Mitochondrial Arabidopsis thaliana TRXo Isoforms Bind an Iron–Sulfur Cluster and Reduce NFU Proteins In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Cloning and Site-Directed Mutagenesis
2.2. Heterologous Expression in Escherichia coli and Purification of Recombinant Proteins
2.3. Preparation of Apo-TRXs o and IscS-Mediated In Vitro Fe-S Cluster Reconstitution of TRXs o
2.4. Determination of the Oligomerization State
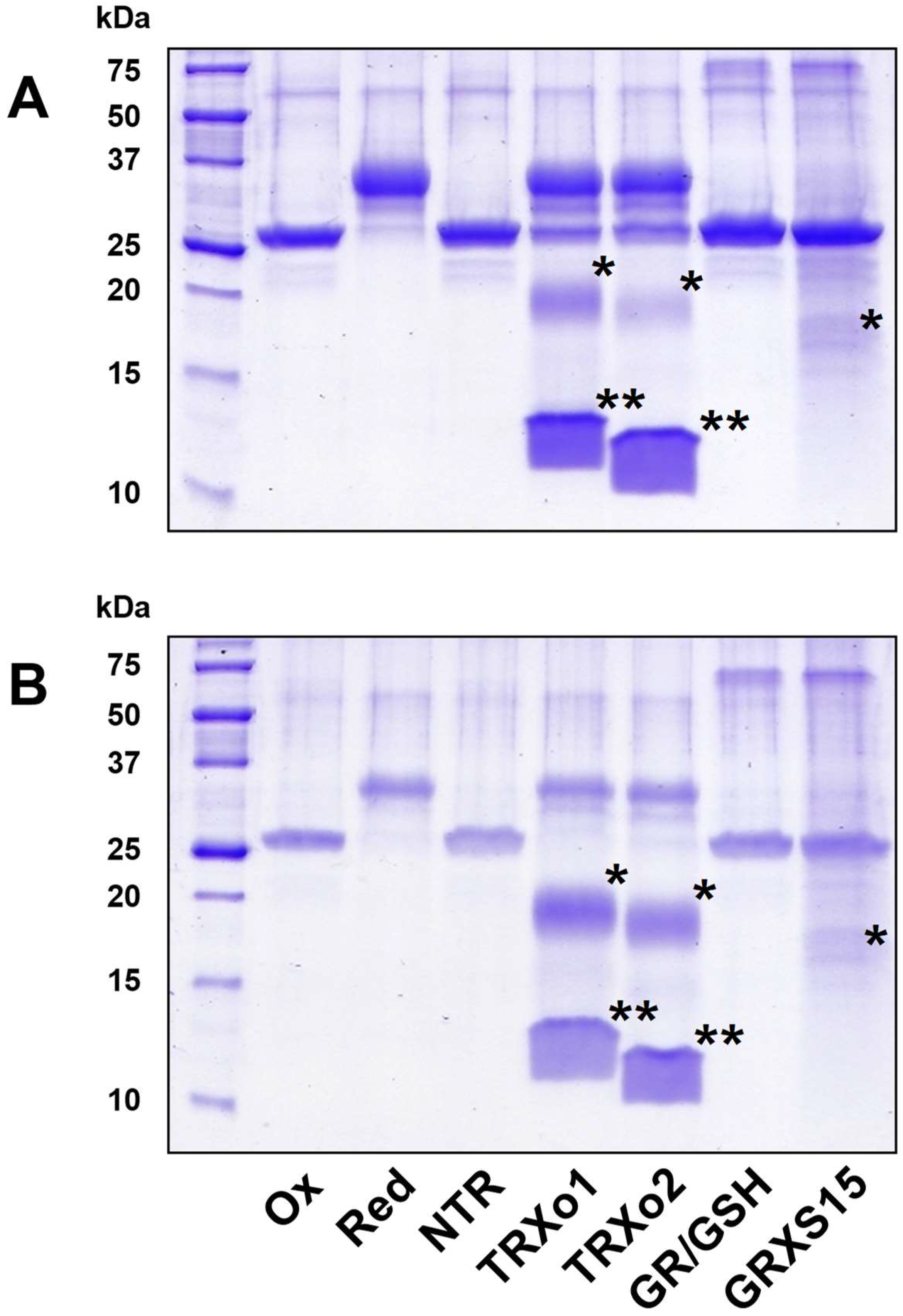
2.5. GRX- and TRX-Mediated Reduction of NFUs
2.6. Crystallization, Diffraction Data Collection, Processing, Structure Solution, and Refinement
3. Results
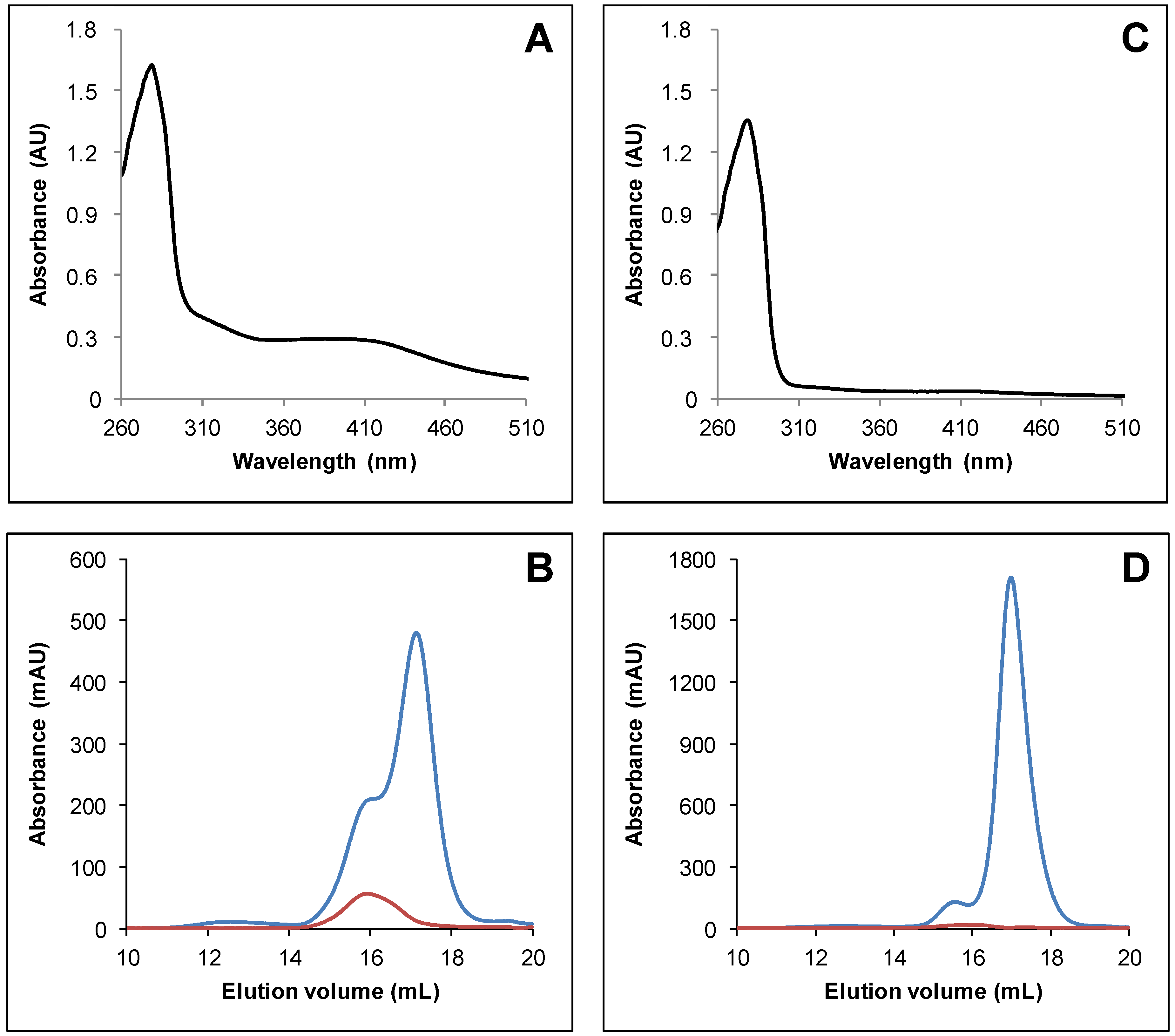
3.1. Arabidopsis thaliana TRXo Isoforms Exist in Two Forms upon Expression in E. coli
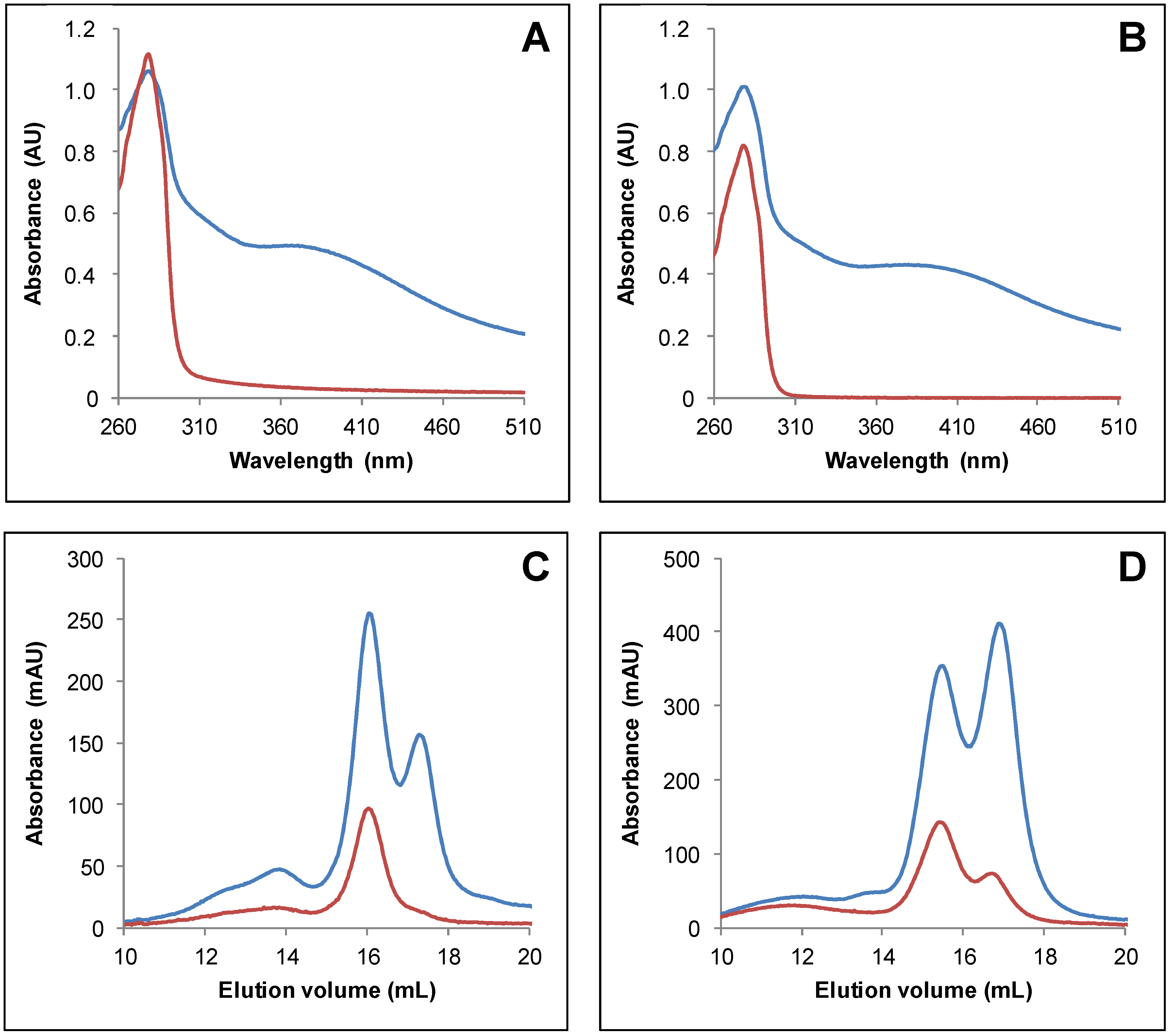
3.2. IscS-Mediated In Vitro Fe-S Cluster Reconstitution in Arabidopsis TRXo Isoforms
3.3. Both Active Site Cysteines of TRXo2 Are Required for Fe-S Cluster Incorporation
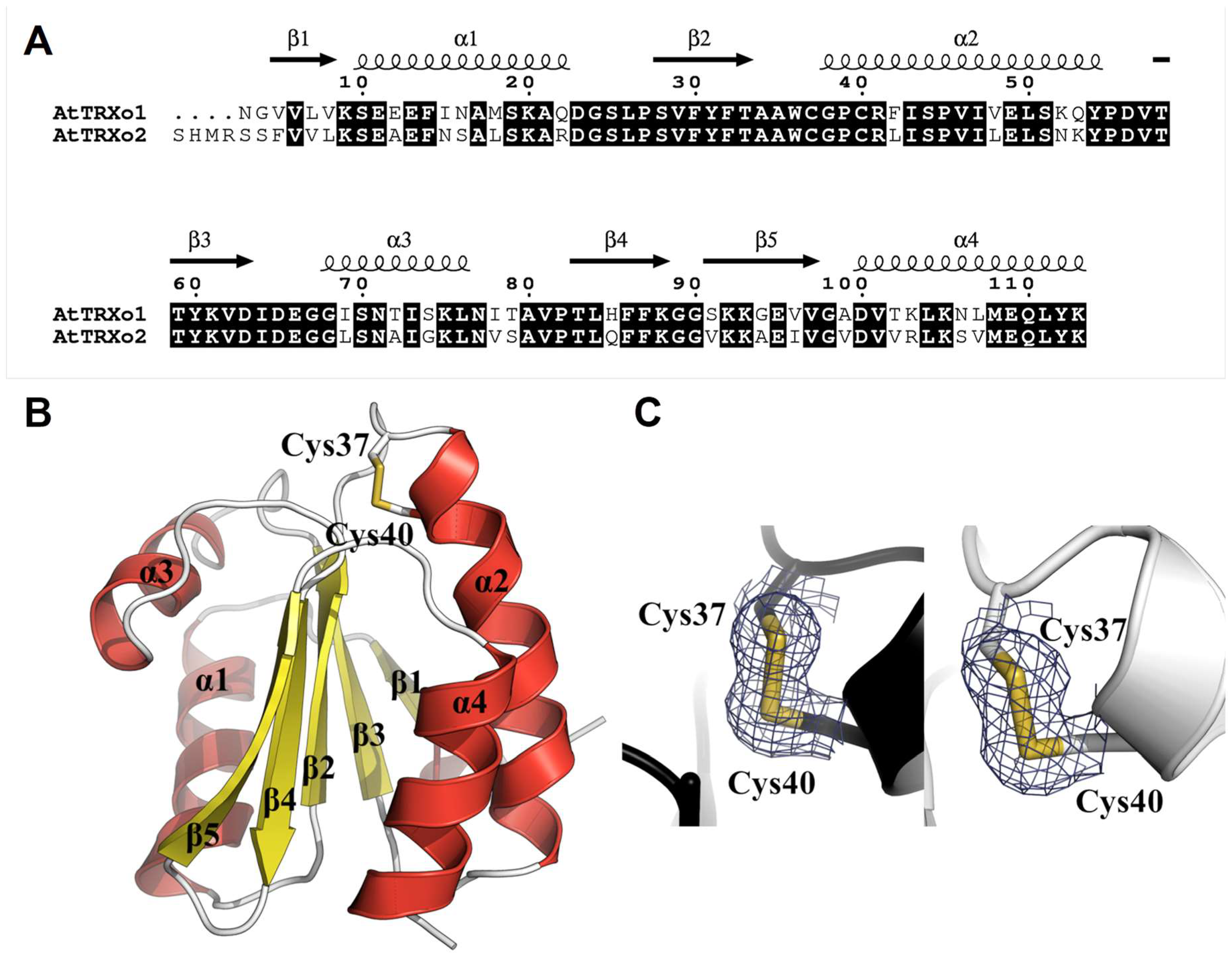
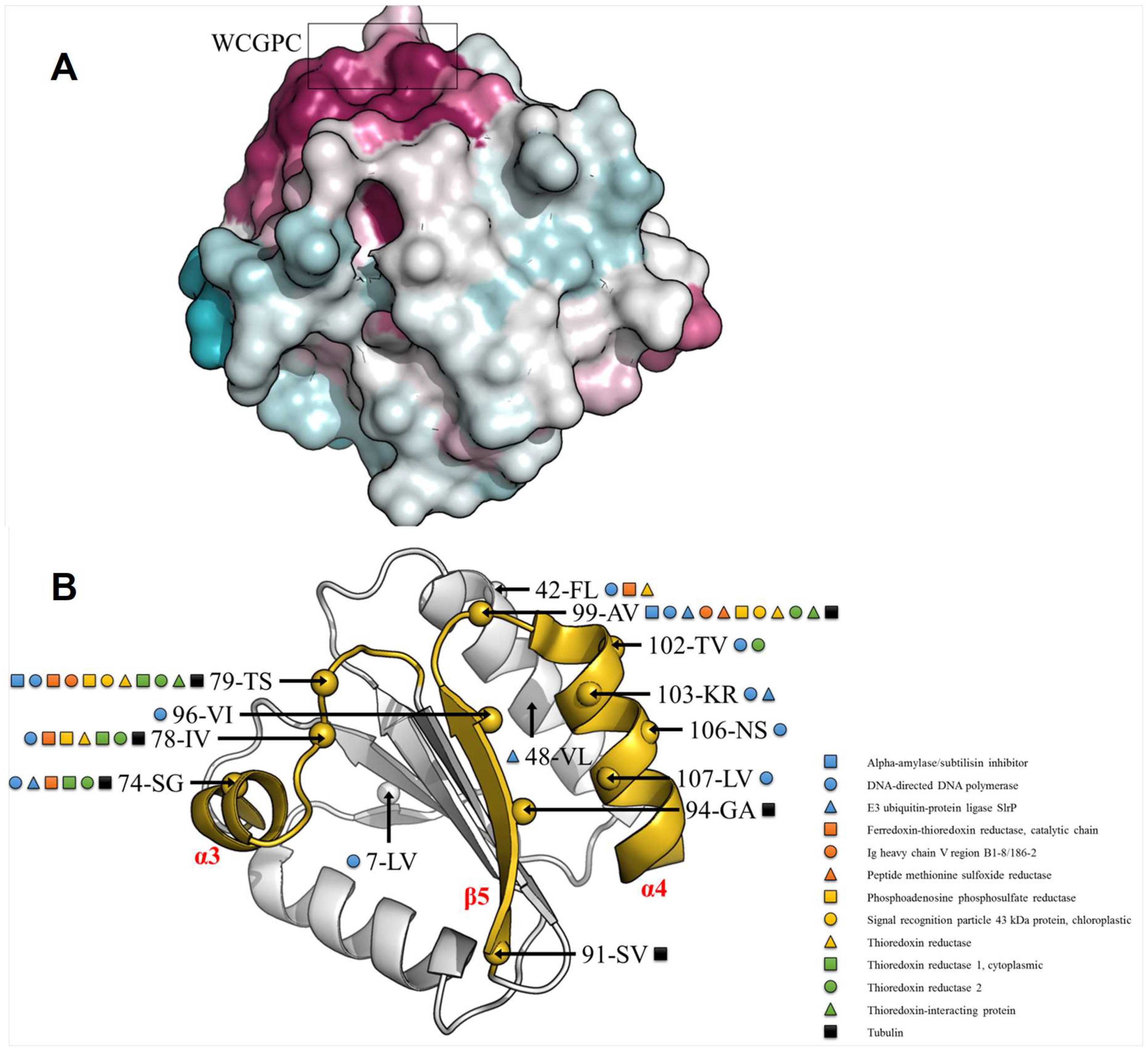
3.4. TRXo1 and TRXo2 Possess the Structural Properties of TRX Family Members
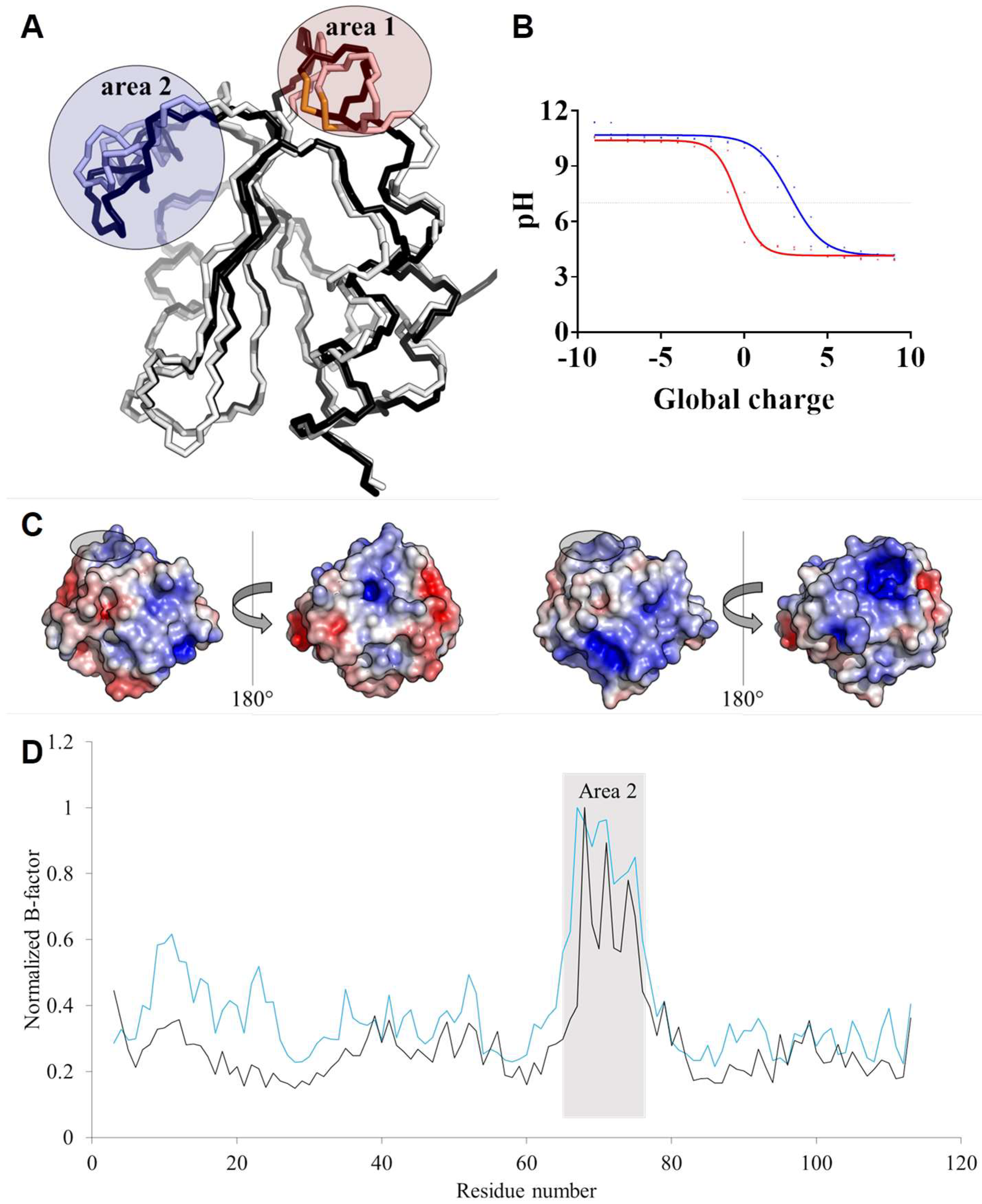
3.5. TRXo1 and TRXo2 Structures Are Not Strictly Superimposable
3.6. Oxidized NFU4 and NFU5 Are Reduced by TRXo Isoforms but Not by GRXS15
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Couturier, J.; Touraine, B.; Briat, J.F.; Gaymard, F.; Rouhier, N. The iron–sulfur cluster assembly machineries in plants: Current knowledge and open questions. Front. Plant Sci. 2013, 4, 259. [Google Scholar] [CrossRef] [PubMed]
- Braymer, J.J.; Lill, R. Iron–sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 2017, 292, 12754–12763. [Google Scholar] [CrossRef] [PubMed]
- Ciofi-Baffoni, S.; Nasta, V.; Banci, L. Protein networks in the maturation of human iron–sulfur proteins. Metallomics 2018, 10, 4972. [Google Scholar] [CrossRef] [PubMed]
- Navrot, N.; Rouhier, N.; Gelhaye, E.; Jacquot, J.P. Reactive oxygen species generation and antioxidant systems in plant mitochondria. Physiol. Plant. 2007, 129, 185–195. [Google Scholar] [CrossRef]
- Navrot, N.; Collin, V.; Gualberto, J.; Gelhaye, E.; Hirasawa, M.; Rey, P.; Knaff, D.B.; Issakidis, E.; Jacquot, J.P.; Rouhier, N. Plant glutathione peroxidases are functional peroxiredoxins distributed in several subcellular compartments and regulated during biotic and abiotic stresses. Plant Physiol. 2006, 142, 1364–1379. [Google Scholar] [CrossRef] [PubMed]
