Antiradical and Xanthine Oxidase Inhibitory Activity Evaluations of Averrhoa bilimbi L. Leaves and Tentative Identification of Bioactive Constituents through LC-QTOF-MS/MS and Molecular Docking Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plant Collection and Processing
2.3. Extraction and Fractionation
2.4. Preliminary Phytochemical Screening
2.5. LC-QTOF-MS and LC-QTOF-MS/MS Analyses of Bioactive Constituents of n-Butanol Fraction
2.6. DPPH Free Radical Scavenging Activity
2.7. Xanthine Oxidase Inhibitory Activity Assay
2.8. Molecular Docking
2.9. Statistical Analysis
3. Results and Discussion
3.1. DPPH Radical Scavenging Activity
3.2. Xanthine Oxidase Inhibitory Activity
3.3. LC-QTOF-MS Analysis of n-Butanol Fraction of Averrhoa bilimbi Crude Methanolic Leaves Extract
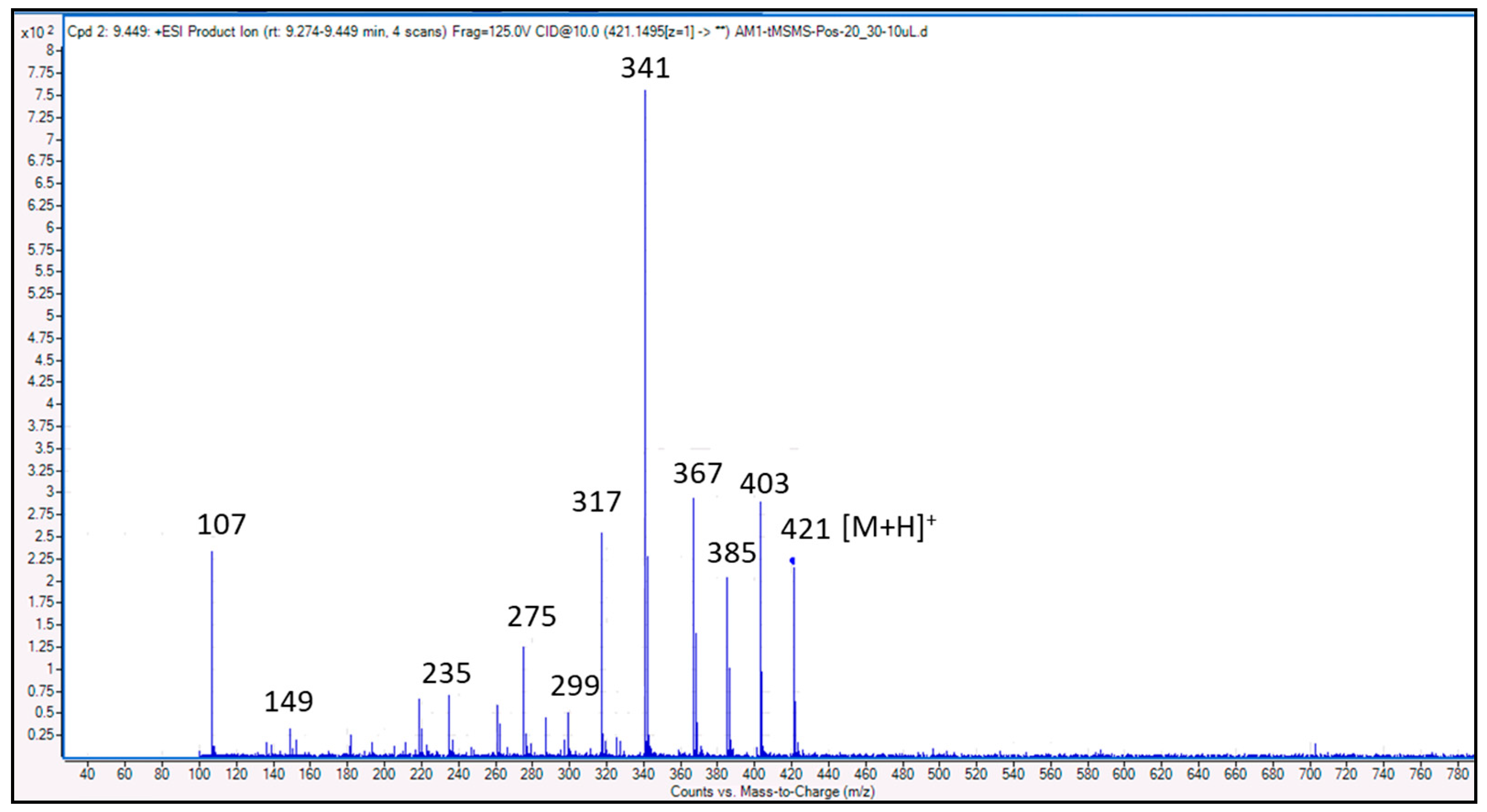
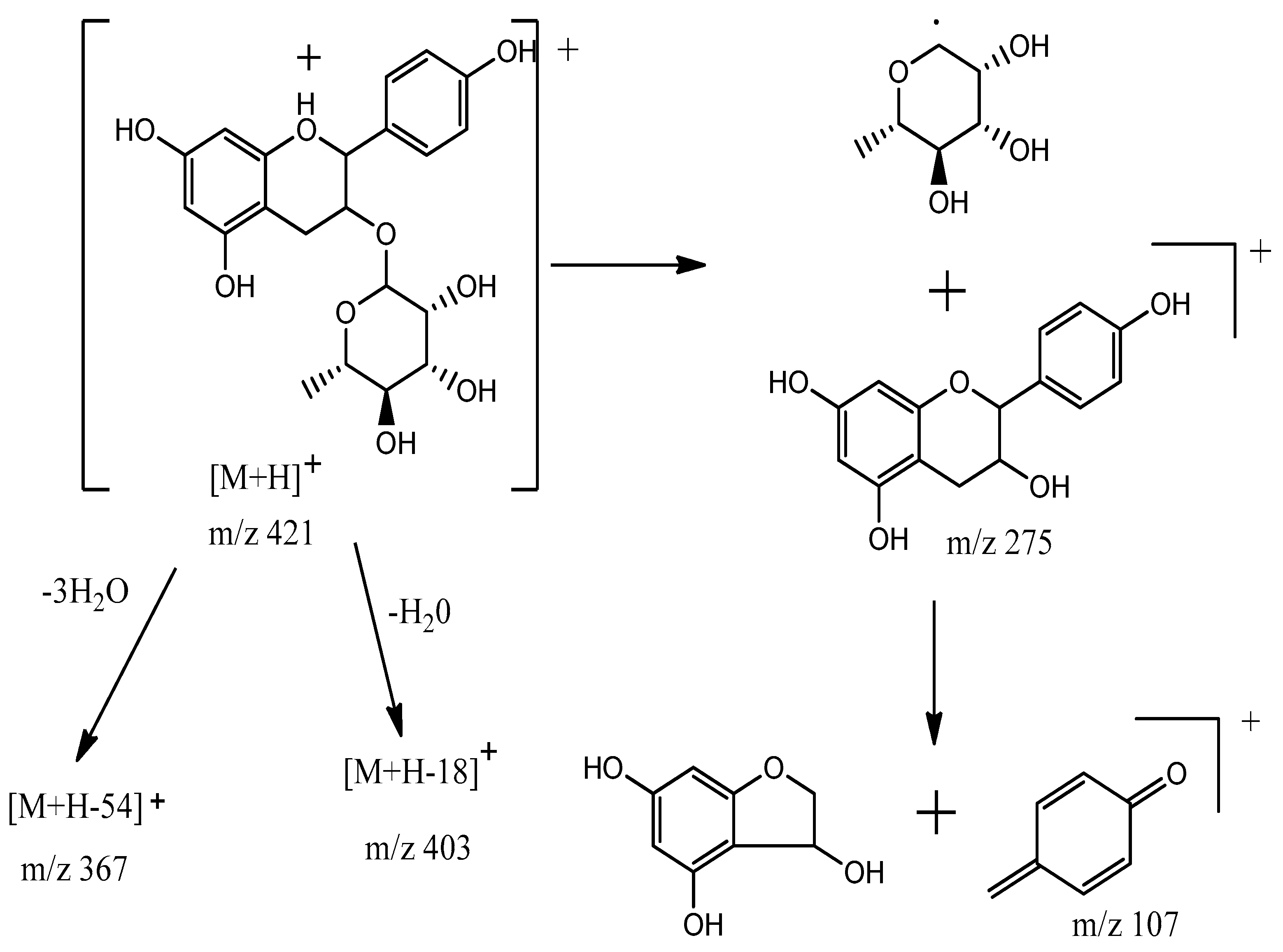
3.4. Identification of Compounds Structures through Fragmentation Analysis (LC-QTOF-MS/MS)
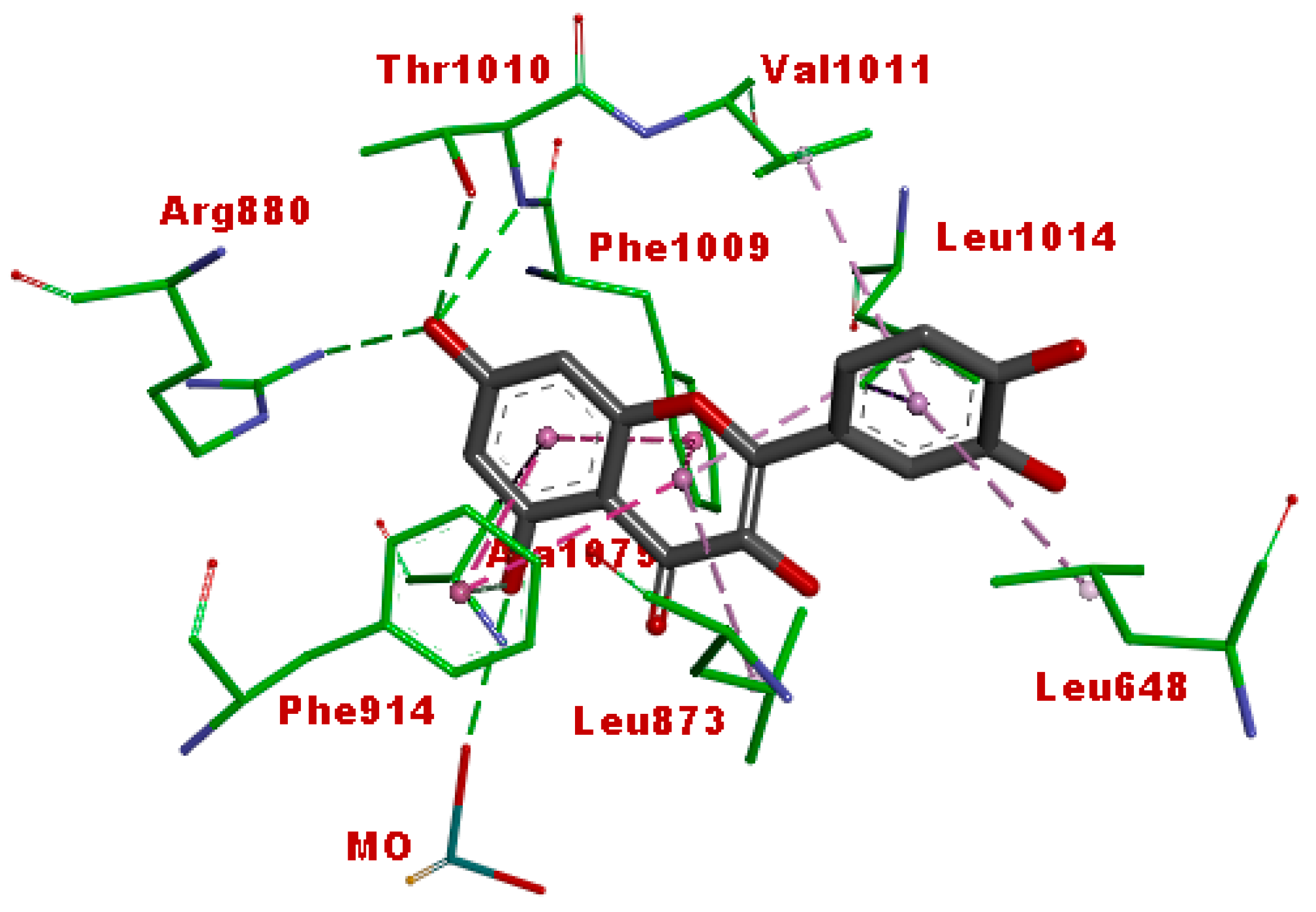
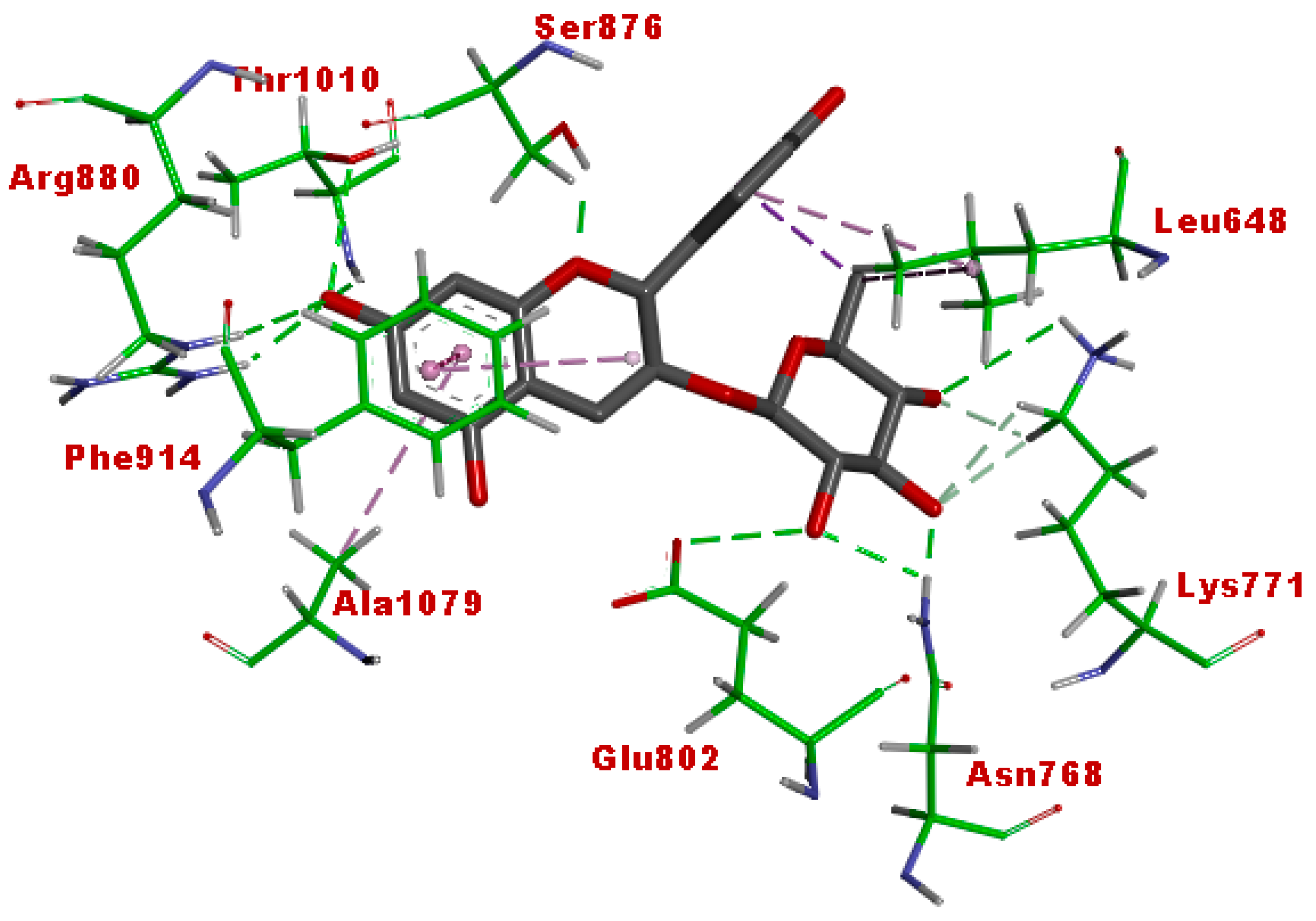
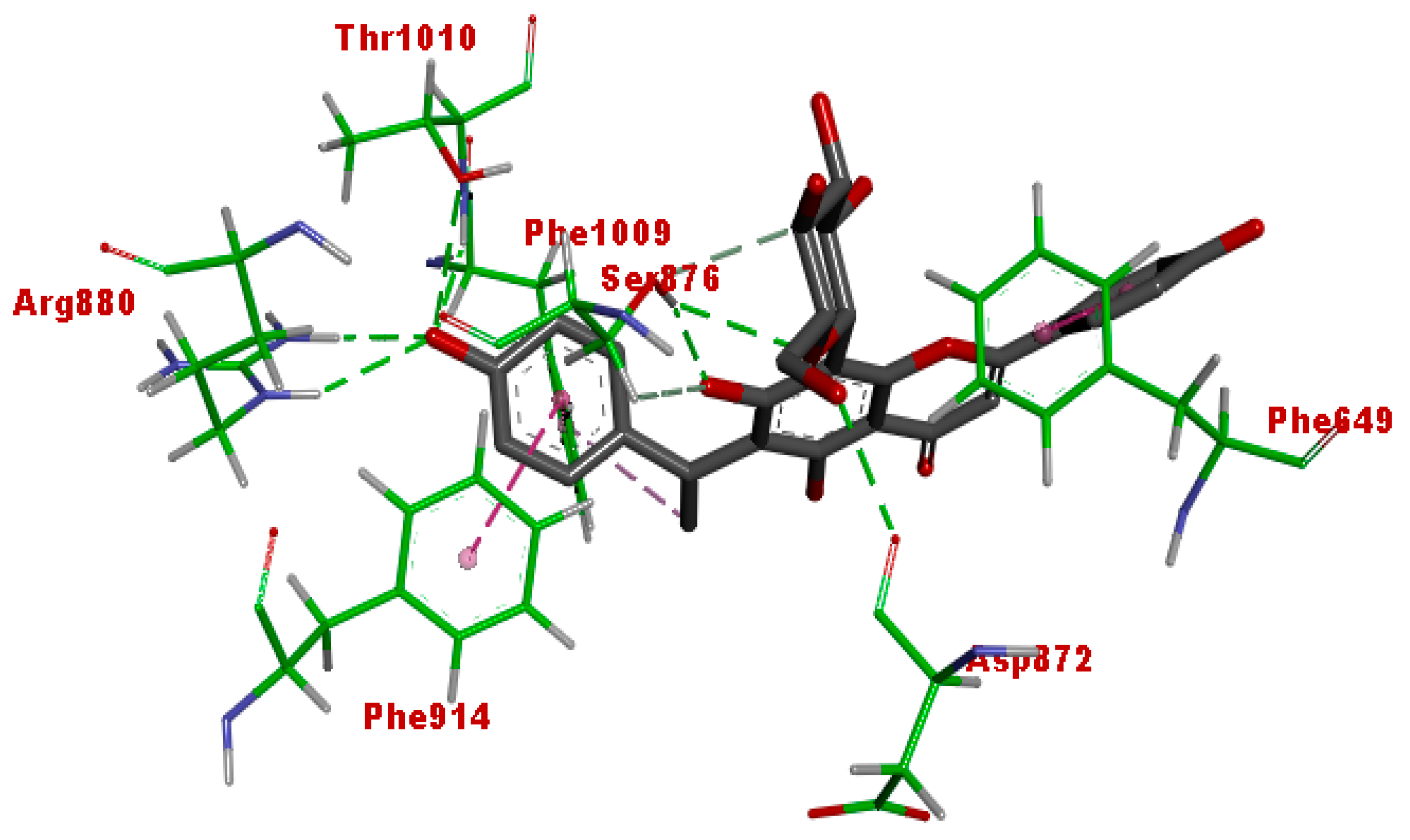
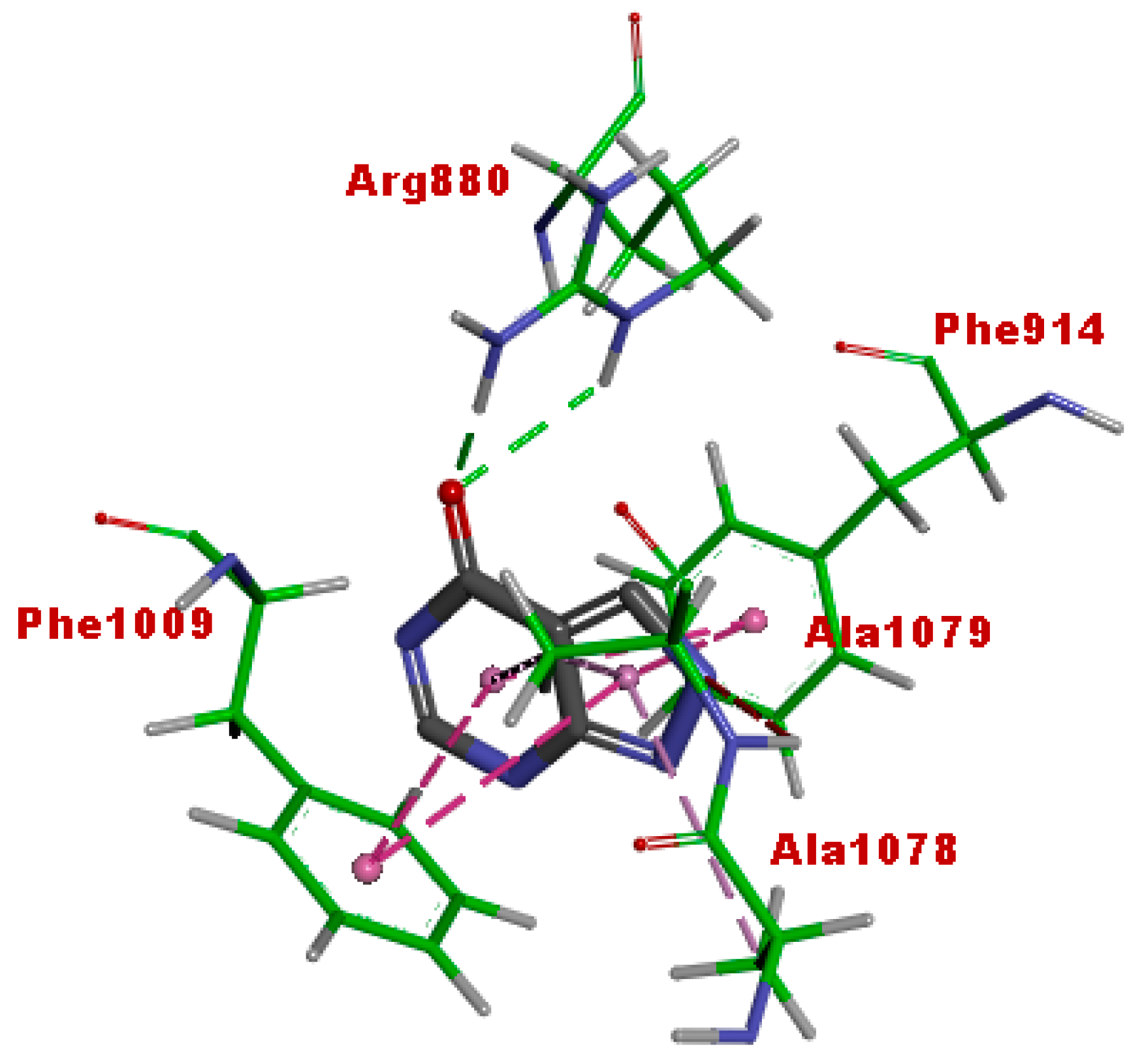
3.5. Molecular Docking
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rahal, A.; Kumar, A.; Singh, V.; Yadav, B.; Tiwari, R.; Chakraborty, S.; Dhama, K. Oxidative stress, prooxidants, and antioxidants: The interplay. BioMed Res. Int. 2014, 2014, 761264. [Google Scholar] [CrossRef] [PubMed]
