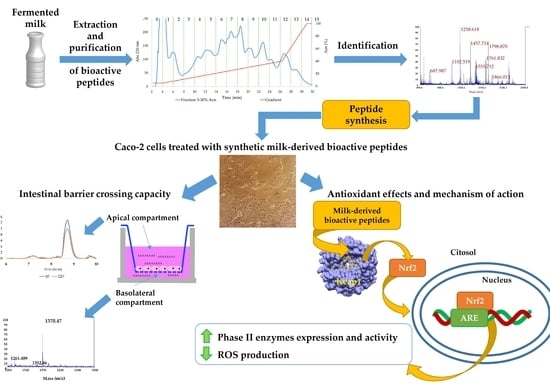
Identification of New Peptides from Fermented Milk Showing Antioxidant Properties: Mechanism of Action
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Fermented Milk
2.3. Aqueous Extract of Samples
2.4. Solid Phase Extraction of Bioactive Fractions from Aqueous Preparations
2.5. Purification of Peptide Fractions
2.6. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis
2.7. Database Search and Peptide Identification
2.8. Peptide Synthesis
2.9. ABTS and DPPH Scavenging
2.10. Caco-2 Cell Culture
2.11. Transepithelial Transport of Fractions or Peptides Through Caco-2 Cell Monolayers
RP-HPLC and MS Analyses
2.12. Cell Viability
2.13. ROS Production Estimation
2.14. Nrf2 Translocation to the Nucleus
2.15. Cell Lysates
2.15.1. Antioxidant Enzymes Detection
2.15.2. TrxR1 and GR Activities
2.16. Gene Expression Analysis
2.17. Molecular Docking Analysis
2.18. Statistical Analysis
3. Results
3.1. Analysis of 5–30% ACN Fraction in Caco-2 Cells: Evaluation of Cell Viability
3.2. HPLC Analysis, Antioxidant Properties In Vitro and In a Cellular Model of the Purified Fractions Obtained From the 5–30% ACN Pool
3.3. Identification of the Peptides with Mass Spectrometry Analysis
3.4. New Identified Peptides
3.4.1. Antioxidant Properties In Vitro and In Caco-2 Cells
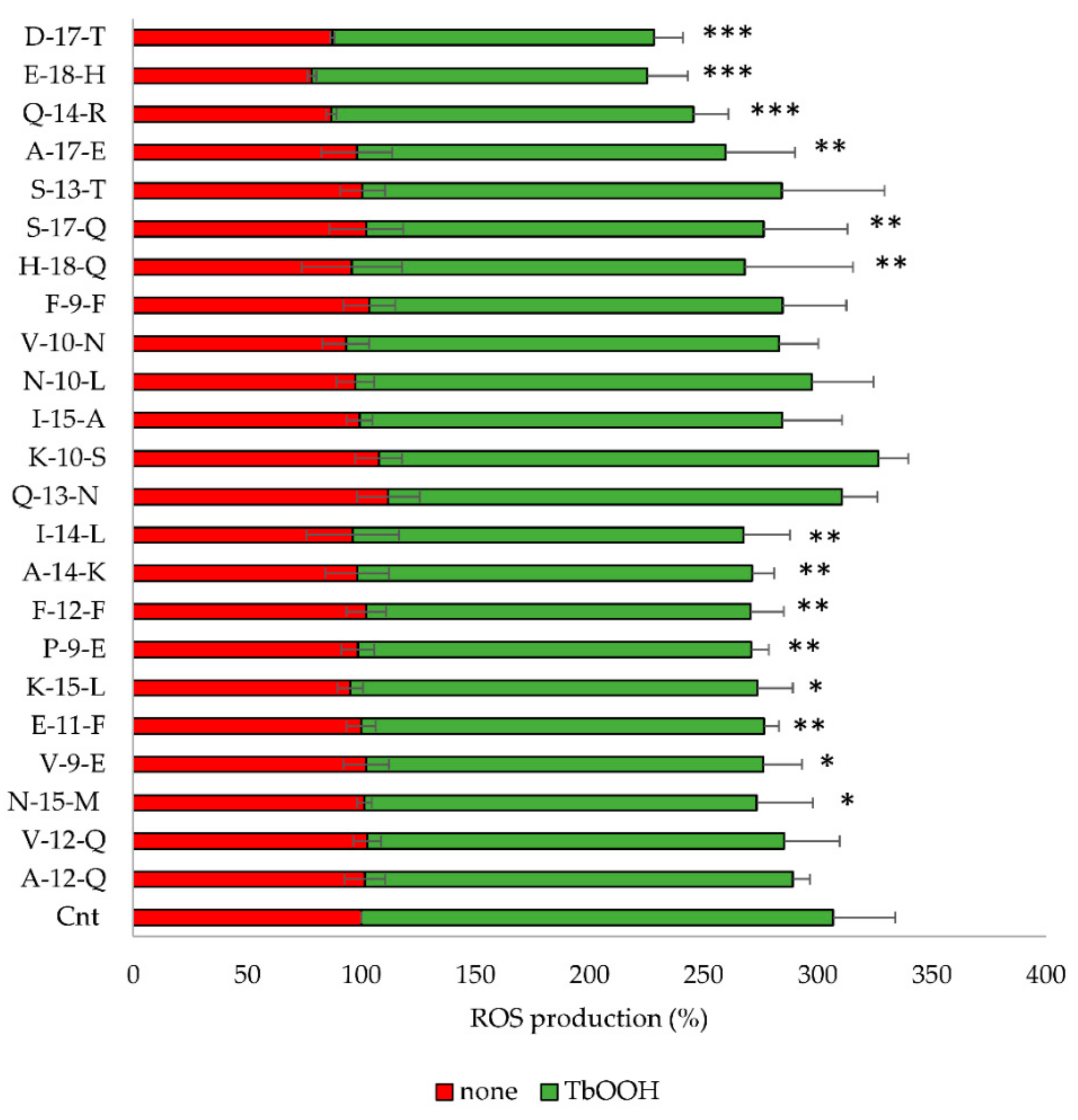
3.4.2. Inhibition of ROS Production by Bioactive Peptides
3.5. Analysis of the Mechanism of Action of the Antioxidant Peptides
3.5.1. Nrf2 Translocation to the Nucleus
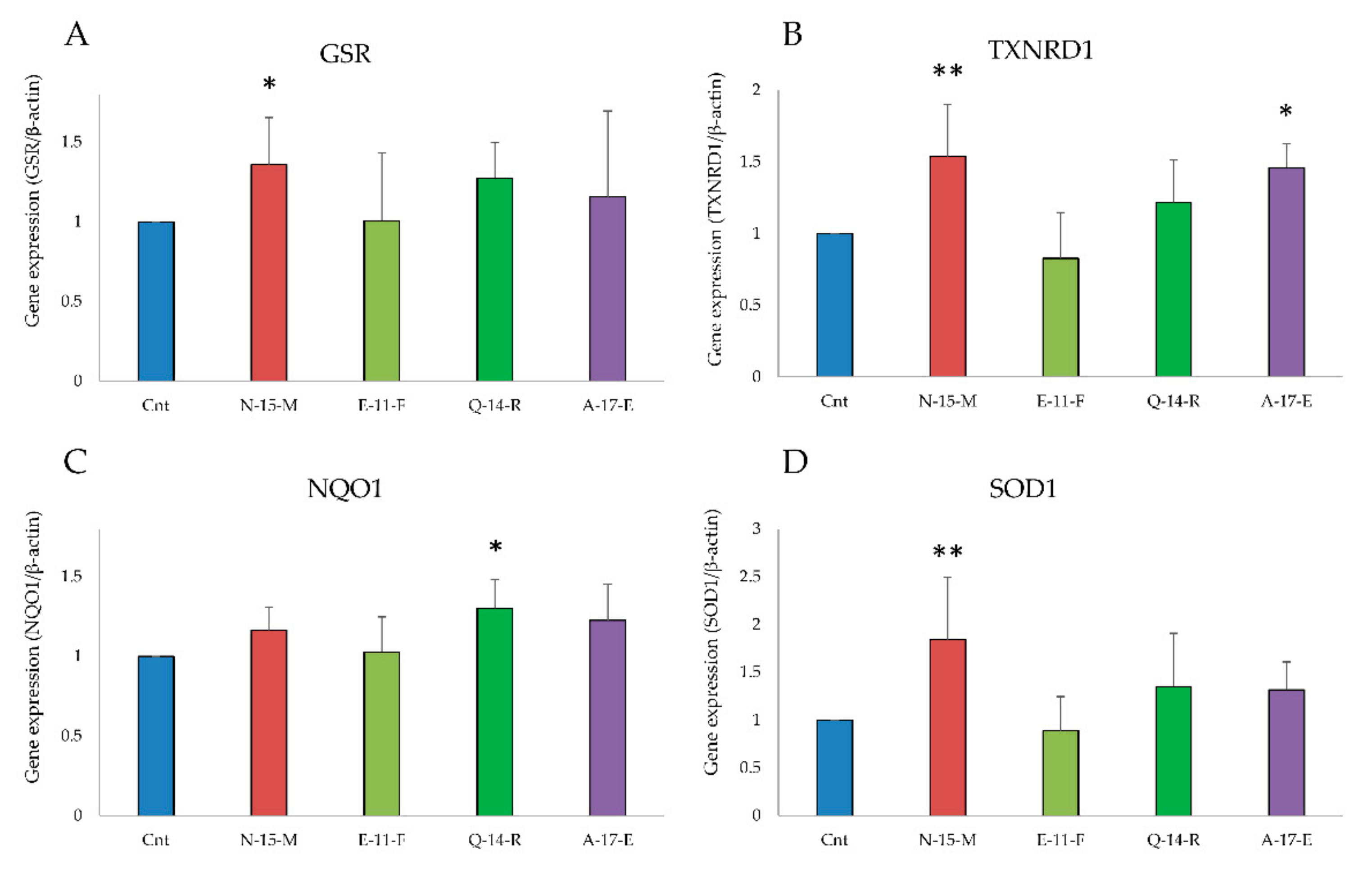
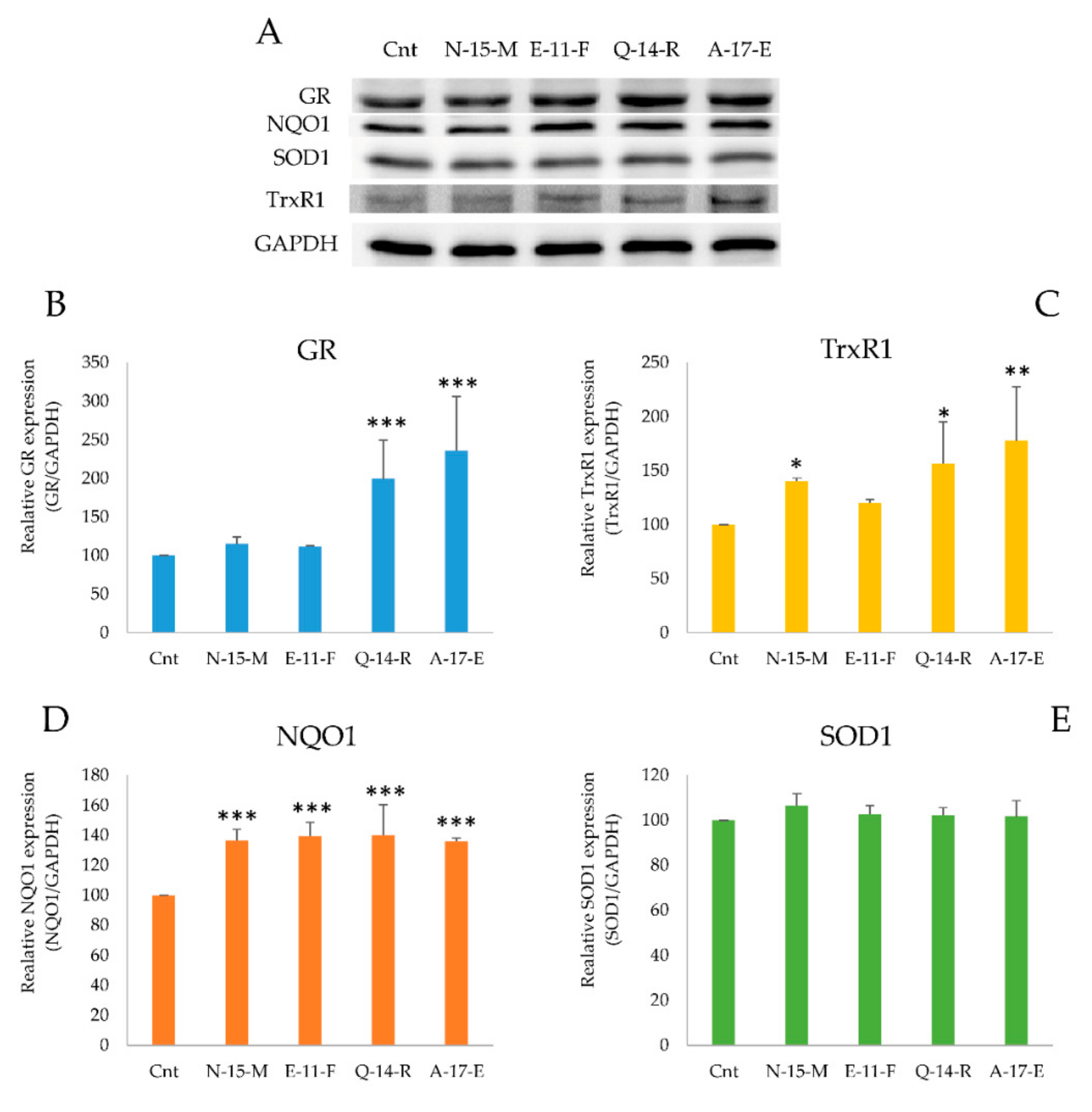
3.5.2. Antioxidant Enzymes Gene Expression Analysis
3.5.3. Antioxidant Enzymes Detection in the Presence of the Four Peptides
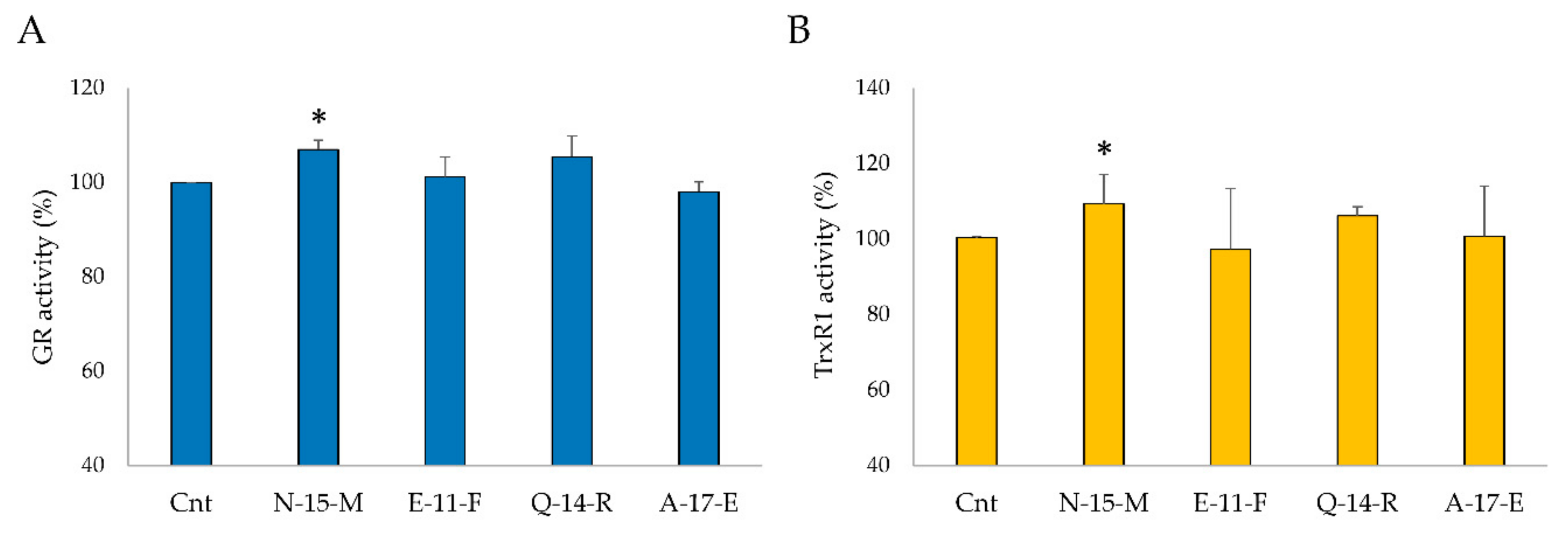
3.5.4. TrxR1 and GR Activities in Cell Lysates
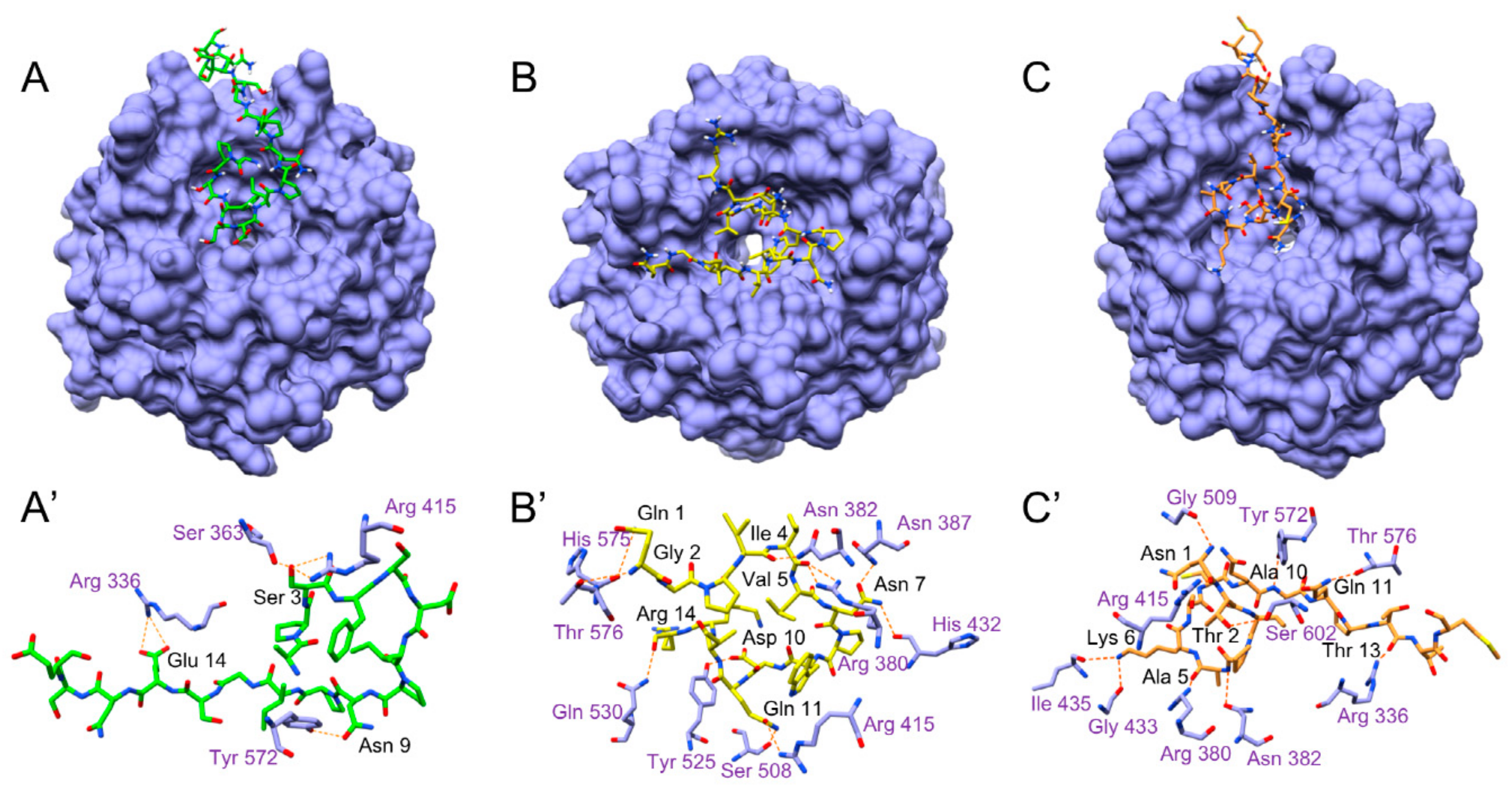
3.5.5. Molecular Docking Analysis
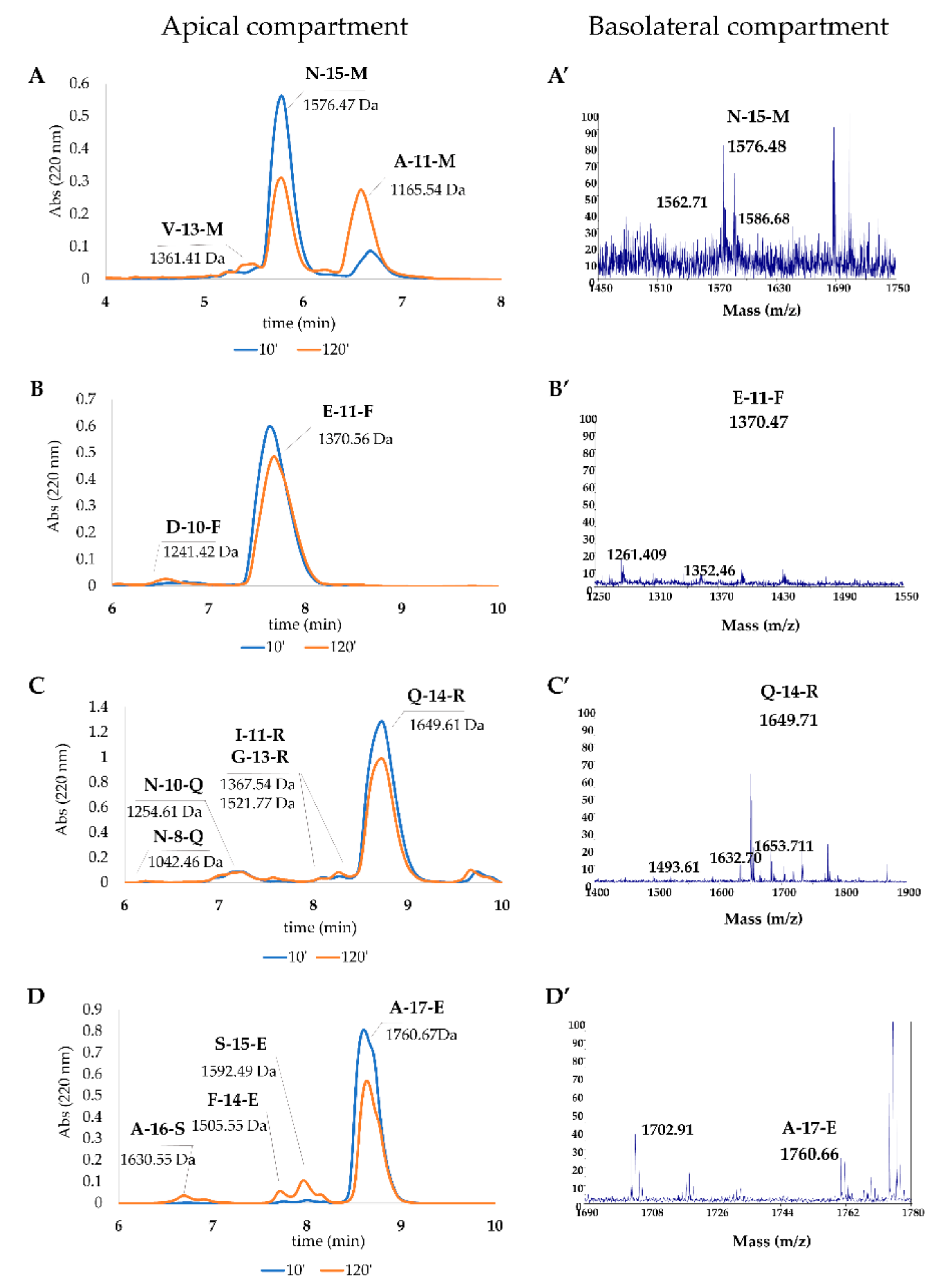