- Gama, F.; Keech, O.; Eymery, F.; Finkemeier, I.; Gelhaye, E.; Gardeström, P.; Dietz, K.J.; Rey, P.; Jacquot, J.P.; Rouhier, N. The mitochondrial type II peroxiredoxin from poplar. Physiol. Plant 2007, 129, 196–206. [Google Scholar] [CrossRef]
- Finkemeier, I.; Goodman, M.; Lamkemeyer, P.; Kandlbinder, A.; Sweetlove, L.J.; Dietz, K.J. The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 2005, 280, 12168–12180. [Google Scholar] [CrossRef] [PubMed]
- Barranco-Medina, S.; Krell, T.; Bernier-Villamor, L.; Sevilla, F.; Lázaro, J.J.; Dietz, K.J. Hexameric oligomerization of mitochondrial peroxiredoxin PrxIIF and formation of an ultrahigh affinity complex with its electron donor thioredoxin Trx-o. J. Exp. Bot. 2008, 59, 3259–3269. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Jacquot, J.P.; Rouhier, N. Evolution and diversity of glutaredoxins in photosynthetic organisms. Cell. Mol. Life Sci. 2009, 66, 2539–2557. [Google Scholar] [CrossRef] [PubMed]
- Chibani, K.; Wingsle, G.; Jacquot, J.P.; Gelhaye, E.; Rouhier, N. Comparative genomic study of the thioredoxin family in photosynthetic organisms with emphasis on Populus trichocarpa. Mol. Plant 2009, 2, 308–322. [Google Scholar] [CrossRef] [PubMed]
- Plomion, C.; Aury, J.M.; Amselem, J.; Leroy, T.; Murat, F.; Duplessis, S.; Faye, S.; Francillonne, N.; Labadie, K.; Le Provost, G.; et al. Oak genome reveals facets of long lifespan. Nat. Plants 2018, 1. [Google Scholar] [CrossRef] [PubMed]
- Moseler, A.; Aller, I.; Wagner, S.; Nietzel, T.; Przybyla-Toscano, J.; Mühlenhoff, U.; Lill, R.; Berndt, C.; Rouhier, N.; Schwarzländer, M.; et al. The mitochondrial monothiol glutaredoxin S15 is essential for iron–sulfur protein maturation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2015, 112, 13735–13740. [Google Scholar] [CrossRef] [PubMed]
- Laloi, C.; Rayapuram, N.; Chartier, Y.; Grienenberger, J.M.; Bonnard, G.; Meyer, Y. Identification and characterization of a mitochondrial thioredoxin system in plants. Proc. Natl. Acad. Sci. USA 2001, 98, 14144–14149. [Google Scholar] [CrossRef] [PubMed]
