of the transcriptional oxidative stress response. Redox Biol. 2019, 21, 101104. [Google Scholar] [CrossRef]
- Attarian, R.; Hu, G.; Sánchez-León, E.; Caza, M.; Croll, D.; Do, E.; Bach, H.; Missall, T.; Lodge, J.; Jung, W.H.; et al. The monothiol glutaredoxin Grx4 regulates iron homeostasis and virulence in Cryptococcus neoformans. MBio 2018, 9, e02377-18. [Google Scholar] [CrossRef]
- Meyer, Y.; Buchanan, B.B.; Vignols, F.; Reichheld, J.-P. Thioredoxins and glutaredoxins: Unifying elements in redox biology. Annu. Rev. Genet. 2009, 43, 335–367. [Google Scholar] [CrossRef]
- Negri, A.; Javidnia, P.; Mu, R.; Zhang, X.; Vendome, J.; Gold, B.; Roberts, J.; Barman, D.; Ioerger, T.; Sacchettini, J.C.; et al. Identification of a mycothiol-dependent nitroreductase from Mycobacterium tuberculosis. ACS Infect. Dis. 2018, 4, 771–787. [Google Scholar] [CrossRef]
- Gaballa, A.; Chi, B.K.; Roberts, A.A.; Becher, D.; Hamilton, C.J.; Antelmann, H.; Helmann, J.D. Redox regulation in Bacillus subtilis: The bacilliredoxins brxa(YPHP) and brxb(YQIW) function in de-bacillithiolation of s-bacillithiolated ohrr and mete. Antioxid. Redox Signal. 2014, 21, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Chandrangsu, P.; Loi, V.V.; Antelmann, H.; Helmann, J.D. The role of bacillithiol in Gram-positive firmicutes. Antioxid. Redox Signal. 2018, 28, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Antelmann, H.; Hamilton, C.J. Bacterial mechanisms of reversible protein S-thiolation: Structural and mechanistic insights into mycoredoxins. Mol. Microbiol. 2012, 86, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, U.R.; Balasubramanian, S.; Oelschlaeger, T.A.; Grkovic, T.; Pham, N.B.; Quinn, R.J.; Hentschel, U. Potential of marine natural products against drug-resistant fungal, viral, and parasitic infections. Lancet Infect. Dis. 2017, 17, e30–e41. [Google Scholar] [CrossRef]
- Andersson, D.I.; Hughes, D. Antibiotic resistance and its cost: Is it possible to reverse resistance? Nat. Rev. Microbiol. 2010, 8, 260–271. [Google Scholar] [CrossRef]
- Hughes, D.; Andersson, D.I. Evolutionary trajectories to antibiotic resistance. Annu. Rev. Microbiol. 2017, 71, 579–596. [Google Scholar] [CrossRef]
- Abed, N.; Couvreur, P. Nanocarriers for antibiotics: A promising solution to treat intracellular bacterial infections. Int. J. Antimicrob. Agents 2014, 43, 485–496. [Google Scholar] [CrossRef]
- Ren, X.; Zou, L.; Holmgren, A. Targeting bacterial antioxidant systems for antibiotics development. Curr. Med. Chem. 2019, 26. [Google Scholar] [CrossRef]
- Zhao, R.; Masayasu, H.; Holmgren, A. Ebselen: A substrate for human thioredoxin reductase strongly stimulating its hydroperoxide reductase activity and a superfast thioredoxin oxidant. Proc. Natl. Acad. Sci. USA 2002, 99, 8579–8584. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Zhou, J.; Wang, P.; Li, T.; Zhao, Y.; Ren, X.; Lu, J.; Wang, J.; Holmgren, A.; Zou, L. Topical therapeutic efficacy of ebselen against multidrug-resistant Staphylococcus aureus LT-1. Front. Microbiol. 2020, 10, 3016. [Google Scholar] [CrossRef] [PubMed]
- Schewe, T. Molecular Actions of Ebselen-an Antiinflammatory Antioxidant. Gen. Pharmacol. 1995, 26, 1153–1169. [Google Scholar] [CrossRef]
- Carcinogenesis, I.F.O.R.I.; Nakamura, Y.; Feng, Q.; Kumagai, T.; Torikai, K.; Ohigashi, H.; Osawa, T.; Noguchi, N.; Niki, E.; Uchida, K. Ebselen, a Glutathione Peroxidase Mimetic Seleno-organic Compound, as a Multifunctional Antioxidant. J. Biol. Chem. 2002, 277, 2687–2694. [Google Scholar]
- Lu, J.; Vlamis-gardikas, A.; Kandasamy, K.; Zhao, R.; Gustafsson, T.N.; Engstrand, L.; Hoffner, S.; Engman, L.; Holmgren, A. Inhibition of bacterial thioredoxin reductase: An antibiotic mechanism targeting bacteria lacking glutathione. FASEB J. 2013, 27, 1394–1403. [Google Scholar] [CrossRef]