3.5.6. Peptide Absorption Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Şanlier, N.; Gökcen, B.B.; Sezgin, A.C. Health benefits of fermented foods. Crit. Rev. Food Sci. Nutr. 2019, 59, 506–527. [Google Scholar] [CrossRef] [PubMed]
- Rezac, S.; Kok, C.R.; Heermann, M.; Hutkins, R. Fermented Foods as a Dietary Source of Live Organisms. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Savijoki, K.; Ingmer, H.; Varmanen, P. Proteolytic systems of lactic acid bacteria. Appl. Microbiol. Biotechnol. 2006, 71, 394–406. [Google Scholar] [CrossRef]
- Shu, G.; Shi, X.; Chen, L.; Kou, J.; Meng, J.; Chen, H. Antioxidant Peptides from Goat Milk Fermented by Lactobacillus casei L61: Preparation, Optimization, and Stability Evaluation in Simulated Gastrointestinal Fluid. Nutrients 2018, 10, 797. [Google Scholar] [CrossRef] [PubMed]
- Solieri, L.; Rutella, G.S.; Tagliazucchi, D. Impact of non-starter lactobacilli on release of peptides with angiotensin-converting enzyme inhibitory and antioxidant activities during bovine milk fermentation. Food Microbiol. 2015, 51, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Pihlanto, A. Lactic Fermentation and Bioactive Peptides. In Lact. Acid Bact.—R Food Health Livest. Purp.; InTech: Rijeka, Croatia, 2013; pp. 309–332. [Google Scholar] [CrossRef]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; et al. Food fermentations: Microorganisms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef]
- Vargas-Bello-Pérez, E.; Márquez-Hernández, R.I.; Hernández-Castellano, L.E. Bioactive peptides from milk: animal determinants and their implications in human health. J. Dairy Res. 2019, 86, 136–144. [Google Scholar] [CrossRef]
- Dziuba, B.; Dziuba, M. Milk proteins-derived bioactive peptides in dairy products: molecular, biological and methodological aspects. Acta Sci. Pol. Technol. Aliment. 2014, 13, 5–26. [Google Scholar] [CrossRef]
- Pihlanto, A. Antioxidative peptides derived from milk proteins. Int. Dairy J. 2006, 16, 1306–1314. [Google Scholar] [CrossRef]
- Sánchez, A.; Vázquez, A. Bioactive peptides: A review. Food Qual. Saf. 2017, 1, 29–46. [Google Scholar] [CrossRef]