- Gelhaye, E.; Rouhier, N.; Jacquot, J.P. The thioredoxin h system of higher plants. Plant Physiol. Biochem. 2004, 42, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Wong, J.H.; Feldman, L.J.; Lemaux, P.G.; Buchanan, B.B. A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc. Natl. Acad. Sci. USA 2010, 107, 3900–3905. [Google Scholar] [CrossRef] [PubMed]
- Reichheld, J.P.; Meyer, E.; Khafif, M.; Bonnard, G.; Meyer, Y. AtNTRB is the major mitochondrial thioredoxin reductase in Arabidopsis thaliana. FEBS Lett. 2005, 579, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Begas, P.; Liedgens, L.; Moseler, A.; Meyer, A.J.; Deponte, M. Glutaredoxin catalysis requires two distinct glutathione interaction sites. Nat. Commun. 2017, 8, 14835. [Google Scholar] [CrossRef] [PubMed]
- Ströher, E.; Grassl, J.; Carrie, C.; Fenske, R.; Whelan, J.; Millar, A.H. Glutaredoxin S15 Is Involved in Fe-S Cluster Transfer in Mitochondria Influencing Lipoic Acid-Dependent Enzymes, Plant Growth, and Arsenic Tolerance in Arabidopsis. Plant Physiol. 2016, 170, 1284–1299. [Google Scholar] [CrossRef] [PubMed]
- Bedhomme, M.; Adamo, M.; Marchand, C.H.; Couturier, J.; Rouhier, N.; Lemaire, S.D.; Zaffagnini, M.; Trost, P. Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem. J. 2012, 445, 337–347. [Google Scholar] [CrossRef] [PubMed]
- Kneeshaw, S.; Gelineau, S.; Tada, Y.; Loake, G.J.; Spoel, S.H. Selective protein denitrosylation activity of thioredoxin-h5 modulates plant immunity. Mol. Cell 2014, 56, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Schmidtmann, E.; König, A.C.; Orwat, A.; Leister, D.; Hartl, M.; Finkemeier, I. Redox regulation of Arabidopsis mitochondrial citrate synthase. Mol. Plant 2014, 7, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Noguchi, K.; Motohashi, K.; Hisabori, T. Systematic exploration of thioredoxin target proteins in plant mitochondria. Plant Cell Physiol. 2013, 54, 875–892. [Google Scholar] [CrossRef] [PubMed]
- Martí, M.C.; Olmos, E.; Calvete, J.J.; Díaz, I.; Barranco-Medina, S.; Whelan, J.; Lázaro, J.J.; Sevilla, F.; Jiménez, A. Mitochondrial and nuclear localization of a novel pea thioredoxin: Identification of its mitochondrial target proteins. Plant Physiol. 2009, 150, 646–657. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Hisabori, T. Mitochondrial isocitrate dehydrogenase is inactivated upon oxidation and reactivated by thioredoxin-dependent reduction in Arabidopsis. Front. Environ. Sci. 2014, 2, 38. [Google Scholar] [CrossRef]
