Energy Intake-Dependent Genetic Associations with Obesity Risk: BDNF Val66Met Polymorphism and Interactions with Dietary Bioactive Compounds
Abstract
1. Introduction
2. Methods
2.1. Participants
2.2. Demographic, Anthropometric, and Biochemical Data Collection
2.3. Obesity Definition and Grouping According to Energy Intake
2.4. Dietary Assessment and Nutritional Analysis
2.5. Genotyping and Genetic Variant Analysis
2.6. Genetic Interaction and Risk Score Modeling
2.7. Collection and Screening of Bioactive Compounds from Foods with Low Binding Energy with Brain-Derived Neurotrophic Factor (BDNF) rs6265
2.8. Clustering of the Natural Compounds with Lower Binding Energy
2.9. Statistical Analysis
3. Results
3.1. Baseline Characteristics of the Participants
3.2. Anthropometric Association
3.3. Risk of Metabolic Syndrome
3.4. Intake of Antioxidant Nutrients and Bioactive Compounds
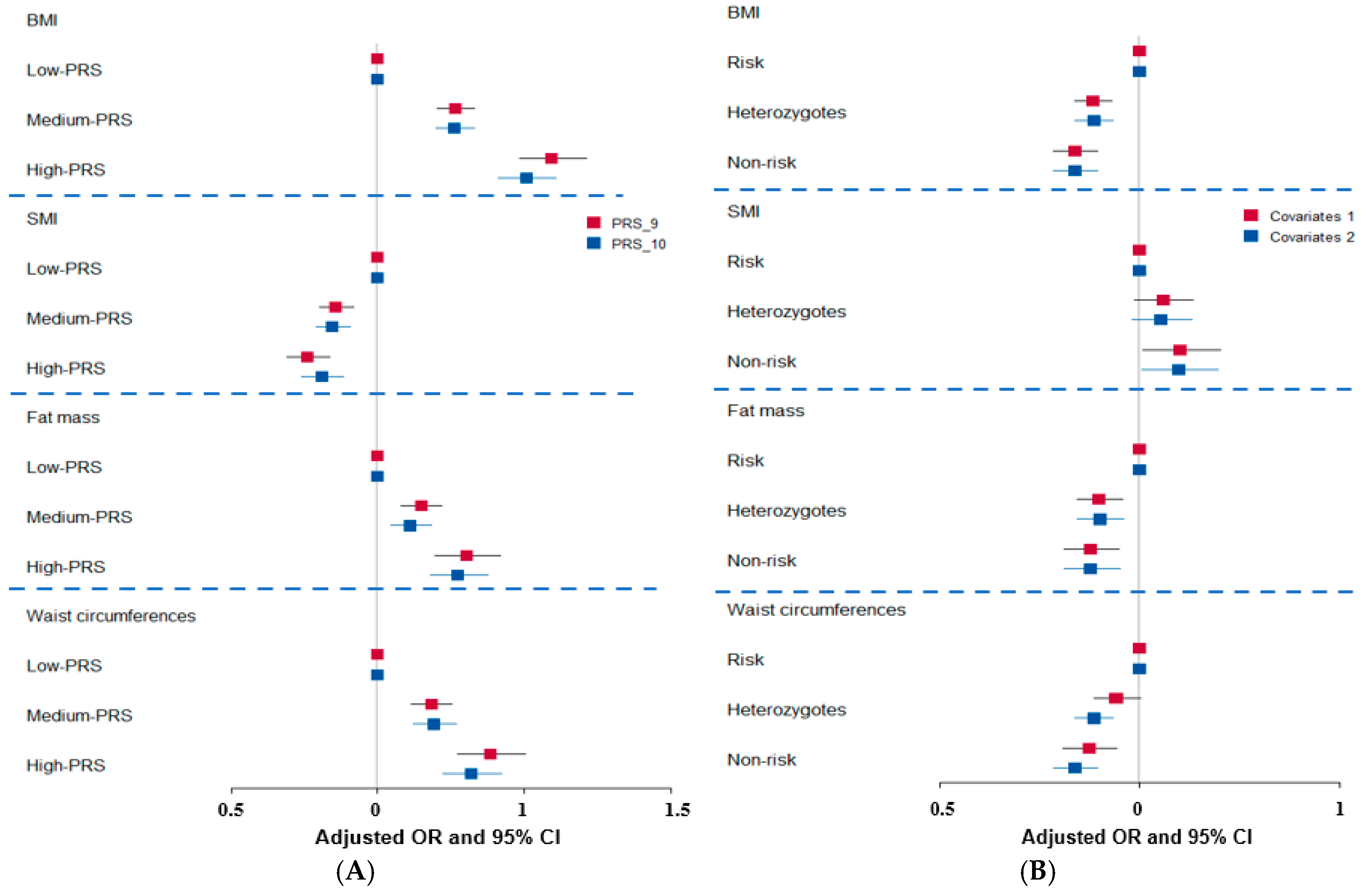
3.5. Genetic Impact
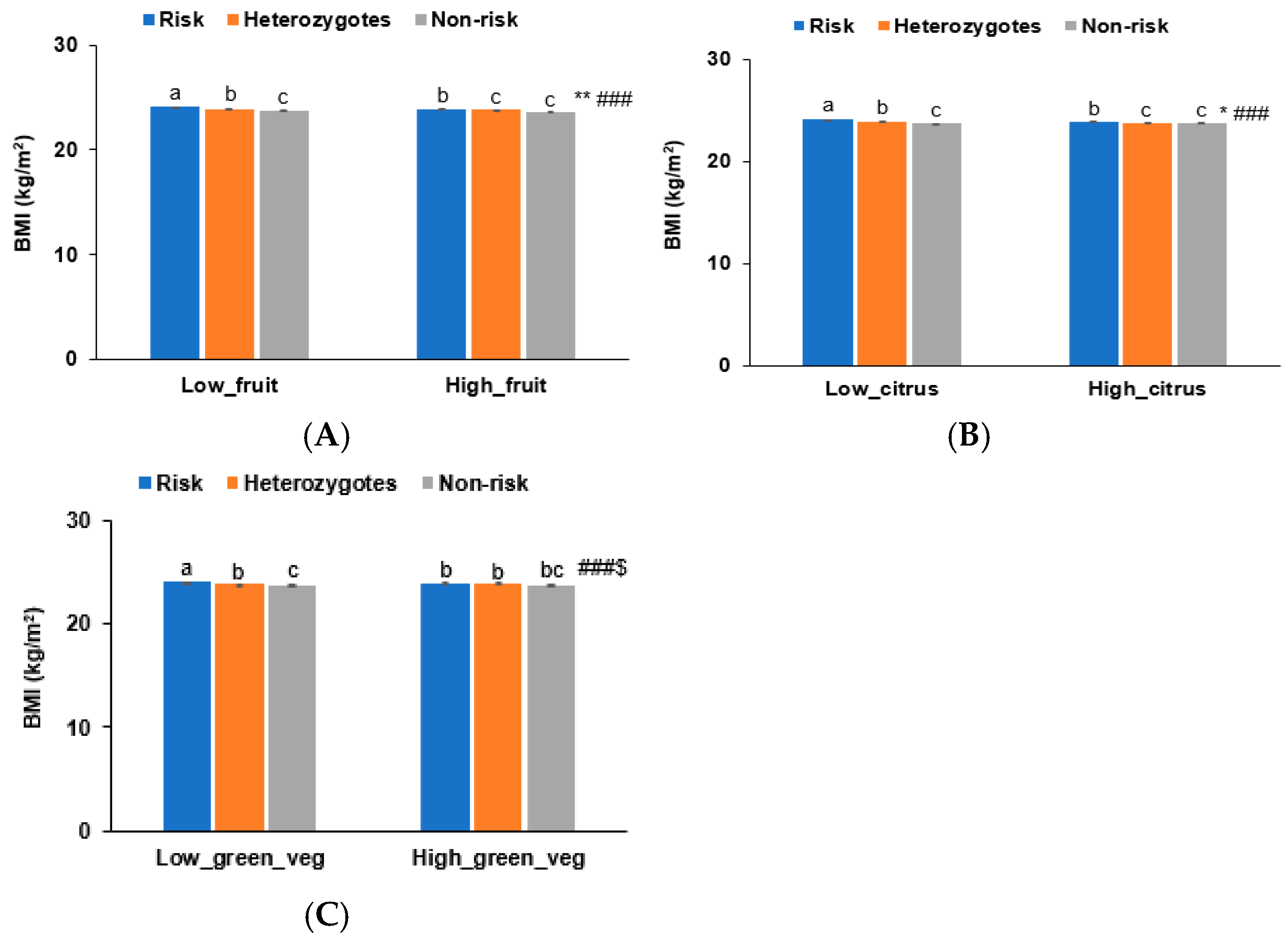
3.6. Interaction of BNDF Val66Met Polymorphisms with Food Intake Influencing Obesity
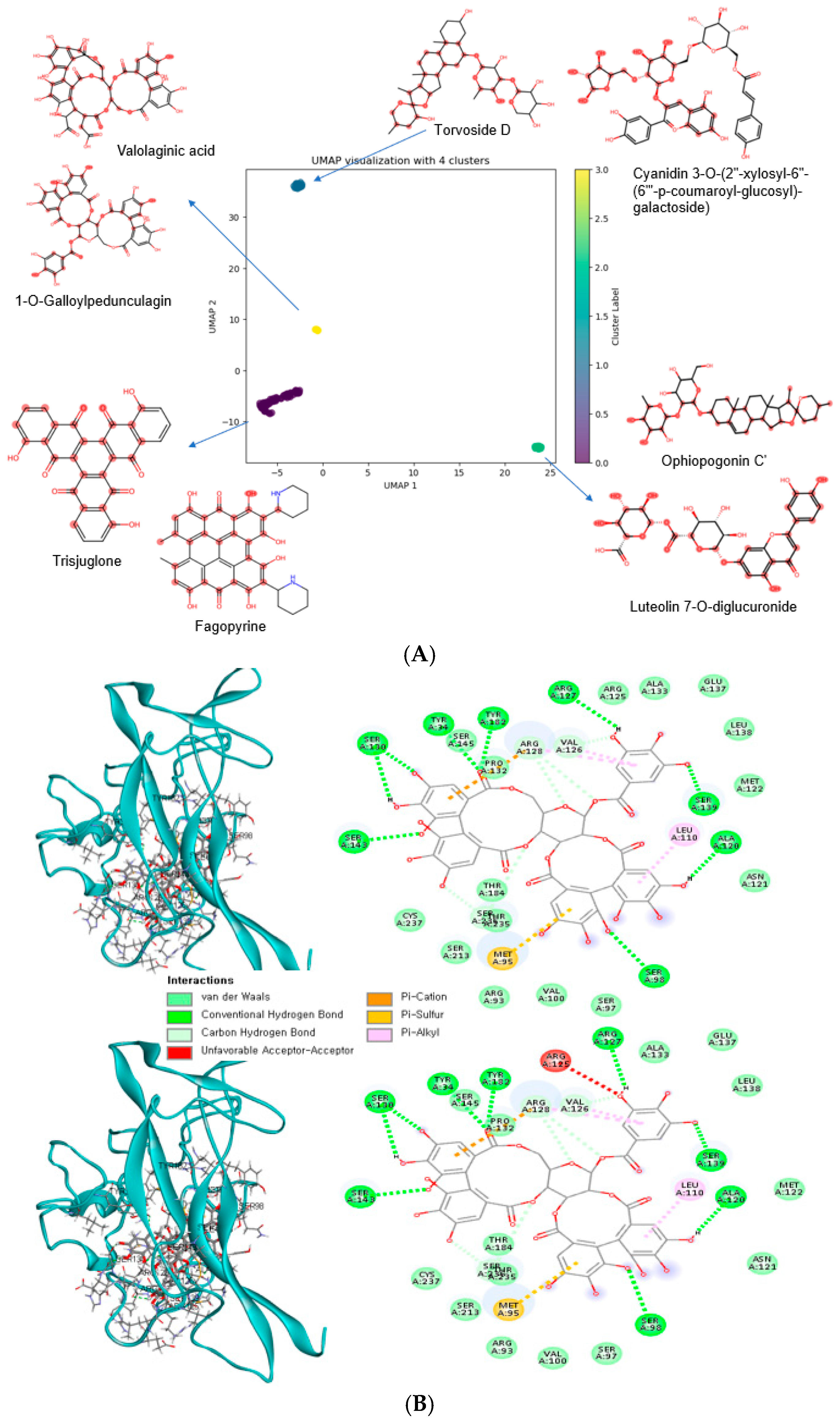
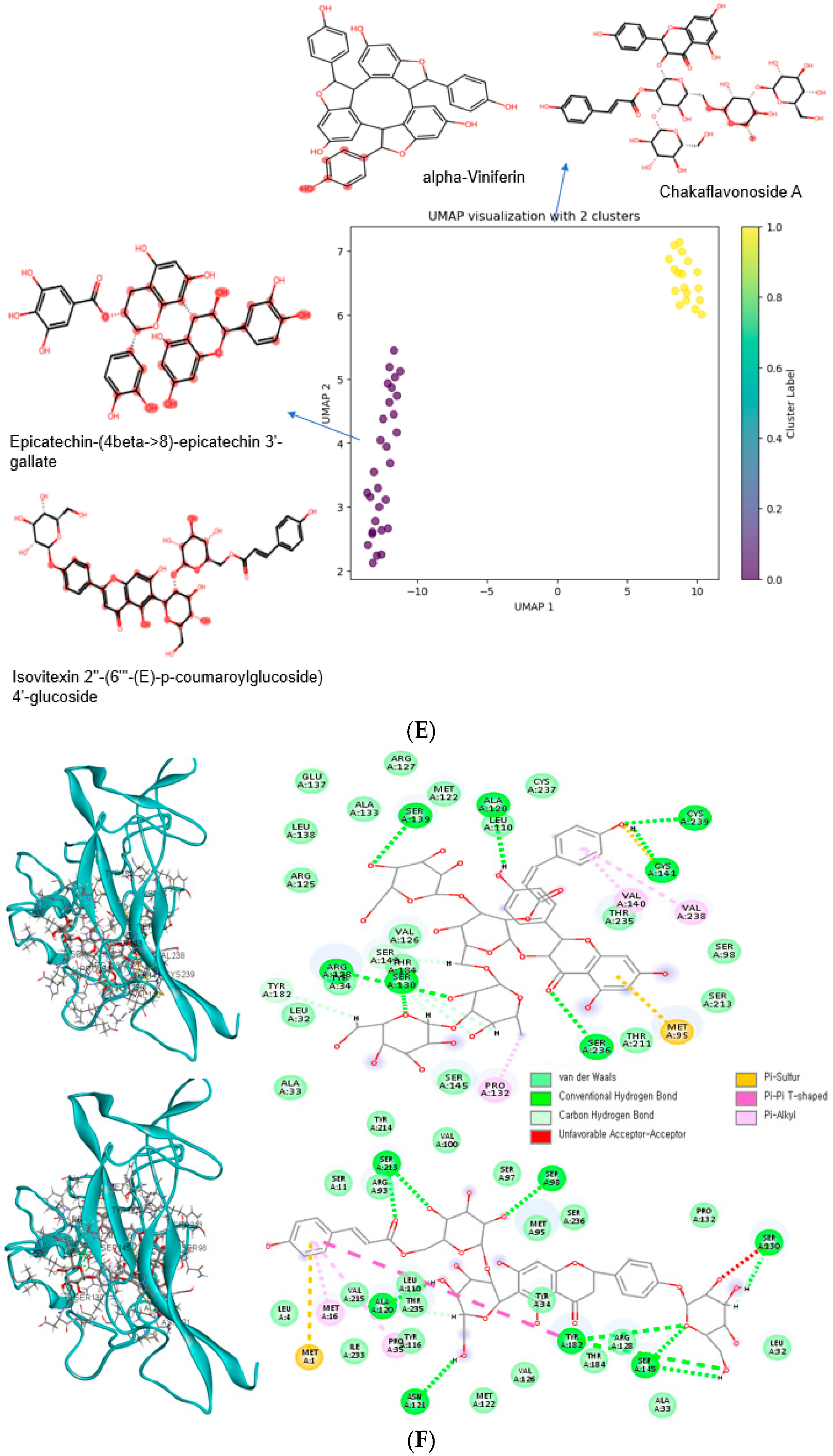
3.7. Bioactive Compounds with Low Binding Energy with Wild-Type (WT) and Mutant-Type (MT) BDNF Val66Met (rs6265)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wen, X.; Zhang, B.; Wu, B.; Xiao, H.; Li, Z.; Li, R.; Xu, X.; Li, T. Signaling pathways in obesity: Mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2022, 7, 298. [Google Scholar] [CrossRef] [PubMed]
- Hall, K.D.; Farooqi, I.S.; Friedman, J.M.; Klein, S.; Loos, R.J.F.; Mangelsdorf, D.J.; O’Rahilly, S.; Ravussin, E.; Redman, L.M.; Ryan, D.H.; et al. The energy balance model of obesity: Beyond calories in, calories out. Am. J. Clin. Nutr. 2022, 115, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Duarte, M.K.R.N.; Leite-Lais, L.; Agnez-Lima, L.F.; Maciel, B.L.L.; Morais, A.H.d.A. Obesity and Nutrigenetics Testing: New Insights. Nutrients 2024, 16, 607. [Google Scholar] [CrossRef] [PubMed]
- Bonomini, F. Antioxidants and Obesity. Int. J. Mol. Sci. 2023, 24, 12832. [Google Scholar] [CrossRef]
- Manna, P.; Jain, S.K. Obesity, Oxidative Stress, Adipose Tissue Dysfunction, and the Associated Health Risks: Causes and Therapeutic Strategies. Metab. Syndr. Relat. Disord. 2015, 13, 423–444. [Google Scholar] [CrossRef]
- Wadden, T.A.; Tronieri, J.S.; Butryn, M.L. Lifestyle modification approaches for the treatment of obesity in adults. Am. Psychol. 2020, 75, 235–251. [Google Scholar] [CrossRef]
- Qualls-Creekmore, E.; Marlatt, K.L.; Aarts, E.; Bruce-Keller, A.; Church, T.S.; Clément, K.; Fisher, J.O.; Gordon-Larsen, P.; Morrison, C.D.; Raybould, H.E.; et al. What Should I Eat and Why? The Environmental, Genetic, and Behavioral Determinants of Food Choice: Summary from a Pennington Scientific Symposium. Obesity 2020, 28, 1386–1396. [Google Scholar] [CrossRef]
- Li, J.; Song, F. A causal relationship between antioxidants, minerals and vitamins and metabolic syndrome traits: A Mendelian randomization study. Diabetol. Metab. Syndr. 2023, 15, 194. [Google Scholar] [CrossRef]