- Toldrá, F.; Reig, M.; Aristoy, M.-C.; Mora, L. Generation of bioactive peptides during food processing. Food Chem. 2018, 267, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, J.M.; Munekata, P.E.S.; Gómez, B.; Barba, F.J.; Mora, L.; Pérez-Santaescolástica, C.; Toldrá, F. Bioactive peptides as natural antioxidants in food products—A review. Trends Food Sci. Technol. 2018, 79, 136–147. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, A.; Yue, T.; Wang, J.; Wang, Y.; Su, J. Purification and identification of five novel antioxidant peptides from goat milk casein hydrolysates. J. Dairy Sci. 2013, 96, 4242–4251. [Google Scholar] [CrossRef] [PubMed]
- Karami, Z.; Akbari-adergani, B. Bioactive food derived peptides: a review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Technol. 2019, 56, 535–547. [Google Scholar] [CrossRef]
- Bindoli, A.; Rigobello, M.P. Principles in Redox Signaling: From Chemistry to Functional Significance. Antioxid. Redox Signal. 2012, 18, 1557–1593. [Google Scholar] [CrossRef]
- Kurutas, E.B. The importance of antioxidants which play the role in cellular response against oxidative/nitrosative stress: current state. Nutr. J. 2016, 15, 71. [Google Scholar] [CrossRef]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell Survival Responses to Environmental Stresses Via the Keap1-Nrf2-ARE Pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2017, 29, 1727–1745. [Google Scholar] [CrossRef]
- Baird, L.; Dinkova-Kostova, A.T. The cytoprotective role of the Keap1-Nrf2 pathway. Arch. Toxicol. 2011, 85, 241–272. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Kostov, R.V.; Canning, P. Keap1, the cysteine-based mammalian intracellular sensor for electrophiles and oxidants. Arch. Biochem. Biophys. 2017, 617, 84–93. [Google Scholar] [CrossRef]
- Tonolo, F.; Moretto, L.; Folda, A.; Scalcon, V.; Bindoli, A.; Bellamio, M.; Feller, E.; Rigobello, M.P. Antioxidant Properties of Fermented Soy during Shelf Life. Plant Foods Hum. Nutr. 2019, 74, 287–292. [Google Scholar] [CrossRef]