- Daloso, D.M.; Müller, K.; Obata, T.; Florian, A.; Tohge, T.; Bottcher, A.; Riondet, C.; Bariat, L.; Carrari, F.; Nunes-Nesi, A.; et al. Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proc. Natl. Acad. Sci. USA 2015, 112, E1392–E1400. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Hisabori, T. Adenine nucleotide-dependent and redox-independent control of mitochondrial malate dehydrogenase activity in Arabidopsis thaliana. Biochim. Biophys. Acta 2016, 1857, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Espín, A.; Iglesias-Fernández, R.; Calderón, A.; Carbonero, P.; Sevilla, F.; Jiménez, A. Mitochondrial AtTrxo1 is transcriptionally regulated by AtbZIP9 and AtAZF2 and affects seed germination under saline conditions. J. Exp. Bot. 2017, 68, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Balmer, Y.; Vensel, W.H.; Tanaka, C.K.; Hurkman, W.J.; Gelhaye, E.; Rouhier, N.; Jacquot, J.P.; Manieri, W.; Schürmann, P.; Droux, M.; et al. Thioredoxin links redox to the regulation of fundamental processes of plant mitochondria. Proc. Natl. Acad. Sci. USA 2004, 101, 2642–2647. [Google Scholar] [CrossRef] [PubMed]
- Winger, A.M.; Taylor, N.L.; Heazlewood, J.L.; Day, D.A.; Millar, A.H. Identification of intra- and intermolecular disulphide bonding in the plant mitochondrial proteome by diagonal gel electrophoresis. Proteomics 2007, 7, 4158–4170. [Google Scholar] [CrossRef] [PubMed]
- Keech, O.; Gardeström, P.; Kleczkowski, L.A.; Rouhier, N. The redox control of photorespiration: From biochemical and physiological aspects to biotechnological considerations. Plant Cell Environ. 2017, 40, 553–569. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R.; Krummel, B.; Saiki, R.K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 1988, 16, 7351–7367. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.N.; Hunt, H.D.; Horton, R.M.; Pullen, J.K.; Pease, L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 1989, 77, 51–59. [Google Scholar] [CrossRef]
- Schenk, P.M.; Baumann, S.; Mattes, R.; Steinbiss, H.H. Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. Biotechniques 1995, 19, 196–200. [Google Scholar] [PubMed]