- Franco-San Sebastián, D.; Alaniz-Monreal, S.; Rabadán-Chávez, G.; Vázquez-Manjarrez, N.; Hernández-Ortega, M.; Gutiérrez-Salmeán, G. Anthocyanins: Potential Therapeutic Approaches towards Obesity and Diabetes Mellitus Type 2. Molecules 2023, 28, 1237. [Google Scholar] [CrossRef]
- Park, S. Interaction of polygenic variants specific for abdominal obesity risk with energy metabolism in large Korean cohorts. Nutr. Bull. 2022, 47, 307–321. [Google Scholar] [CrossRef]
- Daily, J.W.; Park, S. Association of Plant-Based and High-Protein Diets with a Lower Obesity Risk Defined by Fat Mass in Middle-Aged and Elderly Persons with a High Genetic Risk of Obesity. Nutrients 2023, 15, 1063. [Google Scholar] [CrossRef] [PubMed]
- Escalante-Aburto, A.; Mendoza-Córdova, M.Y.; Mahady, G.B.; Luna-Vital, D.A.; Gutiérrez-Uribe, J.A.; Chuck-Hernández, C. Consumption of dietary anthocyanins and their association with a reduction in obesity biomarkers and the prevention of obesity. Trends Food Sci. Technol. 2023, 140, 104140. [Google Scholar] [CrossRef]
- Mahboob, A.; Samuel, S.M.; Mohamed, A.; Wani, M.Y.; Ghorbel, S.; Miled, N.; Büsselberg, D.; Chaari, A. Role of flavonoids in controlling obesity: Molecular targets and mechanisms. Front. Nutr. 2023, 10, 1177897. [Google Scholar] [CrossRef] [PubMed]
- Park, S. Association of polygenic risk scores for insulin resistance risk and their interaction with a plant-based diet, especially fruits, vitamin C, and flavonoid intake, in Asian adults. Nutrition 2023, 111, 112007. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, C.; Wu, X. Development and Validation of an Insulin Resistance Predicting Model Using a Machine-Learning Approach in a Population-Based Cohort in Korea. Diagnostics 2022, 12, 212. [Google Scholar] [CrossRef]
- Wu, X.; Park, S. An Inverse Relation between Hyperglycemia and Skeletal Muscle Mass Predicted by Using a Machine Learning Approach in Middle-Aged and Older Adults in Large Cohorts. J. Clin. Med. 2021, 10, 2133. [Google Scholar] [CrossRef]
- Kim, E.K.; Kim, J.H.; Kim, M.H.; Ndahimana, D.; Yean, S.E.; Yoon, J.S.; Kim, J.H.; Park, J.; Ishikawa-Takata, K. Validation of dietary reference intake equations for estimating energy requirements in Korean adults by using the doubly labeled water method. Nutr. Res. Pract. 2017, 11, 300–306. [Google Scholar] [CrossRef]
- Liu, M.; Park, S. The Role of PNPLA3_rs738409 Gene Variant, Lifestyle Factors, and Bioactive Compounds in Nonalcoholic Fatty Liver Disease: A Population-Based and Molecular Approach towards Healthy Nutrition. Nutrients 2024, 16, 1239. [Google Scholar] [CrossRef]
- Zhou, J.; Kim, Y.K.; Li, C.; Park, S. Natural compounds for Alzheimer’s prevention and treatment: Integrating SELFormer-based computational screening with experimental validation. Comput. Biol. Med. 2024, 185, 109523. [Google Scholar] [CrossRef]
- Kumar, S.; Malviya, R.; Sundram, S. Nutritional neurology: Unraveling cellular mechanisms of natural supplements in brain health. Hum. Nutr. Metab. 2024, 35, 200232. [Google Scholar] [CrossRef]