- Fields, G.B.; Noble, R.L. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 1990, 35, 161–214. [Google Scholar] [CrossRef] [PubMed]
- Carpino, L.A.; Imazumi, H.; El-Faham, A.; Ferrer, F.J.; Zhang, C.; Lee, Y.; Foxman, B.M.; Henklein, P.; Hanay, C.; Mügge, C.; et al. The Uronium/Guanidinium Peptide Coupling Reagents: Finally the True Uronium Salts. Angew. Chem. Int. Ed. 2002, 41, 441–445. [Google Scholar] [CrossRef]
- Tonolo, F.; Moretto, L.; Ferro, S.; Folda, A.; Scalcon, V.; Sandre, M.; Fiorese, F.; Marin, O.; Bindoli, A.; Rigobello, M.P. Insight into antioxidant properties of milk-derived bioactive peptides in vitro and in a cellular model. J. Pept. Sci. 2019, 25. [Google Scholar] [CrossRef] [PubMed]
- Tonolo, F.; Sandre, M.; Ferro, S.; Folda, A.; Scalcon, V.; Scutari, G.; Feller, E.; Marin, O.; Bindoli, A.; Rigobello, M.P. Milk-derived bioactive peptides protect against oxidative stress in a Caco-2 cell model. Food Funct. 2018, 9, 1245–1253. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Ge, C.; Duan, D.; Zhang, B.; Cui, X.; Peng, S.; Liu, Y.; Fang, J. Activation of the Phase II Enzymes for Neuroprotection by Ginger Active Constituent 6-Dehydrogingerdione in PC12 Cells. J. Agric. Food Chem. 2014, 62, 5507–5518. [Google Scholar] [CrossRef]
- Tonolo, F.; Folda, A.; Cesaro, L.; Scalcon, V.; Marin, O.; Ferro, S.; Bindoli, A.; Rigobello, M.P. Milk-derived bioactive peptides exhibit antioxidant activity through the Keap1-Nrf2 signaling pathway. J. Funct. Food 2019. [Google Scholar] [CrossRef]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar]
- Kurcinski, M.; Jamroz, M.; Blaszczyk, M.; Kolinski, A.; Kmiecik, S. CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res. 2015, 43. [Google Scholar] [CrossRef]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A Protein-Protein Docking Approach Based on Biochemical or Biophysical Information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; de Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of Macromolecular Assemblies from Crystalline State. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM Database of Bioactive Peptides: Current Opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.-C.; Li, X.; Henzl, M.T.; Beamer, L.J.; Hannink, M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006, 25, 3605–3617. [Google Scholar] [CrossRef] [PubMed]
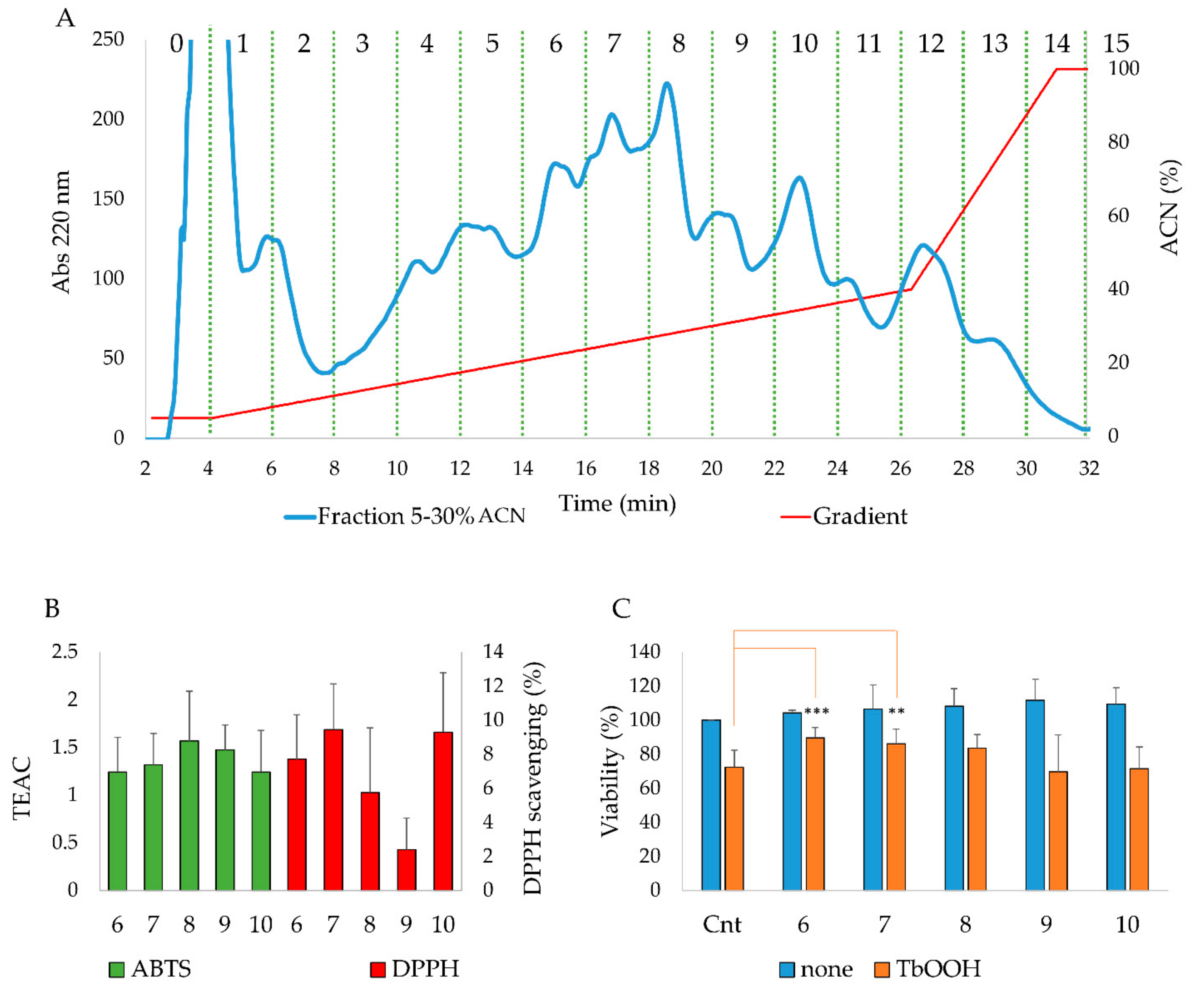

| Fraction | Percentage of Cell Viability | |
|---|---|---|
| None | TbOOH | |
| Control | 100 | 73.11 ± 8.22 |
| 5–30% ACN | 103.59 ± 5.20 | 83.60 ± 2.06 * |
| 30–50% ACN | 108.67 ± 10.15 | 77.80 ± 3.27 |
| Native Protein | Peptide Sequence | Name | Number of Residues | Peptide Fragment | Monoisotopic MW (Da) | Presence on BIOPEP Database [36] |
|---|---|---|---|---|---|---|
| κ-casein | QYVLSRYPSYGIN | Q-13-N | 13 | 50–62 | 1558.76 | NO |
| KYIPIQYVLS | K-10-S | 10 | 45–54 | 1222.68 | NO | |
| INNQFLPYPYYAKPA | I-15-A | 15 | 72–86 | 1797.89 | NO | |
| DKTEIPTINTIASGEPT | D-17-T | 17 | 136–152 | 1785.88 | YES (ID 8194—Kappacin; activity: Antibacterial) | |
| AVRSPAQILQWQ | A-12-Q | 12 | 87–98 | 1395.75 | NO | |
| VIESPPEINTVQ | V-12-Q | 12 | 173–184 | 1324.67 | YES (ID 8194—Kappacin; activity: Antibacterial) | |
| NTVPAKSCQAQPTTm * | N-15-M | 15 | 102–116 | 1591.72 | NO | |
| β-casein | NVPGEIVESL | N-10-L | 10 | 22–31 | 1055.53 | YES (ID: 8173—peptide derived from bovine β-casein (1–28); activity: Immunomodulating) |
| VYPFPGPIPN | V-10-N | 10 | 74–83 | 1099.55 | YES (ID: 7564 e 9240; ACE inhibitor; activity: ACE inhibitor) | |
| HKEMPFPKYPVEPFTESQ | H-18-Q | 18 | 121–138 | 2190.03 | NO | |
| SQSKVLPVPQKAVPYPQ | S-17-Q | 17 | 181–197 | 1865.03 | NO | |
| SWMHQPHQPLPPT | S-13-T | 13 | 157–169 | 1554.72 | NO | |
| VVPPFLQPE | V-9-E | 9 | 98–106 | 1024.54 | NO | |
| EDELQDKIHPF | E-11-F | 11 | 57–67 | 1369.64 | NO | |
| FPKYPVEPF | F-9-F | 9 | 126–134 | 1122.56 | NO | |
| αS1-casein | APSFSDIPNPIGSENSE | A-17-E | 17 | 191–207 | 1759.77 | NO |