- Phaniendra, A.; Jestadi, D.B.; Periyasamy, L. Free radicals: Properties, sources, targets, and their implication in various diseases. Indian J. Clin. Biochem. 2015, 30, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Lobo, V.; Patil, A.; Phatak, A.; Chandra, N. Free radicals, antioxidants and functional foods: Impact on human health. Pharmacogn. Rev. 2010, 4, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Matough, F.A.; Budin, S.B.; Hamid, Z.A.; Alwahaibi, N.; Mohamed, J. The role of oxidative stress and antioxidants in diabetic complications. Sult. Qaboos Univ. Med. J. 2012, 12, 5–18. [Google Scholar] [CrossRef]
- Asmat, U.; Abad, K.; Ismail, K. Diabetes mellitus and oxidative stress-a concise review. Saudi Pharm. J. 2016, 24, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Feoli, A.M.P.; Macagnan, F.E.; Piovesan, C.H.; Bodanese, L.C.; Siqueira, I.R. Xanthine oxidase activity is associated with risk factors for cardiovascular disease and inflammatory and oxidative status markers in metabolic syndrome: Effects of a single exercise session. Oxid. Med. Cell. Longev. 2014, 2014, 587083. [Google Scholar] [CrossRef] [PubMed]
- Tam, H.K.; Kelly, A.S.; Metzig, A.M.; Steinberger, J.; Johnson, L.A. Xanthine oxidase and cardiovascular risk in obese children. Child. Obes. 2014, 10, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Sarian, M.N.; Ahmed, Q.U.; Siti Zaiton, M.S.; Alhassan, M.A.; Suganya, M.; Vikneswari, P.; Sharifah, N.A.S.M.; Alfi, K.; Latip, J. Antioxidant and antidiabetic effects of flavonoids: A structure-activity relationship based study. BioMed Res. Int. 2017, 2017, 8386065. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.M.; Ahmed, Q.U.; Soad, S.Z.M.; Tunna, T.S. Animal models and natural products to investigate in vivo and in vitro antidiabetic activity. Biomed. Pharmacother. 2018, 101, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Alhassan, A.M.; Ahmed, Q.U. Averrhoa bilimbi L.: A review of its ethnomedicinal uses, phytochemistry, and pharmacology. J. Pharm. Bioallied Sci. 2016, 8, 265–271. [Google Scholar] [PubMed]
- Precious, L.A.; Coren, J.P.; Celestine, L.A.; Mary, R.M.; Janina, C.E.; Rheinmark, L.S. Topical administration of Averrhoa bilimbi Linn. leaves crude extract prevents UVB-induced oxidative damage in albino mice. The STETH 2012, 6, 29–41. [Google Scholar]
- Nagmoti, D.M.; Yeshwante, S.B.; Wankhede, S.S.; Juvekar, A.R. Hepatoprotective effect of Averrhoa bilimbi Linn. against carbon tetrachloride induced hepatic damage in rats. Pharmacologyonline 2010, 3, 1–6. [Google Scholar]
- Pushparaj, P.; Tan, C.H.; Tan, B.K.H. Effects of Averrhoa bilimbi leaf extract on blood glucose and lipids in streptozotocin-diabetic rats. J. Ethnopharmacol. 2000, 72, 69–76. [Google Scholar] [CrossRef]
- Pushparaj, P.N.; Tan, B.K.H.; Tan, C.H. The mechanism of hypoglycemic action of the semi-purified fractions of Averrhoa bilimbi in streptozotocin-diabetic rats. Life Sci. 2001, 70, 535–547. [Google Scholar] [CrossRef]
- Tan, B.K.H.; Tan, C.H.; Pushparaj, P.N. Anti-diabetic activity of the semi-purified fractions of Averrhoa bilimbi in high fat diet fed-streptozotocin-induced diabetic rats. Life Sci. 2005, 76, 2827–2839. [Google Scholar] [CrossRef] [PubMed]