- Thangamani, S.; Younis, W.; Seleem, M.N. Repurposing Clinical Molecule Ebselen to Combat Drug Resistant Pathogens. PLoS ONE 2015, 10, e0133877. [Google Scholar] [CrossRef]
- Zou, L.; Wang, J.; Gao, Y.; Ren, X.; Rottenberg, M.E.; Lu, J.; Holmgren, A. Synergistic antibacterial activity of silver with antibiotics correlating with the upregulation of the ROS production. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Zou, L.; Lu, J.; Wang, J.; Ren, X.; Zhang, L.; Gao, Y.; Rottenberg, M.E.; Holmgren, A. Synergistic antibacterial effect of silver and ebselen against multidrug-resistant Gram-negative bacterial infections. EMBO Mol. Med. 2017, 9, 1165–1178. [Google Scholar] [CrossRef]
- Loi, V.V.; Busche, T.; Preuß, T.; Kalinowski, J.; Bernhardt, J.; Antelmann, H. The AGXX® antimicrobial coating causes a thiol-specific oxidative stress response and protein s-bacillithiolation in Staphylococcus aureus. Front. Microbiol. 2018, 9, 3037. [Google Scholar] [CrossRef] [PubMed]
- Kadiyala, U.; Kotov, N.A.; VanEpps, J.S. Antibacterial metal oxide nanoparticles: Challenges in interpreting the literature. Curr. Pharm. Des. 2019, 24, 896–903. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Lee, D.G. Gold nanoparticles induce a reactive oxygen species-independent apoptotic pathway in Escherichia coli. Colloids Surf. B Biointerfaces 2018, 167, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Wu, Y.; Wang, H.; Han, H. Synergistic antibacterial effects of curcumin modified silver nanoparticles through ROS-mediated pathways. Mater. Sci. Eng. C 2019, 99, 255–263. [Google Scholar] [CrossRef]
- Teow, S.Y.; Wong, M.M.T.; Yap, H.Y.; Peh, S.C.; Shameli, K. Bactericidal properties of plants-derived metal and metal oxide nanoparticles (NPs). Molecules 2018, 23, 1366. [Google Scholar] [CrossRef]
- Lipovsky, A.; Gedanken, A.; Nitzan, Y.; Lubart, R. Enhanced inactivation of bacteria by metal-oxide nanoparticles combined with visible light irradiation. Lasers Surg. Med. 2011, 43, 236–240. [Google Scholar] [CrossRef]
- Wang, D.; Zhao, L.; Ma, H.; Zhang, H.; Guo, L.H. Quantitative Analysis of Reactive Oxygen Species Photogenerated on Metal Oxide Nanoparticles and Their Bacteria Toxicity: The Role of Superoxide Radicals. Environ. Sci. Technol. 2017, 51, 10137–10145. [Google Scholar] [CrossRef]
- Pati, R.; Mehta, R.K.; Mohanty, S.; Goswami, C.; Sonawane, A. Topical application of zinc oxide nanoparticles reduces bacterial skin infection in mice and exhibits antibacterial activity by inducing oxidative stress response and cell membrane disintegration in macrophages. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 1195–1208. [Google Scholar] [CrossRef]
- Kashef, N.; Hospital, M.G.; Huang, Y.; Hospital, M.G.; Hamblin, M.R.; Hospital, M.G. Advances in antimicrobial photodynamic inactivation at the nanoscale. Nanophotonics 2017, 6, 853–879. [Google Scholar] [CrossRef]
- Ozkan, E.; Allan, E.; Parkin, I.P. White-Light-Activated Antibacterial Surfaces Generated by Synergy between Zinc Oxide Nanoparticles and Crystal Violet. ACS Omega 2018, 3, 3190–3199. [Google Scholar] [CrossRef]
- Aboelzahab, A.; Azad, A.; Dolan, S.; Goel, V. Mitigation of Staphylococcus aureus-Mediated Surgical Site Infections with IR Photoactivated TiO 2 coatings on Ti Implants. Adv. Healthc. Mater. 2012, 1, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, D.J.; Kohanski, M.A.; Hayete, B.; Collins, J.J. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol. Syst. Biol. 2007, 3, 91. [Google Scholar] [CrossRef] [PubMed]