| KHQGLPQEVLNENLL | K-15-L | 15 | 22–36 | 1730.92 | YES (ID: 8171—Isracidin-peptide derived from αS1-casein (1–23); activity: Immunomodulating) | |
| PFPEVFGKE | P-9-E | 9 | 42–50 | 1048.51 | NO | |
| αS2-casein | QGPIVLNPWDQVKR | Q-14-R | 14 | 116–129 | 1648.89 | NO |
| ALPQYLKTVYQHQK | A-14-K | 14 | 190–203 | 1715.92 | YES (ID: 8257, 8258, 8259; fragments of bovine αS2-casein; activity: Antibacterial) | |
| IQPKTKVIPYVRYL | I-14-L | 14 | 209–222 | 1717.01 | YES (ID: 8255; 8256; 8257; 8258; 8259; fragments of bovine αS2-casein; activity: Antibacterial) | |
| FLKKISQRYQKF | F-12-F | 12 | 163–174 | 1584.90 | YES (ID: Casocidin-I f(150–188); activity: antibacterial) | |
| NaPi2B ** | EKDDTGTPITKIELVPSH | E-18-H | 18 | 36–53 | 1979.01 | NO |
| Samples | In Vitro Antioxidant Tests | Percentage of Cell Viability | ||
|---|---|---|---|---|
| DPPH (%) Scavenging (a) | ABTS TEAC (b) | None (c) | TbOOH (d) | |
| Cnt | n.d. | n.d. | 100 | 73.11 ± 8.22 |
| Q-13 N | 24.44 ± 7.07 | 6.09 ± 1.08 | 104.10 ± 6.64 | 64.03 ± 10.15 |
| K-10-S | 19.09 ± 3.35 | 4.14 ± 0.82 | 97.27 ± 6.69 | 55.32 ± 10.01 |
| I-15-A | 19.40 ± 3.64 | 5.71 ± 1.09 | 104.37 ± 5.12 | 76.19 ± 13.27 |
| N-10-L | 13.19 ± 5.50 | n. d. | 119.52 ± 15.74 | 77.26 ± 12.44 |
| V-10-N | 12.24 ± 5.94 | 0.97 ± 0.48 | 110.89 ± 9.93 | 65.40 ± 17.60 |
| F-9-F | 12.69 ± 7.02 | 2.45 ± 0.54 | 109.47 ± 12.69 | 69.30 ± 12.78 |
| H-18-Q | 19.93 ± 5.13 | 1.30 ± 0.69 | 91.92 ± 8.99 | 52.61 ± 16.83 |
| S-13-T | 4.93 ± 3.45 | 2.96 ± 0.59 | 107.33 ± 12.39 | 71.60 ± 6.90 |
| S-17-Q | 3.74 ± 0.55 | 0.83 ± 0.36 | 101.47 ± 7.46 | 74.12 ± 11.76 |
| Q-14-R | 12.22 ± 4.72 | 1.28 ± 0.1 | 114.52 ± 15.55 | 81.47 ± 11.28 |
| A-17-E | 1.02 ± 0.15 | 0.64 ± 0.13 | 108.13 ± 13.19 | 78.33 ± 15.44 |
| D-17-T | 5.75 ± 0.38 | 0.84 ± 0.14 | 104.19 ± 5.62 | 73.36 ± 12.10 |
| E-18-H | n. d.a | n. d. | 103.64 ± 5.01 | 74.70 ± 10.52 |
| A-12-Q | 8.6± 5.98 | 1.39 ± 0.11 | 101.85 ± 4.69 | 78.89 ± 8.75 |
| V-12-Q | n. d. | n. d. | 112.70 ± 3.52 | 93.74 ± 10.68 |