- Firdouse, S.; Alam, P. Phytochemical investigation of extract of Amorphophallus campanulatus tubers. Int. J. Phytomed. 2011, 3, 32–35. [Google Scholar]
- Ayoola, G.A.; Coker, H.A.; Adesegun, S.A.; Adepoju-Bello, A.A.; Obaweya, K.; Ezennia, E.C.; Atangbayila, T.O. Phytochemical screening and antioxidant activities of some selected medicinal plants used for malaria therapy in Southwestern Nigeria. Trop. J. Pharm. Res. 2008, 7, 1019–1024. [Google Scholar]
- Iqbal, E.; Salim, K.A.; Lim, L.B. Phytochemical screening, total phenolics and antioxidant activities of bark and leaf extracts of Goniothalamus velutinus (Airy Shaw) from Brunei Darussalam. J. King Saud Univ. Sci. 2015, 27, 224–232. [Google Scholar] [CrossRef]
- Nickavar, B.; Kamalinejad, M.; Izadpanah, H. In vitro free radical scavenging activity of five Salvia species. Pak. J. Pharm. Sci. 2007, 20, 291–294. [Google Scholar] [PubMed]
- Barontini, M.; Bernini, R.; Carastro, I.; Gentili, P.; Romani, A. Synthesis and DPPH radical scavenging activity of novel compounds obtained from tyrosol and cinnamic acid derivatives. New J. Chem. 2014, 38, 809–816. [Google Scholar] [CrossRef]
- Thiombiano, A.M.E.; Adama, H.; Jean, B.M.; Bayala, B.; Nabèrè, O.; Samson, G.; Roland, M.N.T.; Moussa, C.; Martin, K.; Millogo, F.; et al. In vitro antioxidant, lipoxygenase and xanthine oxidase inhibitory activity of fractions and macerate from Pandiaka angustifolia (Vahl) Hepper. J. Appl. Pharm. Sci. 2014, 4, 9–13. [Google Scholar]
- Protein Data Bank. Available online: http://www.rcsb.org/ (accessed on 3 July 2018).