- Rodriguez, R.L.; Albeck, J.G.; Taha, A.Y.; Ori-McKenney, K.M.; Recanzone, G.H.; Stradleigh, T.W.; Hernandez, B.C.; Tang, F.-Y.V.; Chiang, E.-P.I.; Cruz-Orengo, L. Impact of diet-derived signaling molecules on human cognition: Exploring the food–brain axis. npj Sci. Food 2017, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.; Marroni, F.; Hayward, C.; Franklin, C.S.; Kirichenko, A.V.; Jonasson, I.; Hicks, A.A.; Vitart, V.; Isaacs, A.; Axenovich, T.; et al. Linkage and genome-wide association analysis of obesity-related phenotypes: Association of weight with the MGAT1 gene. Obesity 2010, 18, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Loos, R.J.F.; Yeo, G.S.H. The genetics of obesity: From discovery to biology. Nat. Rev. Genet. 2022, 23, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Rupérez, A.I.; Gil, A.; Aguilera, C.M. Genetics of Oxidative Stress in Obesity. Int. J. Mol. Sci. 2014, 15, 3118–3144. [Google Scholar] [CrossRef]
- Hernández-Guerrero, C.; Parra-Carriedo, A.; Ruiz-de-Santiago, D.; Galicia-Castillo, O.; Buenrostro-Jáuregui, M.; Díaz-Gutiérrez, C. Genetic polymorphisms of antioxidant enzymes CAT and SOD affect the outcome of clinical, biochemical, and anthropometric variables in people with obesity under a dietary intervention. Genes Nutr. 2018, 13, 1. [Google Scholar] [CrossRef]
- Jiménez-Ortega, R.F.; Aparicio-Bautista, D.I.; Becerra-Cervera, A.; López-Montoya, P.; León-Reyes, G.; Flores-Morales, J.; Castillejos-López, M.; Hidalgo-Bravo, A.; Salmerón, J.; Rivera-Paredez, B.; et al. Association Study between Antioxidant Nutrient Intake and Low Bone Mineral Density with Oxidative Stress-Single Nucleotide Variants: GPX1 (rs1050450 and rs17650792), SOD2 (rs4880) and CAT (rs769217) in Mexican Women. Antioxidants 2023, 12, 2089. [Google Scholar] [CrossRef]
- Rahal, D.; Chiang, J.J.; Huynh, V.W.; Bower, J.E.; McCreath, H.; Fuligni, A.J. Low subjective social status is associated with daily selection of fewer healthy foods and more high-fat/high sugar foods. Appetite 2023, 180, 106338. [Google Scholar] [CrossRef]
- Feraco, A.; Gorini, S.; Camajani, E.; Filardi, T.; Karav, S.; Cava, E.; Strollo, R.; Padua, E.; Caprio, M.; Armani, A.; et al. Gender differences in dietary patterns and physical activity: An insight with principal component analysis (PCA). J. Transl. Med. 2024, 22, 1112. [Google Scholar] [CrossRef]
- Lundsgaard, A.M.; Kiens, B. Gender differences in skeletal muscle substrate metabolism-molecular mechanisms and insulin sensitivity. Front. Endocrinol. 2014, 5, 195. [Google Scholar] [CrossRef]
- Speakman, J.R. The ‘Fat Mass and Obesity-Related’ (FTO) gene: Mechanisms of Impact on Obesity and Energy Balance. Curr. Obes. Rep. 2015, 4, 73–91. [Google Scholar] [CrossRef]