- Gill, S.C.; von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef]
- Jacquot, J.P.; Rivera-Madrid, R.; Marinho, P.; Kollarova, M.; Le Marechal, P.; Miginiac-Maslow, M.; Meyer, Y. Arabidopsis thaliana NAPHP thioredoxin reductase. cDNA characterization and expression of the recombinant protein in Escherichia coli. J. Mol. Biol. 1994, 235, 1357–1363. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Gama, F.; Molina-Navarro, M.M.; Gualberto, J.M.; Claxton, R.; Naik, S.G.; Huynh, B.H.; Herrero, E.; Jacquot, J.P.; Johnson, M.K.; et al. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 2008, 27, 1122–1133. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Stroher, E.; Albetel, A.N.; Roret, T.; Muthuramalingam, M.; Tarrago, L.; Seidel, T.; Tsan, P.; Jacquot, J.P.; Johnson, M.K.; et al. Arabidopsis chloroplastic glutaredoxin C5 as a model to explore molecular determinants for iron–sulfur cluster binding into glutaredoxins. J. Biol. Chem. 2011, 286, 27515–27527. [Google Scholar] [CrossRef] [PubMed]
- Zannini, F.; Couturier, J.; Keech, O.; Rouhier, N. In Vitro Alkylation Methods for Assessing the Protein Redox State. Methods Mol. Biol. 2017, 1653, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Langer, G.; Cohen, S.X.; Lamzin, V.S.; Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008, 3, 1171–1179. [Google Scholar] [CrossRef] [PubMed]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 22–25. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Rouhier, N.; Unno, H.; Bandyopadhyay, S.; Masip, L.; Kim, S.K.; Hirasawa, M.; Gualberto, J.M.; Lattard, V.; Kusunoki, M.; Knaff, D.B.; et al. Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc. Natl. Acad. Sci. USA 2007, 104, 7379–7384. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Subramanian, S.; Couturier, J.; Naik, S.G.; Kim, S.K.; Leustek, T.; Knaff, D.B.; Wu, H.C.; Vignols, F.; Huynh, B.H.; et al. Arabidopsis thaliana Nfu2 accommodates [2Fe-2S] or [4Fe-4S] clusters and is competent for in vitro maturation of chloroplast [2Fe-2S] and [4Fe-4S] cluster-containing proteins. Biochemistry 2013, 52, 6633–6645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Bandyopadhyay, S.; Shakamuri, P.; Naik, S.G.; Huynh, B.H.; Couturier, J.; Rouhier, N.; Johnson, M.K. Monothiol glutaredoxins can bind linear [Fe3S4]+ and [Fe4S4]2+ clusters in addition to [Fe2S2]2+ clusters: spectroscopic characterization and functional implications. J. Am. Chem. Soc. 2013, 135, 15153–15164. [Google Scholar] [CrossRef] [PubMed]