- Molecular Graphics Laboratory, The Scripps Research Institute, San Diego, USA. Available online: www.scripps.edu (accessed on 3 July 2018).
- Discovery Studio Visualizer 4.0, Bovia, San Diego, USA. Available online: www.accelrys.com (accessed on 3 July 2018).
- Zhang, H.J.; Hu, Y.J.; Xu, P.; Liang, W.Q.; Zhou, J.; Liu, P.G.; Cheng, L.; Pu, J.B. Screening of potential xanthine oxidase inhibitors in Gnaphalium hypoleucum DC. by immobilized metal affinity chromatography and ultrafiltration-ultra performance liquid chromatography-Mass spectrometry. Molecules 2016, 21, 1242. [Google Scholar] [CrossRef] [PubMed]
- Mishra, K.; Ojha, H.; Chaudhury, N.K. Estimation of antiradical properties of antioxidants using DPPH assay: A critical review and results. Food Chem. 2012, 130, 1036–1043. [Google Scholar] [CrossRef]
- Złotek, U.; Mikulska, S.; Nagajek, M.; Świeca, M. The effect of different solvents and number of extraction steps on the polyphenol content and antioxidant capacity of basil leaves (Ocimum basilicum L.) extracts. Saudi J. Biol. Sci. 2016, 23, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Maiuolo, J.; Oppedisano, F.; Gratteri, S.; Muscoli, C.; Mollace, V. Regulation of uric acid metabolism and excretion. Int. J. Cardiol. 2015, 213, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Dalbeth, N.; Lauterio, T.J.; Wolfe, H.R. Mechanism of action of colchicine in the treatment of gout. Clin. Ther. 2014, 36, 1465–1479. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Zhang, C.; Song, H. Natural products improving hyperuricemia with hepatorenal dual effects. Evid. Based Complement. Altern. Med. 2016, 2016, 7390504. [Google Scholar] [CrossRef] [PubMed]
- Tatsimo, S.; Tamokou, J.; Havyarimana, L.; Csupor, D.; Forgo, P.; Hohmann, J.; Kuiate, J.R.; Tane, P. Antimicrobial and antioxidant activity of kaempferol rhamnoside derivatives from Bryophyllum pinnatum. BMC Res. Notes 2012, 5, 158. [Google Scholar] [CrossRef] [PubMed]