- Wang, P.; Loh, K.H.; Wu, M.; Morgan, D.A.; Schneeberger, M.; Yu, X.; Chi, J.; Kosse, C.; Kim, D.; Rahmouni, K.; et al. A leptin-BDNF pathway regulating sympathetic innervation of adipose tissue. Nature 2020, 583, 839–844. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, A.; Renström, F.; Lyssenko, V.; Brito, E.C.; Isomaa, B.; Berglund, G.; Nilsson, P.M.; Groop, L.; Franks, P.W. Assessing the effect of interaction between an FTO variant (rs9939609) and physical activity on obesity in 15,925 Swedish and 2511 Finnish adults. Diabetologia 2009, 52, 1334–1338. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xie, X.; Houtz, J.; Liao, G.Y.; Chen, Y.; Xu, B. Genetic Val66Met BDNF Variant Increases Hyperphagia on Fat-rich Diets in Mice. Endocrinology 2023, 164, bqad008. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Lu, W.; Tian, Y.; Wang, B. Intestinal SEC16B modulates obesity by regulating chylomicron metabolism. Mol. Metab. 2023, 70, 101693. [Google Scholar] [CrossRef]
- Kim, M.S.; Shim, I.; Fahed, A.C.; Do, R.; Park, W.Y.; Natarajan, P.; Khera, A.V.; Won, H.H. Association of genetic risk, lifestyle, and their interaction with obesity and obesity-related morbidities. Cell Metab. 2024, 36, 1494–1503.e3. [Google Scholar] [CrossRef]
- Motsinger-Reif, A.A.; Reif, D.M.; Akhtari, F.S.; House, J.S.; Campbell, C.R.; Messier, K.P.; Fargo, D.C.; Bowen, T.A.; Nadadur, S.S.; Schmitt, C.P.; et al. Gene-environment interactions within a precision environmental health framework. Cell Genom. 2024, 4, 100591. [Google Scholar] [CrossRef]
- Emami Kazemabad, M.J.; Asgari Toni, S.; Tizro, N.; Dadkhah, P.A.; Amani, H.; Akhavan Rezayat, S.; Sheikh, Z.; Mohammadi, M.; Alijanzadeh, D.; Alimohammadi, F.; et al. Pharmacotherapeutic potential of pomegranate in age-related neurological disorders. Front. Aging Neurosci. 2022, 14, 955735. [Google Scholar] [CrossRef]
- Gao, A.X.; Xia, T.C.; Lin, L.S.; Dong, T.T.; Tsim, K.W. The neurotrophic activities of brain-derived neurotrophic factor are potentiated by binding with apigenin, a common flavone in vegetables, in stimulating the receptor signaling. CNS Neurosci. Ther. 2023, 29, 2787–2799. [Google Scholar] [CrossRef]
- Martínez-Coria, H.; Serrano-García, N.; López-Valdés, H.E.; López-Chávez, G.S.; Rivera-Alvarez, J.; Romero-Hernández, Á.; Valverde, F.F.; Orozco-Ibarra, M.; Torres-Ramos, M.A. Morin improves learning and memory in healthy adult mice. Brain Behav. 2024, 14, e3444. [Google Scholar] [CrossRef]
- Subash, S.; Essa, M.M.; Al-Adawi, S.; Memon, M.A.; Manivasagam, T.; Akbar, M. Neuroprotective effects of berry fruits on neurodegenerative diseases. Neural Regen. Res. 2014, 9, 1557–1566. [Google Scholar]
- Marosi, K.; Mattson, M.P. BDNF mediates adaptive brain and body responses to energetic challenges. Trends Endocrinol. Metab. 2014, 25, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Sharebiani, H.; Mokaram, M.; Mirghani, M.; Fazeli, B.; Stanek, A. The Effects of Antioxidant Supplementation on the Pathologic Mechanisms of Metabolic Syndrome and Cardiovascular Disease Development. Nutrients 2024, 16, 1641. [Google Scholar] [CrossRef] [PubMed]