- Droux, M.; Jacquot, J.P.; Miginac-Maslow, M.; Gadal, P.; Huet, J.C.; Crawford, N.A.; Yee, B.C.; Buchanan, B.B. Ferredoxin-thioredoxin reductase, an iron–sulfur enzyme linking light to enzyme regulation in oxygenic photosynthesis: Purification and properties of the enzyme from C3, C4, and cyanobacterial species. Arch. Biochem. Biophys. 1987, 252, 426–439. [Google Scholar] [CrossRef]
- Bisio, H.; Bonilla, M.; Manta, B.; Graña, M.; Salzman, V.; Aguilar, P.S.; Gladyshev, V.N.; Comini, M.A.; Salinas, G. A new class of thioredoxin-related protein able to bind iron–sulfur clusters. Antioxid. Redox Signal. 2015, 24, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Masip, L.; Pan, J.L.; Haldar, S.; Penner-Hahn, J.E.; DeLisa, M.P.; Georgiou, G.; Bardwell, J.C.; Collet, J.F. An engineered pathway for the formation of protein disulfide bonds. Science 2004, 303, 1185–1189. [Google Scholar] [CrossRef] [PubMed]
- Saarinen, M.; Gleason, F.K.; Eklund, H. Crystal structure of thioredoxin-2 from Anabaena. Structure 1995, 3, 1097–1108. [Google Scholar] [CrossRef]
- Shen, J.; Zeng, Y.; Zhuang, X.; Sun, L.; Yao, X.; Pimpl, P.; Jiang, L. Organelle pH in the Arabidopsis endomembrane system. Mol. Plant 2013, 6, 1419–1437. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, 522–525. [Google Scholar] [CrossRef] [PubMed]
- Selles, B.; Zannini, F.; Couturier, J.; Jacquot, J.P.; Rouhier, N. Atypical protein disulfide isomerases (PDI): Comparison of the molecular and catalytic properties of poplar PDI-A and PDI-M with PDI-L1A. PLoS ONE 2017, 12, e0174753. [Google Scholar] [CrossRef] [PubMed]
- Remelli, W.; Santabarbara, S.; Carbonera, D.; Bonomi, F.; Ceriotti, A.; Casazza, A.P. Iron Binding Properties of Recombinant Class A Protein Disulfide Isomerase from Arabidopsis thaliana. Biochemistry 2017, 56, 2116–2125. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhong, N.; Rouhier, N.; Hase, T.; Kusunoki, M.; Jacquot, J.P.; Jin, C.; Xia, B. Structural insight into poplar glutaredoxin C1 with a bridging iron–sulfur cluster at the active site. Biochemistry 2006, 45, 7998–8008. [Google Scholar] [CrossRef] [PubMed]