- Spanou, C.; Veskoukis, A.S.; Kerasioti, T.; Kontou, M.; Angelis, A.; Aligiannis, N.; Skaltsounis, A.L.; Kouretas, D. Flavonoid glycosides isolated from unique legume plant extracts as novel inhibitors of xanthine oxidase. PLoS ONE 2012, 7, e32214. [Google Scholar] [CrossRef] [PubMed]
- Lawal, U.; Sze, W.L.; Khozirah, S.; Intan, S.I.; Alfi, K.; Faridah, A. α-Glucosidase inhibitory and antioxidant activities of different Ipomoea aquatica cultivars and LC–MS/MS profiling of the active cultivar. J. Food Biochem. 2016, 41, e12303. [Google Scholar] [CrossRef]
- Colombo, R.; Yariwake, J.H.; McCullagh, M. Study of C-and O-glycosylflavones in sugarcane extracts using liquid chromatography: Exact mass measurement mass spectrometry. J. Braz. Chem. Soc. 2008, 19, 483–490. [Google Scholar] [CrossRef]
- Martucci, M.E.P.; De Vos, R.C.; Carollo, C.A.; Gobbo-Neto, L. Metabolomics as a potential chemotaxonomical tool: Application in the genus Vernonia Schreb. PLoS ONE 2014, 9, e93149. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Pauff, J.M.; Hille, R. X-ray crystal structure of a xanthine oxidase complex with the flavonoid inhibitor quercetin. J. Nat. Prod. 2014, 77, 1693–1699. [Google Scholar] [CrossRef] [PubMed]

| Bioactive | Test/Procedure | Observation | Inference |
|---|---|---|---|
| Alkaloid | Dragendorff’s reagent | Orange red ppt. | + |
| Mayer’s reagent | Cream ppt. | + | |
| Saponin | Frothing test | Frothing | ++ |
| Terpenoid | Salkowski test: | ||
| Chloroform + H2SO4 | Brownish red ring at the junction | +++ | |
| Flavonoid | Shinoda test: | ||
| Flavones: Mg filling | Reddish brown | ++ | |
| Flavonols: Zn pellet | Amber | ++ | |
| Free anthraquinone | Chloroform + NH3 (10%) | No change from dirty green | − |
| Combined anthraquinone | 10% HCl + Chloroform | No change from colourless | − |
| Sample | IC50 (μg/mL) |
|---|---|
| Crude methanolic extract | 10.53 ± 0.72 * |
| Hexane fraction | >1000 |
| Chloroform fraction | 13.44 ± 1.00 * |
| n-Butanol fraction | 4.14 ± 0.21 * |
| Ascorbic acid (positive control) | 5.52 ± 0.29 |
| Sample | IC50 μg/mL |
|---|---|