- Farhangi, M.A. Dietary total antioxidant capacity significantly interacts with 6-P21 rs2010963 gene polymorphisms in terms of cardio-metabolic risk factors in patients with metabolic syndrome. BMC Res. Notes 2020, 13, 145. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.T.; Hill, B.; Sims, N.; Heck, A.; Negron, M.; Lusk, C.; Galindo, C.L. Brain-derived neurotrophic factor rs6265 (Val66Met) single nucleotide polymorphism as a master modifier of human pathophysiology. Neural Regen. Res. 2023, 18, 102–106. [Google Scholar]
- Pandit, M.; Behl, T.; Sachdeva, M.; Arora, S. Role of brain-derived neurotropic factor in obesity. Obes. Med. 2020, 17, 100189. [Google Scholar] [CrossRef]
- Miksza, U.; Adamska-Patruno, E.; Bauer, W.; Fiedorczuk, J.; Czajkowski, P.; Moroz, M.; Drygalski, K.; Ustymowicz, A.; Tomkiewicz, E.; Gorska, M.; et al. Obesity-related parameters in carriers of some BDNF genetic variants may depend on daily dietary macronutrients intake. Sci. Rep. 2023, 13, 6585. [Google Scholar] [CrossRef]
- Khutami, C.; Sumiwi, S.A.; Khairul Ikram, N.K.; Muchtaridi, M. The Effects of Antioxidants from Natural Products on Obesity, Dyslipidemia, Diabetes and Their Molecular Signaling Mechanism. Int. J. Mol. Sci. 2022, 23, 2056. [Google Scholar] [CrossRef]
- Mangge, H.; Ciardi, C.; Becker, K.; Strasser, B.; Fuchs, D.; Gostner, J.M. Influence of Antioxidants on Leptin Metabolism and its Role in the Pathogenesis of Obesity. Adv. Exp. Med. Biol. 2017, 960, 399–413. [Google Scholar]


| L-EN-Lean (n = 24,483) | L-EN-Obs (n = 11,729) | H-EN-Lean (n = 11,382) | H-EN-Obs (n = 5523) | |
|---|---|---|---|---|
| BMI (kg/m2) | 22.4 ± 0.01 c | 27 ± 0.02 b | 22.4 ± 0.02 c | 27.2 ± 0.03 a**+++## |
| Energy intake (EER %) | 80.3 ± 0.13 d | 81.6 ± 0.19 c | 128.4 ± 0.19 b | 130.2 ± 0.28 a***+++# |
| Age (years) | 52.2 ± 0.05 d | 52.9 ± 0.07 c | 56.2 ± 0.08 b | 57.3 ± 0.11 a*** |
| Gender (male %) | 9599 (39.2) | 6117 (52.2) | 1930 (17.0) | 1510 (27.3) *** |
| Education (N, %) | ||||
| <High school | 3171 (18.1) | 2065 (24.0) | 1294 (15.7) | 1036 (24.2) $$$ |
| High school | 12,978 (74.1) | 5955 (69.3) | 6362 (77.0) | 2942 (69.3) |
| College and over | 1361 (7.77) | 576 (6.7) | 611 (7.39) | 270 (6.36) |
| Monthly income (N, %) | ||||
| ≤USD 2000 | 2416 (10.4) | 1312 (11.9) | 847 (7.89) | 574 (11.2) $$$ |
| USD 2000–4000 | 9882 (42.5) | 4749 (43.0) | 4864 (45.3) | 2333 (44.7) |
| >USD 4000 | 10,967(47.1) | 4975 (45.1) | 5018 (46.8) | 2309 (44.3) |
| Non-Smoking (N, %) | 22,290 (91.1) | 10,338 (88.2) | 10,940 (96.2) | 5169 (93.6) $$$ |
| Former smoking | 1224 (5.0) | 847 (7.22) | 248 (2.18) | 206 (3.73) |
| Current smoking | 964 (3.94) | 541 (4.61) | 188 (1.65) | 145 (2.63) |
| Alcohol (g/week) | 101 ± 2.16 c | 123 ± 3 b | 104 ± 3.48 c | 132 ± 4.6 a+++ |
| Exercise (Yes: N, %) | 13,042 (53.4) | 6254 (53.4) | 6552 (57.7) | 3013 (54.7) $$$ |
| L-EN-Lean (n = 24,483) | L-EN-Obs (n = 11,729) | H-EN-Lean (n = 11,382) | H-EN-Obs (n = 5523) | |
|---|---|---|---|---|
| Skeletal muscle mass index (kg/m2) | 7.16 ± 0.004 b | 6.67 ± 0.005 a | 7.16 ± 0.006 b | 6.63 ± 0.008 a+++# |
| Fat mass index (kg/m2) | 10.4 ± 0.01 c | 12.5 ± 0.01 b | 10.4 ± 0.01 c | 12.7 ± 0.02 a*+++# |