| N-15-M | 16.93 ± 3.42 | 15.18 ± 0.04 | 107.99 ± 5.96 | 83.40 ± 5.51 |
| V-9-E | n. d. | n. d. | 106.59 ± 8.02 | 80.55 ± 6.62 |
| E-11-F | n. d. | n. d. | 111.51 ± 13.50 | 87.77 ± 4.50 |
| K-15-L | 4.14 ± 2.65 | n. d. | 104.70 ± 14.49 | 83.76 ± 6.30 |
| P-9-E | n. d. | n. d. | 100.97 ± 14.49 | 72.56 ± 3.68 |
| F-12-F | 15.29 ± 3.33 | 6.38 ± 0.29 | 97.73 ± 19.21 | 75.91 ± 4.80 |
| A-14-K | 2.16 ± 0.83 | 7.19 ± 0.23 | 84.39 ± 9.69 | 71.51 ± 5.22 |
| I-14-L | 22.9 ± 1.21 | 8.56 ± 0.43 | 92.98 ± 13.61 | 84.48 ± 8.64 |
| KEAP1 | NRF2 | A-17-E | Q-14-R | N-15-M |
|---|---|---|---|---|
| ARG 336 | Glu 14 | Thr 13 | ||
| SER 363 | Glu 82 | Ser 3 | ||
| ARG 380 | Glu 82 | Val 5, Asn 7 | Ala 5 | |
| ASN 382 | Glu 82, Phe 83 | Ile 4 | Ala 5 | |
| ASN 387 | Asn 7 | |||
| ARG 415 | Glu 79, Thr 80 | Ser 3 | Gln 11 | Asn 1 |
| HIS 432 | Asn 7 | |||
| GLY 433 | Lys 6 | |||
| ILE 435 | Lys 6 | |||
| SER 508 | Glu 79 | Gln 11 | ||
| GLY 509 | Asn 1 | |||
| TYR 525 | Glu 79 | Asp 10 | ||
| GLN 530 | Glu 78 | Arg 14 | ||
| TYR 572 | Leu 76, Gly 81 | Asn 9 | Ala 10 | |
| HIS 575 | Gly 2 | |||
| THR 576 | Gln 1 | Gln 11 | ||
| SER 602 | Thr 80 | Thr 2 |
| Peptide | Sequence | Retention Time (min) | Theoretical Mass | Measured MW | RP-HPLC Estimation AP (%) | RP-HPLC Estimation BL (%) |
|---|---|---|---|---|---|---|
| N-15-M | NTVPAKSCQAQPTTM | 5.77 | 1575.73 | 1576.46 | 60.76 ± 8.83 | 0.13 ± 0.04 |
| V-13-M | VPAKSCQAQPTTM | 5.22 | 1360.63 | 1361.41 | 2.21 ± 0.53 | n. d. a |
| A-11-M | AKSCQAQPTTM | 6.47 | 1164.51 | 1165.53 | 44.96 ± 9.09 | 0.16 ± 0.02 |
| S-9-M | SCQAQPTTM | 7.18 | 965.38 | 965.43 | n. d. | 0.12 ± 0.03 |
| E-11-F | EDELQDKIHPF | 7.63 | 1369.64 | 1370.56 | 88.28 ± 5.89 | 0.21 ± 0.01 |
| D-10-F | DELQDKIHPF | 6.75 | 1240.598 | 1241.42 | 4.17 ± 0.27 | n. d. |
| Q-14-R | QGPIVLNPWDQVKR | 8.73 | 1648.90 | 1649.60 | 87.59 ± 12.18 | 0.02 ± 0.01 |
| G-13-R | GPIVLNPWDQVKR | 8.29 | 1520.83 | 1521.76 | 3.19 ± 0.27 | n. d. |
| Q-13-K | QGPIVLNPWDQVK | 5.03 | 1492.79 | 1493.56 | n. d. | 0.07 ± 0.05 |