- Keren, I.; Wu, Y.; Inocencio, J.; Mulcahy, L.R.; Lewis, K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 2013, 339, 1213–1216. [Google Scholar] [CrossRef]
- Bhaskar, A.; Chawla, M.; Mehta, M.; Parikh, P.; Chandra, P.; Bhave, D.; Kumar, D.; Carroll, K.S.; Singh, A. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during Infection. PLoS Pathog. 2014, 10, e1003902. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Belenky, P.A.; Yang, J.H.; Cody MacDonald, I.; Martell, J.D.; Takahashi, N.; Chan, C.T.Y.; Lobritz, M.A.; Braff, D.; Schwarz, E.G.; et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. USA 2014, 111, E2100–E2109. [Google Scholar] [CrossRef]
- Dwyer, D.J.; Collins, J.J.; Walker, G.C. Unraveling the physiological complexities of antibiotic lethality. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 313–332. [Google Scholar] [CrossRef]
- Mourenza, Á.; Gil, J.A.; Mateos, L.M.; Letek, M. A novel screening strategy reveals ROS—Generating antimicrobials that act synergistically against the intracellular veterinary pathogen Rhodococcus equi. Antioxidants 2020, 9, 114. [Google Scholar] [CrossRef]
- Kim, S.Y.; Park, C.; Jang, H.J.; Kim, B.O.; Bae, H.W.; Chung, I.Y.; Kim, E.S.; Cho, Y.H. Antibacterial strategies inspired by the oxidative stress and response networks. J. Microbiol. 2019, 57, 203–212. [Google Scholar] [CrossRef]
- Liebeke, M.; Pöther, D.C.; Van Duy, N.; Albrecht, D.; Becher, D.; Hochgräfe, F.; Lalk, M.; Hecker, M.; Antelmann, H. Depletion of thiol-containing proteins in response to quinones in Bacillus subtilis. Mol. Microbiol. 2008, 69, 1513–1529. [Google Scholar] [CrossRef]
- Hillion, M.; Antelmann, H. Thiol-based redox switches in prokaryotes. Biol. Chem. 2015, 396, 415–444. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.; Cruz, J.; Simone, M.; Bernasconi, A.; Brunati, C.; Sosio, M.; Donadio, S.; Maffioli, S.I. Antibacterial paramagnetic quinones from Actinoallomurus. J. Nat. Prod. 2017, 80, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Xie, X.; Chen, L.; Yan, S.; Ye, X.; Anjum, K.; Huang, H.; Lian, X.; Zhang, Z. Bioactive polycyclic quinones from marine Streptomyces sp. 182SMLY. Mar. Drugs 2016, 14, 10. [Google Scholar] [CrossRef]
- Horn, J.; Stelzner, K.; Rudel, T.; Fraunholz, M. Inside job: Staphylococcus aureus host-pathogen interactions. Int. J. Med. Microbiol. 2018, 308, 607–624. [Google Scholar] [CrossRef] [PubMed]
- Borlinghaus, J.; Albrecht, F.; Gruhlke, M.C.H.; Nwachukwu, I.D.; Slusarenko, A.J. Allicin: Chemistry and biological properties. Molecules 2014, 19, 12591–12618. [Google Scholar] [CrossRef] [PubMed]
- Ankri, S.; Mirelman, D. Antimicrobial properties of allicin from garlic. Microbes Infect. 1999, 1, 125–129. [Google Scholar] [CrossRef]
- Loi, V.V.; Huyen, N.T.T.; Busche, T.; Tung, Q.N.; Gruhlke, M.C.H.; Kalinowski, J.; Bernhardt, J.; Slusarenko, A.J.; Antelmann, H. Staphylococcus aureus responds to allicin by global S-thioallylation—Role of the Brx/BSH/YpdA pathway and the disulfide reductase MerA to overcome allicin stress. Free Radic. Biol. Med. 2019, 139, 55–69. [Google Scholar] [CrossRef]
- Chi, B.K.; Huyen, N.T.T.; Loi, V.V.; Gruhlke, M.C.H.; Schaffer, M.; Mäder, U.; Maaß, S.; Becher, D.; Bernhardt, J.; Arbach, M.; et al. The disulfide stress response and protein s-thioallylation caused by allicin and diallyl polysulfanes in Bacillus subtilis as revealed by transcriptomics and proteomics. Antioxidants 2019, 8, 605. [Google Scholar] [CrossRef]
- Ajiboye, T.O.; Aliyu, M.; Isiaka, I.; Haliru, F.Z.; Ibitoye, O.B.; Uwazie, J.N.; Muritala, H.F.; Bello, S.A.; Yusuf, I.I.; Mohammed, A.O. Contribution of reactive oxygen species to (+)-catechin-mediated bacterial lethality. Chem. Biol. Interact. 2016, 258, 276–287. [Google Scholar] [CrossRef]