| Body fat (%) | 26.6 ± 0.02 c | 32.1 ± 0.02 b | 26.6 ± 0.03 c | 32.3 ± 0.04 a**+++## |
| Waist circumferences (cm) | 77.5 ± 0.04 c | 87.4 ± 0.06 b | 77.6 ± 0.07 c | 87.8 ± 0.09 a**+++# |
| Hip circumferences (cm) | 91.9 ± 0.03 c | 98.5 ± 0.05 b | 92 ± 0.05 c | 98.8 ± 0.07 a**+++## |
| WHR | 0.84 ± 0.0004 | 0.89 ± 0.0005 | 0.844 ± 0.0001 | 0.89 ± 0.0008 +++ |
| L-EN-Lean (n = 24,483) | L-EN-Obs (n = 11,729) | H-EN-Lean (n = 11,382) | H-EN-Obs (n = 5523) | |
|---|---|---|---|---|
| Energy intake (EER %) | 80.3 ± 0.13 d | 81.6 ± 0.19 c | 128.4 ± 0.19 b | 130.2 ± 0.28 a***+++# |
| Plant-based diet (Yes, N, %) | 5892 (24.1) | 2303 (19.6) | 6817 (59.9) | 2857 (51.7) $$$ |
| Dietary fiber (g/day) | 14.6 ± 0.06 b | 14.8 ± 0.08 ab | 14.7 ± 0.09 b | 15.1 ± 0.12 a*++ |
| Vitamin C (mg/day) | 103 ± 0.39 c | 104 ± 0.54 c | 112 ± 0.62 a | 108 ± 0.82 b***++### |
| Vitamin D (μg/day) | 6.33 ± 0.034 c | 6.25 ± 0.048 c | 6.73 ± 0.055 a | 6.53 ± 0.073 b***++ |
| Vitamin E (mg/day) | 8.02 ± 0.054 a | 7.89 ± 0.075 ab | 7.84 ± 0.087 ab | 7.58 ± 0.115 b*# |
| Selenium (μg/day) | 15.1 ± 0.15 a | 15.1 ± 0.2 a | 13.5 ± 0.24 b | 13.4 ± 0.31 b*** |
| Cyanidin (μg/day) | 21.8 ± 0.17 c | 21.4 ± 0.23 c | 24.9 ± 0.27 a | 22.8 ± 0.35 b*** |
| DII | −19.8 ± 0.1 | −20 ± 0.13 | −20.1 ± 0.15 | −20.2 ± 0.2 |
| Total anthocyanins (μg/day) | 24.9 ± 0.2 c | 24.8 ± 0.28 c | 43.1 ± 0.28 a | 41.0 ± 0.40 b***+++### |
| Flavonoids (μg/day) | 8.54 ± 0.05 c | 8.82 ± 0.07 a | 14.6 ± 0.08 a | 14.7 ± 0.11 a***+ |
| Isoflavonoids (μg/day) | 6.70 ± 0.04 b | 6.88 ± 0.07 b | 11.6 ± 0.07 a | 11.5 ± 0.09 b*** |
| CHR | SNP | Gene Names | BP | A1 | A2 | OR | SE | p | MAF | HWE_P | Location |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs506589 | SEC16B | 177894287 | C | T | 1.131 | 0.01786 | 5.56 × 10−12 | 0.2845 | 0.5099 | Intron |
| 2 | rs1965122 | ADCY3 | 25052450 | T | C | 1.085 | 0.01634 | 3.65 × 10−7 | 0.4477 | 0.5591 | Intron |
| 3 | rs73078824 | PBRM1 | 52624387 | T | G | 0.9055 | 0.01989 | 4.07 × 10−7 | 0.2188 | 0.3341 | Nmd transcript |
| 11 | rs925947 | BDNF-AS | 27667367 | T | G | 0.9128 | 0.01637 | 2.47 × 10−8 | 0.457 | 0.2032 | Intron |
| 11 | rs6265 | BDNF | 27679916 | T | C | 0.9121 | 0.01637 | 1.87 × 10−8 | 0.4588 | 0.1481 | Missense |
| 16 | rs1421085 | FTO | 53800954 | C | T | 1.166 | 0.02412 | 2.07 × 10−10 | 0.1245 | 0.4604 | Nmd transcript |
| 19 | rs1444988703 | GIPR | 46175046 | A | T | 1.109 | 0.01649 | 3.76 × 10−10 | 0.4066 | 0.4414 | Nmd transcript |
| 19 | rs9636135 | QPCTL | 46200776 | T | C | 0.9195 | 0.01653 | 3.79 × 10−7 | 0.4197 | 0.5474 | Nmd transcript |
| 19 | rs10408067 | SYMPK | 46363536 | G | A | 0.9216 | 0.0165 | 4.56 × 10−7 | 0.4193 | 0.3214 | Intron |
| 19 | rs796090051 | DPRX | 54111540 | A | C | 0.9015 | 0.02041 | 3.74 × 10−7 | 0.2042 | 0.585 | Intron |
| (A) | |||
| Low PRS (n = 13,638) | Medium PRS (n = 17,698) | High PRS (n = 4876) | |
| BMI (kg/m2) | 23.6 ± 0.02 c | 24 ± 0.02 b | 24.3 ± 0.04 a*** |
| Skeletal muscle mass index (kg/m2) | 7.03 ± 0.005 a | 7 ± 0.004 b | 6.96 ± 0.009 c*** |