- Johansson, C.; Roos, A.K.; Montano, S.J.; Sengupta, R.; Filippakopoulos, P.; Guo, K.; von Delft, F.; Holmgren, A.; Oppermann, U.; Kavanagh, K.L. The crystal structure of human GLRX5: Iron–sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem. J. 2011, 433, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Banci, L.; Brancaccio, D.; Ciofi-Baffoni, S.; Del Conte, R.; Gadepalli, R.; Mikolajczyk, M.; Neri, S.; Piccioli, M.; Winkelmann, J. [2Fe-2S] cluster transfer in iron–sulfur protein biogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 6203–6208. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.E.; Wisz, M.S.; Liu, W.; Hellinga, H.W. Construction of a novel redox protein by rational design: conversion of a disulfide bridge into a mononuclear iron–sulfur center. Biochemistry 1998, 37, 7070–7076. [Google Scholar] [CrossRef] [PubMed]
- Collet, J.F.; Peisach, D.; Bardwell, J.C.; Xu, Z. The crystal structure of TrxA(CACA): Insights into the formation of a [2Fe-2S] iron–sulfur cluster in an Escherichia coli thioredoxin mutant. Protein Sci. 2005, 14, 1863–1869. [Google Scholar] [CrossRef] [PubMed]
- Coldren, C.D.; Hellinga, H.W.; Caradonna, J.P. The rational design and construction of a cuboidal iron–sulfur protein. Proc. Natl. Acad. Sci. USA 1997, 94, 6635–6640. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Berndt, C.; Fomenko, D.E.; Holmgren, A.; Gladyshev, V.N. A conserved cis-proline precludes metal binding by the active site thiolates in members of the thioredoxin family of proteins. Biochemistry 2007, 46, 69036910. [Google Scholar] [CrossRef] [PubMed]
- Berndt, C.; Schwenn, J.D.; Lillig, C.H. The specificity of thioredoxins and glutaredoxins is determined by electrostatic and geometric complementarity. Chem. Sci. 2015, 6, 7049–7058. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, M.E.; Mauriès, A.; Maes, A.; Tourasse, N.J.; Hamon, M.; Lemaire, S.D.; Marchand, C.H. The Deep Thioredoxome in Chlamydomonas reinhardtii: New Insights into Redox Regulation. Mol. Plant 2017, 10, 1107–1125. [Google Scholar] [CrossRef] [PubMed]
- Zannini, F.; Moseler, A.; Bchini, R.; Dhalleine, T.; Meyer, A.J.; Rouhier, N.; Couturier, J. The thioredoxin-mediated recycling of Arabidopsis thaliana GRXS16 relies on a conserved C-terminal cysteine. BBA General Subj. under revision.