| Crude methanolic leaves extract | >1000 |
| Hexane fraction | >1000 |
| Chloroform fraction | >1000 |
| n-Butanol fraction | 64.84 ± 3.93 * |
| Allopurinol (positive control) | 16.21 ± 0.91 |
| Number | Tentative Compounds | Retention Time | Molecular Formula/Molecular Weight (M+) |
|---|---|---|---|
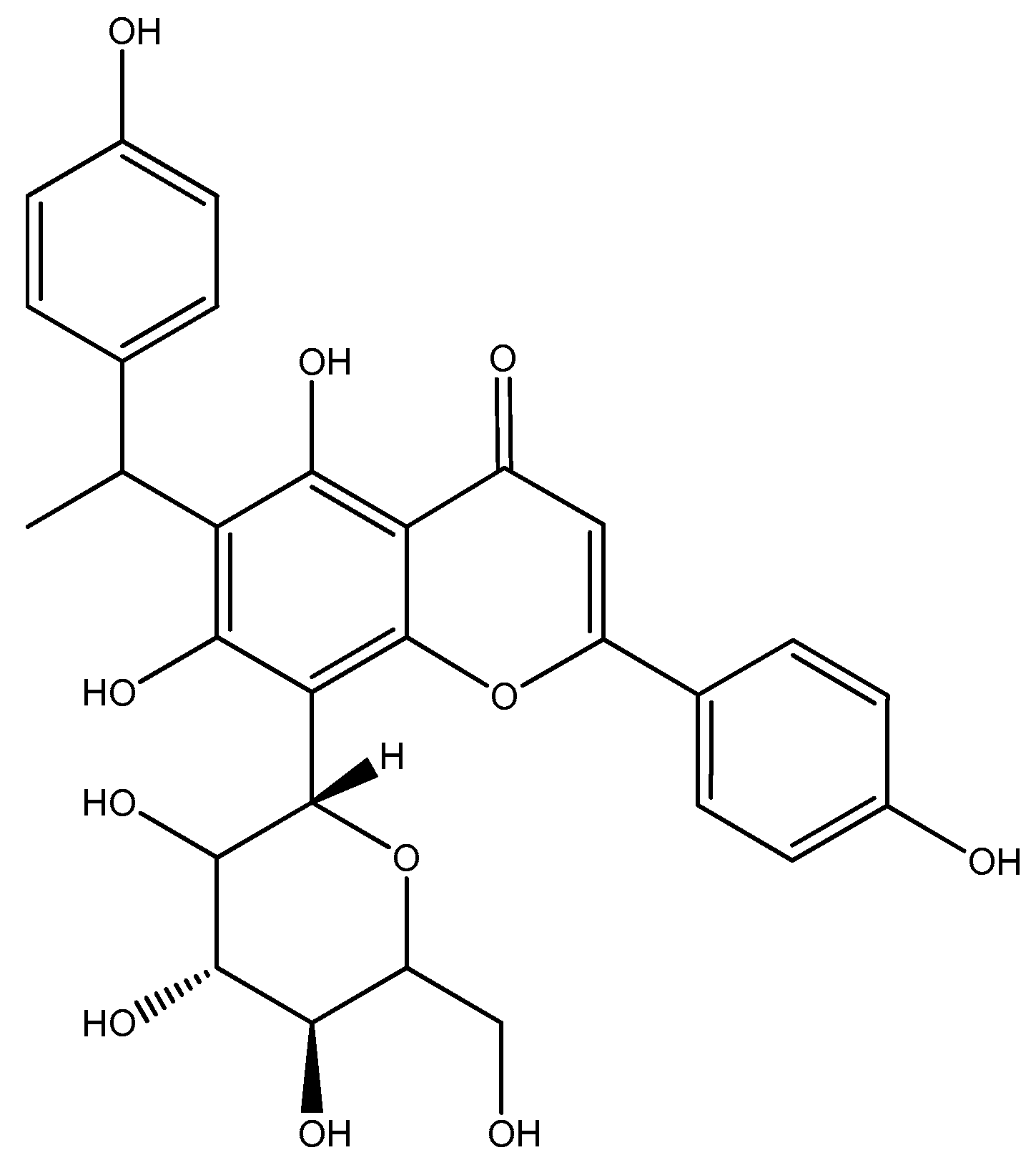
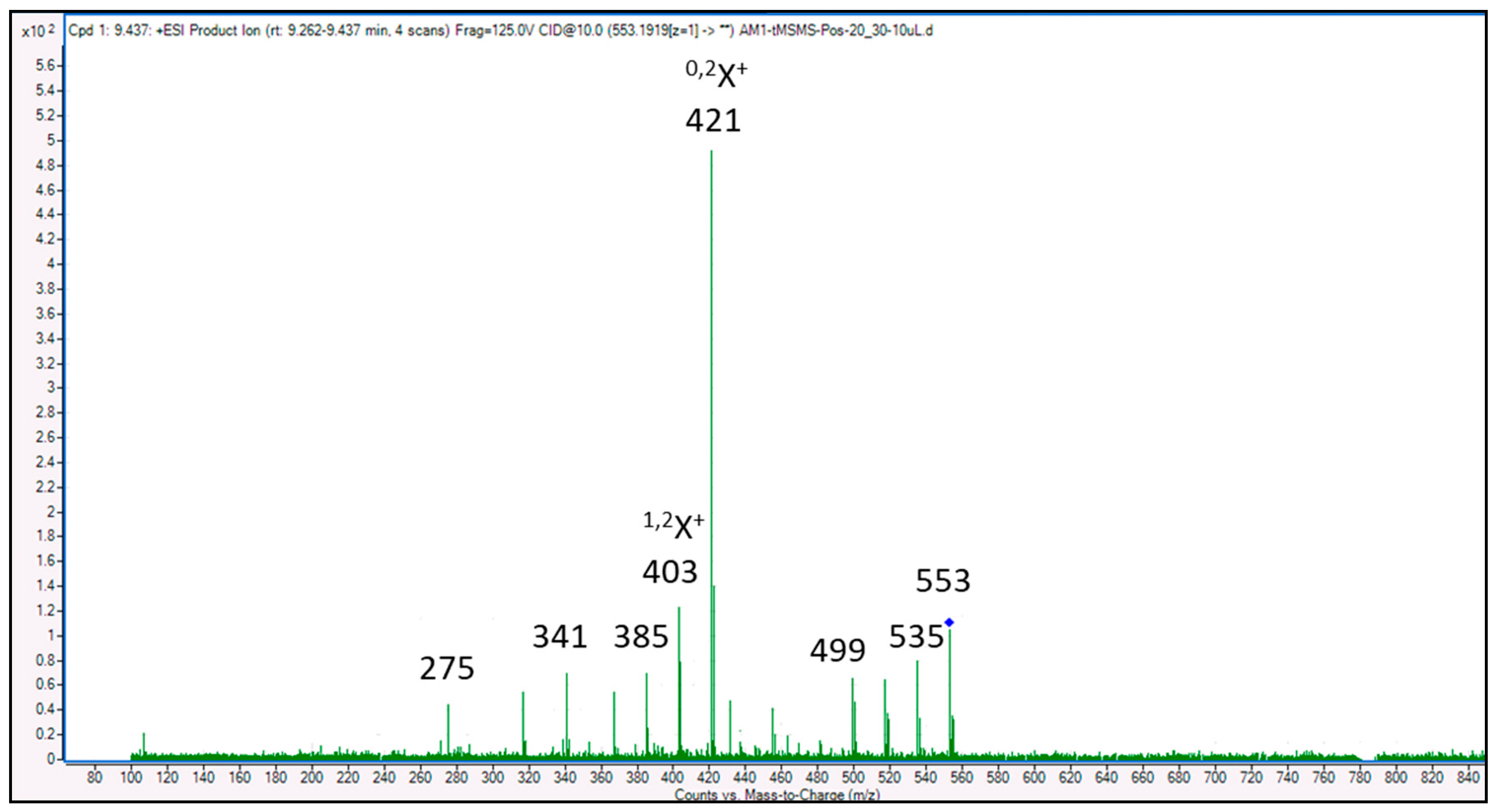
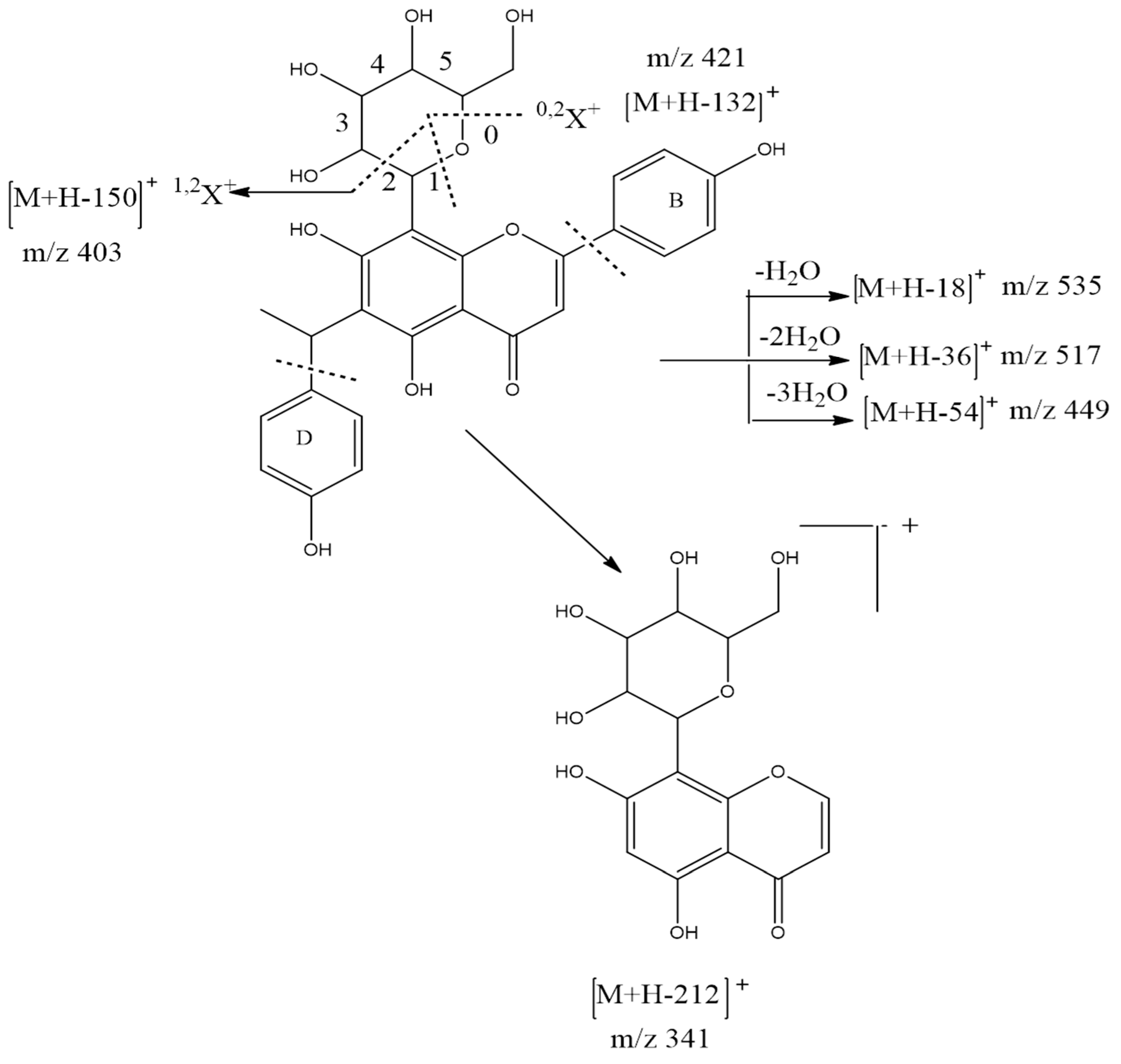
| 1 | 5,7,4′-Trihydroxy-6-(1-ethyl-4-hydroxyphenyl)flavone-8-C-glucoside (Cucumerin A) | 9.305 | C29H28O11/552.185 |
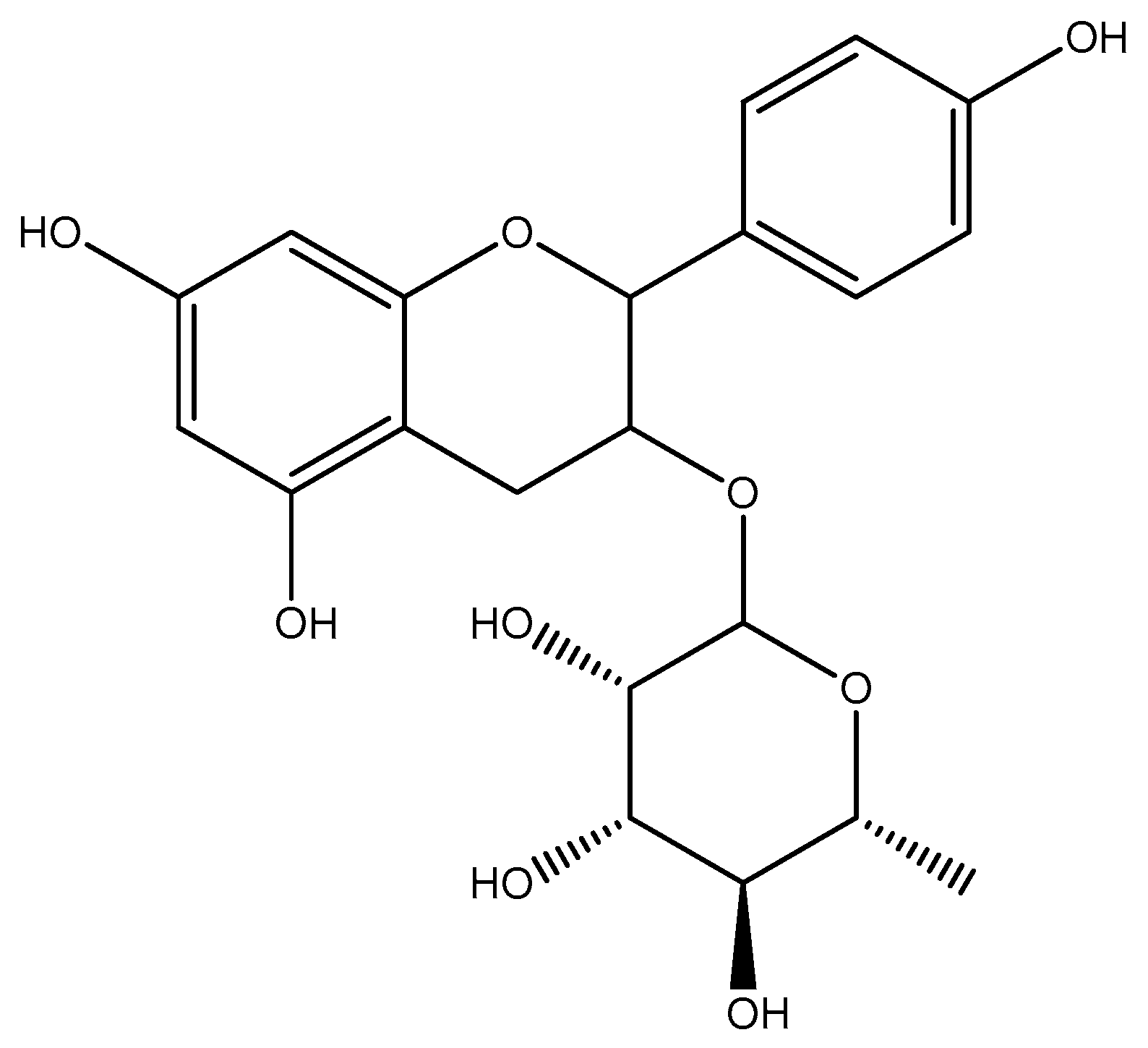
| 2 | Afzelechin 3-O-alpha-l-rhamnopyranoside | 9.305 | C21H24O9/420.142 |
| 3 | Ethyl 3-(N-butylacetamido)propionate | 9.997 | C11H21NO3/215.152 |
| 4 | Elaeokanine C | 10.749 | C12H21NO2/211.156 |
| 5 | 2-Ethyl-dodecanoic acid | 10.958 | C14H28O2/228.208 |
| 6 | Isoavocadienofuran | 11.063 | C17H26O/246.198 |
| 7 | (5alpha,8beta,9beta)-5,9-Epoxy-3,6-megastigmadien-8-ol | 11.451 | C13H20O2/208.144 |
| 8 | Diglycidyl resorcinol ether | 12.120 | C12H14O4/222.090 |
| 9 | 19-Hydroxycinnzeylanol 19-glucoside | 12.212 | C26H42O13/562.264 |
| 10 | Xestoaminol C | 12.236 | C14H31NO/229.240 |
| 11 | Phytosphingosine | 12.266 | C18H39NO3/317.293 |
| 12 | 2-Hydroxyhexadecanoic acid | 12.334 | C16H32O3/272.236 |
| 13 | Pentadecanal | 12.408 | C15H30O/226.230 |
| 14 | Anapheline | 12.707 | C13H24N2O/224.188 |
| 15 | Palmitic amide | 12.713 | C16H33NO/255.256 |
| 16 | Tetradecylamine | 12.769 | C14H31N/213.246 |
| 17 | Pentadecanoyl-EA | 12.825 | C17H35NO2/285.266 |
| 18 | Codonopsine | 12.879 | C14H21NO4/267.147 |
| 19 | Enigmol | 13.365 | C18H39NO2/301.297 |
| 20 | 7-Hexadecen-1-ol | 13.430 | C16H32O/240.245 |
| 21 | (Z)-2-Amino-1-hydroxyoctadec-4-en-3-one | 13.498 | C18H35NO2/297.266 |
| 22 | Dihydroceramide C2 | 13.500 | C20H41NO3/343.308 |
| 23 | 6-Hydroxysphingosine | 13.502 | C18H37NO3/315.277 |
| 24 | Methyl 8-[2-(2-formyl-vinyl)-3-hydroxy-5-oxo-cyclopentyl]-octanoate | 13.585 | C17H26O5/310.179 |
| 25 | Dehydrophytosphingosine | 13.753 | C18H37NO3/315.277 |