| Body fat (%) | 27.3 ± 0.03 c | 27.7 ± 0.03 b | 28.1 ± 0.05 a*** |
| Waist circumferences (cm) | 80.6 ± 0.07 c | 81.3 ± 0.06 b | 82 ± 0.11 a*** |
| Hip circumferences (cm) | 93.8 ± 0.05 c | 94.2 ± 0.04 b | 94.7 ± 0.08 a*** |
| WHR | 0.859 ± 0.0005 c | 0.863 ± 0.0004 b | 0.865 ± 0.0008 a*** |
| Serum triglyceride (mg/dL) | 127.1 ± 0.76 | 128.1 ± 0.67 | 129.7 ± 1.28 |
| Serum glucose (mg/dL) | 95.8 ± 0.18 | 95.8 ± 0.16 | 96.3 ± 0.3 |
| Serum HDL (mg/dL) | 53.2 ± 0.11 | 53 ± 0.1 | 53.1 ± 0.18 |
| SBP (mmHg) | 122.5 ± 0.12 b | 123 ± 0.11 a | 123.4 ± 0.21 a** |
| DBP (mmHg) | 75.9 ± 0.08 b | 76.2 ± 0.07 a | 76.4 ± 0.14 a** |
| MetS (Yes, N, %) | 1822 (13.4) | 2622 (14.8) | 774 (15.9) *** |
| (B) | |||
| Risk Allele (n = 10,579) | Heterozygotes (n = 18,066) | Non-Risk Allele (n = 7567) | |
| Body mass index (kg/m2) | 24.1 ± 0.03 a | 23.9 ± 0.02 b | 23.7 ± 0.03 c*** |
| Skeletal muscle mass index (kg/m2) | 6.99 ± 0.006 b | 7 ± 0.004 ab | 7.02 ± 0.007 a* |
| Body fat (%) | 27.8 ± 0.04 a | 27.6 ± 0.03 b | 27.4 ± 0.04 c*** |
| Waist circumferences (cm) | 81.4 ± 0.08 a | 81.1 ± 0.06 b | 80.8 ± 0.09 c*** |
| Hip circumferences (cm) | 94.3 ± 0.06 a | 94.1 ± 0.04 b | 93.9 ± 0.07 c*** |
| WHR | 0.86 ± 0.001 | 0.86 ± 0 | 0.86 ± 0.001 |
| Serum triglyceride (mg/dL) | 128 ± 0.87 | 128 ± 0.66 | 127.7 ± 1.03 |
| Serum glucose (mg/dL) | 96.1 ± 0.2 | 95.7 ± 0.16 | 95.7 ± 0.24 |
| Serum HDL (mg/dL) | 53 ± 0.12 | 53.1 ± 0.1 | 53.1 ± 0.15 |
| Serum LDL (mg/dL) | 118.9 ± 0.27 | 118.9 ± 0.20 | 118.5 ± 0.32 |
| SBP (mmHg) | 122.9 ± 0.14 | 122.9 ± 0.11 | 122.7 ± 0.17 |
| DBP (mmHg) | 76.1 ± 0.09 | 76.1 ± 0.07 | 76 ± 0.11 |
| MetS (Yes, N, %) | 1559 (14.7) | 2616 (14.5) | 1043 (13.9) |
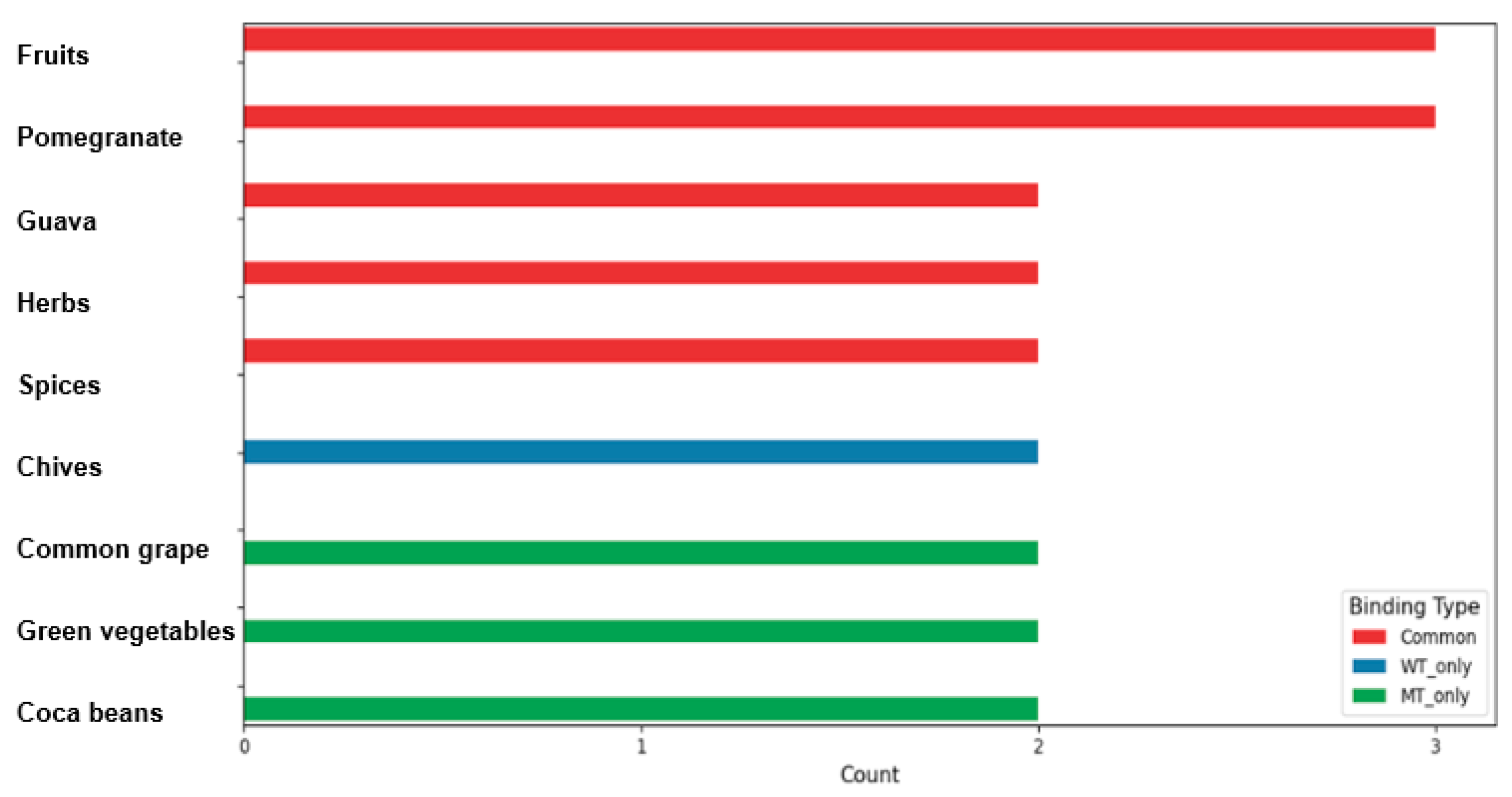
| Name | WT_BE (kcal/mol) | MT_BE (kcal/mol) | Type for Low BE | Cluster by UMP | Food Source |
|---|---|---|---|---|---|
| Valolaginic acid | −11.6 | −11.6 | Common | C−3 | guava |
| Torvoside C | −11.4 | −10.3 | Common | C−3 | fruits |
| Casuariin | −11.3 | −11.3 | Common | C−3 | cloves, pomegranate |
| Pipercyclobutanamide B | −11.3 | −11.3 | Common | C−3 | herbs, spices |
| Putranjivain A | −11.1 | −11 | Common | C−0 | fruits |
| Trisjuglone | −10.7 | −10.7 | Common | C−0 | common walnut, nuts |
| Casuarinin | −10.7 | −10.7 | Common | C−0 | Malabar plum, feijoa, herbs, spices, pomegranate |
| Stachyurin | −10.7 | −10.7 | Common | C−0 | guava |
| Asterlingulatoside D | −10.7 | −10.6 | Common | C−0 | Aster lingulatus |
| Fagopyrin | −10.6 | −10.3 | Common | C−0 | common buckwheat, cereals, cereal products |
| Kaempferol 3-O-rhamnosyl-rhamnosyl-glucoside | −10.1 | −10.1 | Common | C−1 | rosemary, common thyme, capers, common sage |
| Torvoside D | −10.1 | −10.1 | Common | C−1 | fruits |
| Punicafolin | −10.1 | −10.1 | Common | C−1 | pomegranate |
| Luteolin 7-O-diglucuronide | −10 | −10 | Common | C−2 | common verbena, lemon verbena |
| Ophiopogonin C′ | −10 | −10 | Common | C−2 | onion family vegetables |