- Landau, M.; Mayrose, I.; Rosenberg, Y.; Glaser, F.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2005: The projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005, 33, W299–W302. [Google Scholar] [CrossRef] [PubMed]
- Lillig, C.H.; Berndt, C.; Vergnolle, O.; Lönn, M.E.; Hudemann, C.; Bill, E.; Holmgren, A. Characterization of human glutaredoxin 2 as iron–sulfur protein: a possible role as redox sensor. Proc. Natl. Acad. Sci. USA 2005, 102, 8168–8173. [Google Scholar] [CrossRef] [PubMed]

| Data Collection | AtTRXo1 | AtTRXo2 |
|---|---|---|
| Beam line | FIP-BM30A | |
| Space group | P212121 | P65 |
| Cell dimensions a, b, c (Å) α, β, γ (°) | 37.15; 39.24; 79.34 α = β = γ = 90° | 70.81; 70.81; 35.75 α = β = 90° γ = 120° |
| Resolution (Å) | 39.67−1.80 (1.84−1.80) | 35.40−1.50 (1.53−1.50) |
| Rmerge | 0.122 (0.242) | 0.070 (0.432) |
| Rmeas | 0.131 (0.283) | 0.075 (0.500) |
| Rpim | 0.049 (0.145) | 0.028 (0.251) |
| No. unique reflections | 11,316 (653) | 16,473 (814) |
| Mean I/σI | 13.1 (3.7) | 20.0 (3.1) |
| CC1/2 | 0.996 (0.961) | 0.997 (0.942) |
| Completeness (%) | 100.0 (100.0) | 99.4 (100.0) |
| Average redundancy | 11.4 (7.0) | 12.0 (7.3) |
| Refinement | ||
| Resolution (Å) | 39.67−1.80 | 35.40−1.50 |
| Rfree/Rwork | 23.96/21.96 | 17.93/16.74 |
| Total number of atoms | 1848 | 1882 |
| Water | 130 | 92 |
| Crystallographic B-factor | ||
| Overall B-factor (Ų) | 29.16 | 35.36 |
| B-factor: molecule (Ų) | 28.88 | 35.35 |
| B-factor: water (Ų) | 32.89 | 35.47 |
| R.m.s deviations | ||
| Bonds | 0.002 | 0.003 |
| Angles | 0.474 | 0.528 |
| MolProbity analysis | ||
| Clashscore, all atoms | 2.91 (99%) | 0.00 (100%) |
| MolProbity score | 1.40 (96%) | 0.50 (100%) |
| Protein Data Bank entry | 6G61 | 6G62 |
| Protein | Theoretical Size (Da) | Theoretical Size without Met (Da) | Untreated | Treated with DTT | Mass Difference upon Reduction (Da) |
|---|---|---|---|---|---|
| NFU4 | 22,167.9 | 22,036.7 | 22,035.1 | 22,037.4 | +2.3 |
| NFU5 | 21,823.5 | 21,692.3 | 21,690.9 | 21,693.0 | +2.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zannini, F.; Roret, T.; Przybyla-Toscano, J.; Dhalleine, T.; Rouhier, N.; Couturier, J. Mitochondrial Arabidopsis thaliana TRXo Isoforms Bind an Iron–Sulfur Cluster and Reduce NFU Proteins In Vitro. Antioxidants 2018, 7, 142. https://doi.org/10.3390/antiox7100142
Zannini F, Roret T, Przybyla-Toscano J, Dhalleine T, Rouhier N, Couturier J. Mitochondrial Arabidopsis thaliana TRXo Isoforms Bind an Iron–Sulfur Cluster and Reduce NFU Proteins In Vitro. Antioxidants. 2018; 7(10):142. https://doi.org/10.3390/antiox7100142
Chicago/Turabian StyleZannini, Flavien, Thomas Roret, Jonathan Przybyla-Toscano, Tiphaine Dhalleine, Nicolas Rouhier, and Jérémy Couturier. 2018. "Mitochondrial Arabidopsis thaliana TRXo Isoforms Bind an Iron–Sulfur Cluster and Reduce NFU Proteins In Vitro" Antioxidants 7, no. 10: 142. https://doi.org/10.3390/antiox7100142
APA StyleZannini, F., Roret, T., Przybyla-Toscano, J., Dhalleine, T., Rouhier, N., & Couturier, J. (2018). Mitochondrial Arabidopsis thaliana TRXo Isoforms Bind an Iron–Sulfur Cluster and Reduce NFU Proteins In Vitro. Antioxidants, 7(10), 142. https://doi.org/10.3390/antiox7100142