| 26 | 14-Methyl-8-hexadecen-1-ol | 13.795 | C17H34O/254.261 |
| 27 | Nonadecanal | 13.800 | C19H38O/282.291 |
| 28 | Oleoyl Ethanolamide | 15.294 | C20H39NO2/325.296 |
| 29 | Linoleamide | 15.308 | C18H33NO/279.256 |
| Ligand | Free Binding Energy (Kcal/mol) | Estimated Ki |
|---|---|---|
| Afzelechin 3-O-alpha-l-rhamnopyranoside | −11.43 | 4.18 nM |
| Cucumerin A | −12.0 | 1.59 nM |
| Quercetin | −10.18 | 34.57 nM |
| Allopurinol | −5.66 | 70.74 uM |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, Q.U.; Alhassan, A.M.; Khatib, A.; Shah, S.A.A.; Hasan, M.M.; Sarian, M.N. Antiradical and Xanthine Oxidase Inhibitory Activity Evaluations of Averrhoa bilimbi L. Leaves and Tentative Identification of Bioactive Constituents through LC-QTOF-MS/MS and Molecular Docking Approach. Antioxidants 2018, 7, 137. https://doi.org/10.3390/antiox7100137
Ahmed QU, Alhassan AM, Khatib A, Shah SAA, Hasan MM, Sarian MN. Antiradical and Xanthine Oxidase Inhibitory Activity Evaluations of Averrhoa bilimbi L. Leaves and Tentative Identification of Bioactive Constituents through LC-QTOF-MS/MS and Molecular Docking Approach. Antioxidants. 2018; 7(10):137. https://doi.org/10.3390/antiox7100137
Chicago/Turabian StyleAhmed, Qamar Uddin, Alhassan Muhammad Alhassan, Alfi Khatib, Syed Adnan Ali Shah, Muhammad Mahmudul Hasan, and Murni Nazira Sarian. 2018. "Antiradical and Xanthine Oxidase Inhibitory Activity Evaluations of Averrhoa bilimbi L. Leaves and Tentative Identification of Bioactive Constituents through LC-QTOF-MS/MS and Molecular Docking Approach" Antioxidants 7, no. 10: 137. https://doi.org/10.3390/antiox7100137
APA StyleAhmed, Q. U., Alhassan, A. M., Khatib, A., Shah, S. A. A., Hasan, M. M., & Sarian, M. N. (2018). Antiradical and Xanthine Oxidase Inhibitory Activity Evaluations of Averrhoa bilimbi L. Leaves and Tentative Identification of Bioactive Constituents through LC-QTOF-MS/MS and Molecular Docking Approach. Antioxidants, 7(10), 137. https://doi.org/10.3390/antiox7100137