| (3b,16a)-Dihydroxy-12-oleanen-28-oic acid 3-[glucosyl-(1->2)-arabinoside] 28-[rhamnosyl-(1->4)-glucosyl-(1->4)-glucosyl] ester | −10.9 | −8.5 | WT only | C-1 | fruits |
| (Cyanidin 3-O-beta-glucoside)(kaempferol 3-O-(2-O-beta-glucosyl-beta-glucoside)-7-O-beta-glucosiduronic acid) malonate | −10.8 | −8.7 | WT only | C-1 | chives |
| Pitheduloside K | −10.8 | −7.8 | WT only | C-1 | seeds of Pithecellobium dulce |
| (Cyanidin 3-O-(3-O-acetyl-beta-glucoside) (kaempferol 3-O-(2-O-beta-glucosyl-beta-glucoside)-7-O-beta-glucosiduronic acid) malonate | −10.7 | −8.4 | WT only | C-1 | chives |
| Epicatechin-(4beta->8)-epicatechin-(4beta->6)-catechin | −10.7 | −9 | WT only | C-1 | common grape |
| Kaempferol 3-rhamnosyl-(1->3)-rhamnosyl-(1->6)-glucoside | −10.1 | −9.6 | WT only | C-0 | tea |
| Dioscin | −10 | −9 | WT only | C-0 | fenugreek, yam |
| Cyanidin 3-(3-glucosyl-6-malonyl glucoside) 4′-glucoside | −10 | −9.4 | WT only | C-0 | garden onion |
| beta1-Chaconine | −10 | −9 | WT only | C-0 | alcoholic beverages, potato |
| Hesperidin | −10 | −9.6 | WT only | C-0 | citrus fruits, Mentha longifolia |
| alpha-Viniferin | −9.9 | −11.6 | MT only | C-1 | alcoholic beverages, common grape |
| Quillaic acid 3-[galactosyl-(1->2)-[rhamnosyl-(1->3)]-glucuronide] 28-[6-acetyl-glucosyl-(1->3)-[xylosyl-(1->4)-rhamnosyl-(1->2)]-4-acetyl-fucosyl] ester | −8.5 | −10.7 | MT only | C-1 | bark of the Chilean indigenous tree (Quillaja saponaria) |
| Tragopogonsaponin Q | −8.8 | −10.7 | MT only | C-1 | green vegetables |
| Ceposide D | −9.5 | −10.6 | MT only | C-1 | garden onion, onion-family vegetables |
| Eleutheroside M | −8.7 | −10.6 | MT only | C-1 | tea |
| Cinnamtannin A2 | −8.7 | −10.6 | MT only | C-1 | cocoa bean, Chinese cinnamon, chocolate |
| Chakaflavonoside A | −8.9 | −10.4 | MT only | C-1 | tea |
| Epicatechin-(4beta->8)-epicatechin 3′-gallate | −9.6 | −10 | MT only | C-0 | common buckwheat, common grape, tea |
| Epicatechin-(2alpha->7,4alpha->8)-epicatechin 3-galactoside | −9.5 | −10 | MT only | C-0 | cocoa bean |
| Acutoside G | −9.6 | −10 | MT only | C-0 | green vegetables |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Park, S. Energy Intake-Dependent Genetic Associations with Obesity Risk: BDNF Val66Met Polymorphism and Interactions with Dietary Bioactive Compounds. Antioxidants 2025, 14, 170. https://doi.org/10.3390/antiox14020170
Zhang T, Park S. Energy Intake-Dependent Genetic Associations with Obesity Risk: BDNF Val66Met Polymorphism and Interactions with Dietary Bioactive Compounds. Antioxidants. 2025; 14(2):170. https://doi.org/10.3390/antiox14020170
Chicago/Turabian StyleZhang, Ting, and Sunmin Park. 2025. "Energy Intake-Dependent Genetic Associations with Obesity Risk: BDNF Val66Met Polymorphism and Interactions with Dietary Bioactive Compounds" Antioxidants 14, no. 2: 170. https://doi.org/10.3390/antiox14020170
APA StyleZhang, T., & Park, S. (2025). Energy Intake-Dependent Genetic Associations with Obesity Risk: BDNF Val66Met Polymorphism and Interactions with Dietary Bioactive Compounds. Antioxidants, 14(2), 170. https://doi.org/10.3390/antiox14020170