| I-11-R | IVLNPWDQVKR | 8.11 | 1366.76 | 1367.54 | 1.47 ± 0.19 | n. d. |
| Q-11-Q | QGPIVLNPWDQ | 7.19 | 1265.63 | 1266.94 | n. d. | 0.11 ± 0.01 |
| N-10-Q | VLNPWDQVKR | 7.28 | 1253.68 | 1254.60 | 7.44 ± 0.85 | n. d. |
| N-8-R | NPWDQVKR | 6.37 | 1041.52 | 1042.46 | 0.55 ± 0.15 | n. d. |
| A-17-E | APSFSDIPNPIGSENSE | 8.60 | 1759.80 | 1760.66 | 68.4 ± 4.6 | 0.05 ± 0.03 |
| A-16-S | APSFSDIPNPIGSENS | 6.73 | 1630.73 | 1630.45 | 3.22 ± 0.00 | n. d. |
| S-15-E | SFSDIPNPIGSENSE | 8.00 | 1591.69 | 1592.48 | 8.97 ± 0.53 | 0.30 ± 0.07 |
| F-14-E | FSDIPNPIGSENSE | 7.75 | 1504.65 | 1505.54 | 3.07 ± 0.60 | n. d. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tonolo, F.; Fiorese, F.; Moretto, L.; Folda, A.; Scalcon, V.; Grinzato, A.; Ferro, S.; Arrigoni, G.; Bindoli, A.; Feller, E.; et al. Identification of New Peptides from Fermented Milk Showing Antioxidant Properties: Mechanism of Action. Antioxidants 2020, 9, 117. https://doi.org/10.3390/antiox9020117
Tonolo F, Fiorese F, Moretto L, Folda A, Scalcon V, Grinzato A, Ferro S, Arrigoni G, Bindoli A, Feller E, et al. Identification of New Peptides from Fermented Milk Showing Antioxidant Properties: Mechanism of Action. Antioxidants. 2020; 9(2):117. https://doi.org/10.3390/antiox9020117
Chicago/Turabian StyleTonolo, Federica, Federico Fiorese, Laura Moretto, Alessandra Folda, Valeria Scalcon, Alessandro Grinzato, Stefania Ferro, Giorgio Arrigoni, Alberto Bindoli, Emiliano Feller, and et al. 2020. "Identification of New Peptides from Fermented Milk Showing Antioxidant Properties: Mechanism of Action" Antioxidants 9, no. 2: 117. https://doi.org/10.3390/antiox9020117
APA StyleTonolo, F., Fiorese, F., Moretto, L., Folda, A., Scalcon, V., Grinzato, A., Ferro, S., Arrigoni, G., Bindoli, A., Feller, E., Bellamio, M., Marin, O., & Rigobello, M. P. (2020). Identification of New Peptides from Fermented Milk Showing Antioxidant Properties: Mechanism of Action. Antioxidants, 9(2), 117. https://doi.org/10.3390/antiox9020117