- Ibitoye, O.B.; Ajiboye, T.O. Ferulic acid potentiates the antibacterial activity of quinolone-based antibiotics against Acinetobacter baumannii. Microb. Pathog. 2019, 126, 393–398. [Google Scholar] [CrossRef]
- Tune, B.M. Nephrotoxicity of beta-lactam antibiotics: Mechanisms and strategies for prevention. Pediatr. Nephrol. 1997, 11, 768–772. [Google Scholar] [CrossRef] [PubMed]
- Pouzaud, F.; Bernard-Beaubois, K.; Thevenin, M.; Warnet, J.-M.; Hayem, G.; Rat, P. In vitro discrimination of fluoroquinolones toxicity on tendon cells: Involvement of oxidative stress. J. Pharmacol. Exp. Ther. 2004, 308, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Vicente-vicente, L.; Casanova, A.G. A systematic meta-analysis on the efficacy of pre-clinically tested nephroprotectants at preventing aminoglycoside nephrotoxicity. Toxicology 2017, 377, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Randjelovi, P.; Veljkovi, S.; Stojiljkovi, N.; Ili, I. Review article: Gentamicin nephrotoxicity in animals: Current knowledge and future perspectives. EXCLI J. 2017, 16, 388. [Google Scholar]
- Lowes, D.A.; Wallace, C.; Murphy, M.P.; Webster, N.R.; Galley, H.F. The mitochondria targeted antioxidant MitoQ protects against fluoroquinolone-induced oxidative stress and mitochondrial membrane damage in human Achilles tendon cells. Free Radic. Res. 2009, 43, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Sannasimuthu, A.; Sharma, D.; Paray, B.A.; Al-Sadoon, M.K.; Arockiaraj, J. Intracellular oxidative damage due to antibiotics on gut bacteria reduced by glutathione oxidoreductase-derived antioxidant molecule GM15. Arch. Microbiol. 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
| Antibiotic | Primary Mechanism of Action | Microorganism | Reference |
|---|---|---|---|
| Erythromycin | Protein synthesis inhibition | Rhodococcus equi | [109] |
| Rifampicin | RNA synthesis inhibition | Rhodococcus equi | [109] |
| Vancomycin | Cell wall synthesis inhibition | Mycobacterium tuberculosis Rhodococcus equi Staphylococcus aureus | [106] [109] [103] |
| Norfloxacin | DNA gyrase inhibition | Rhodococcus equi Staphylococcus aureus Escherichia coli | [109] [103] [104] |
| Clofazimine | DNA replication inhibition | Mycobacterium tuberculosis | [106] |
| Ethambutol | Cell wall synthesis inhibition | Mycobacterium tuberculosis | [106] |
| Isoniazid | Cell wall synthesis inhibition | Mycobacterium tuberculosis | [106] |
| Quinones | Different cellular targets | Enterococcus sp. Streptococcus sp. Staphylococcus sp. Moraxela catarrhalis | [113] [113] [113] [113] |
| Metal oxide nanoparticles | Undefined | Escherichia coli Staphylococcus aureus Staphylococcus epiderdimis Photobacterium phosphoreum | [91] [97] [97] [98] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mourenza, Á.; Gil, J.A.; Mateos, L.M.; Letek, M. Oxidative Stress-Generating Antimicrobials, a Novel Strategy to Overcome Antibacterial Resistance. Antioxidants 2020, 9, 361. https://doi.org/10.3390/antiox9050361
Mourenza Á, Gil JA, Mateos LM, Letek M. Oxidative Stress-Generating Antimicrobials, a Novel Strategy to Overcome Antibacterial Resistance. Antioxidants. 2020; 9(5):361. https://doi.org/10.3390/antiox9050361
Chicago/Turabian StyleMourenza, Álvaro, José A. Gil, Luís M. Mateos, and Michal Letek. 2020. "Oxidative Stress-Generating Antimicrobials, a Novel Strategy to Overcome Antibacterial Resistance" Antioxidants 9, no. 5: 361. https://doi.org/10.3390/antiox9050361
APA StyleMourenza, Á., Gil, J. A., Mateos, L. M., & Letek, M. (2020). Oxidative Stress-Generating Antimicrobials, a Novel Strategy to Overcome Antibacterial Resistance. Antioxidants, 9(5), 361. https://doi.org/10.3390/antiox9050361








