Olivomycin A Targets Epithelial–Mesenchymal Transition, Apoptosis, and Mitochondrial Quality Control in Renal Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
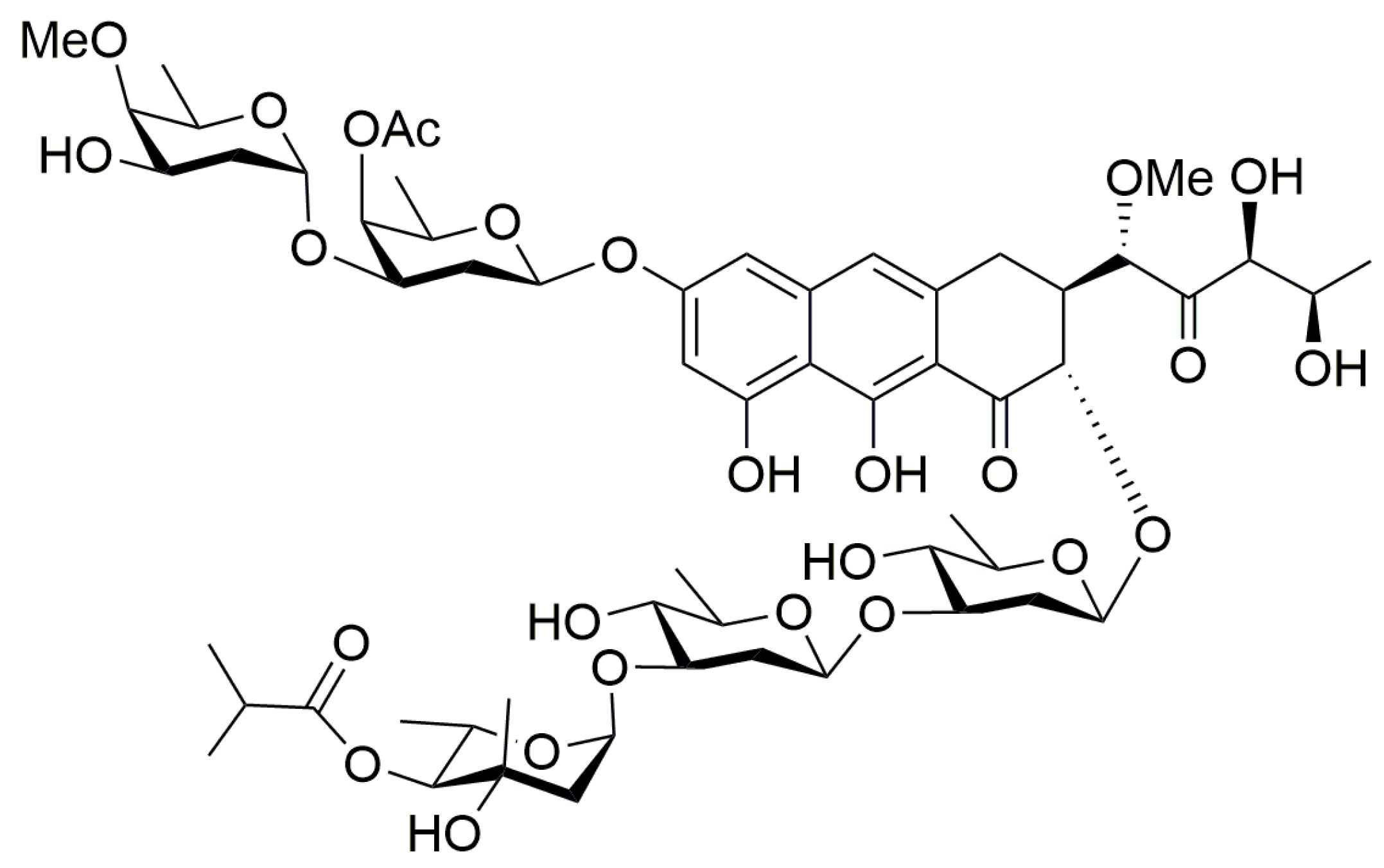
2.1. Chemistry
2.2. Cell Culture and Reagents
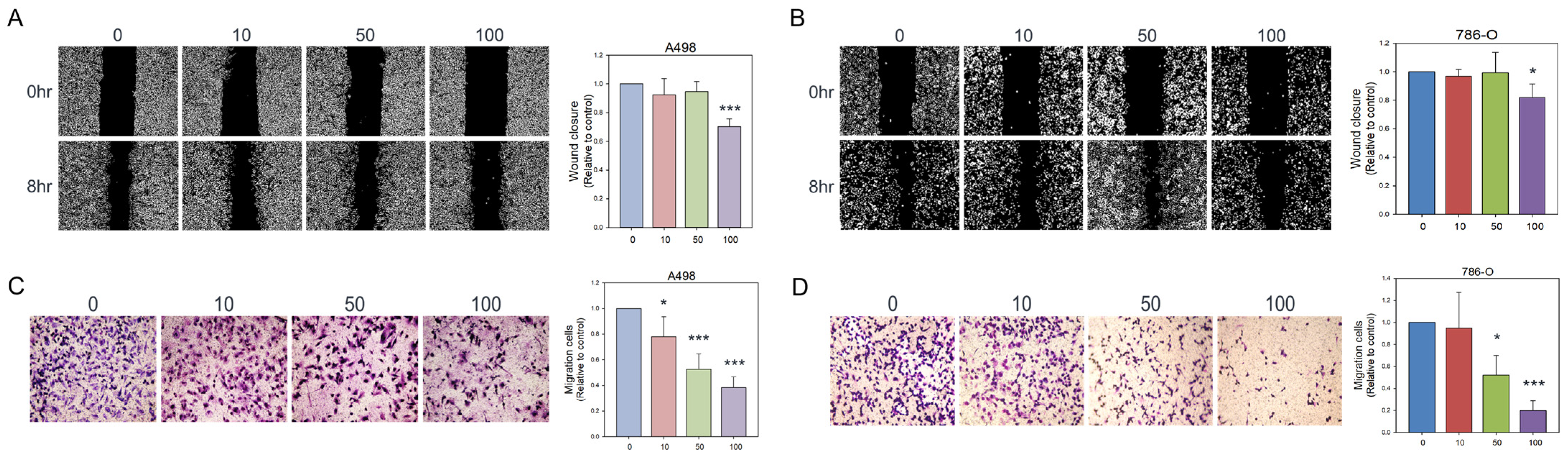
2.3. Wound Healing Assays
2.4. Transwell Assay
2.5. Cell Impedance Measurements
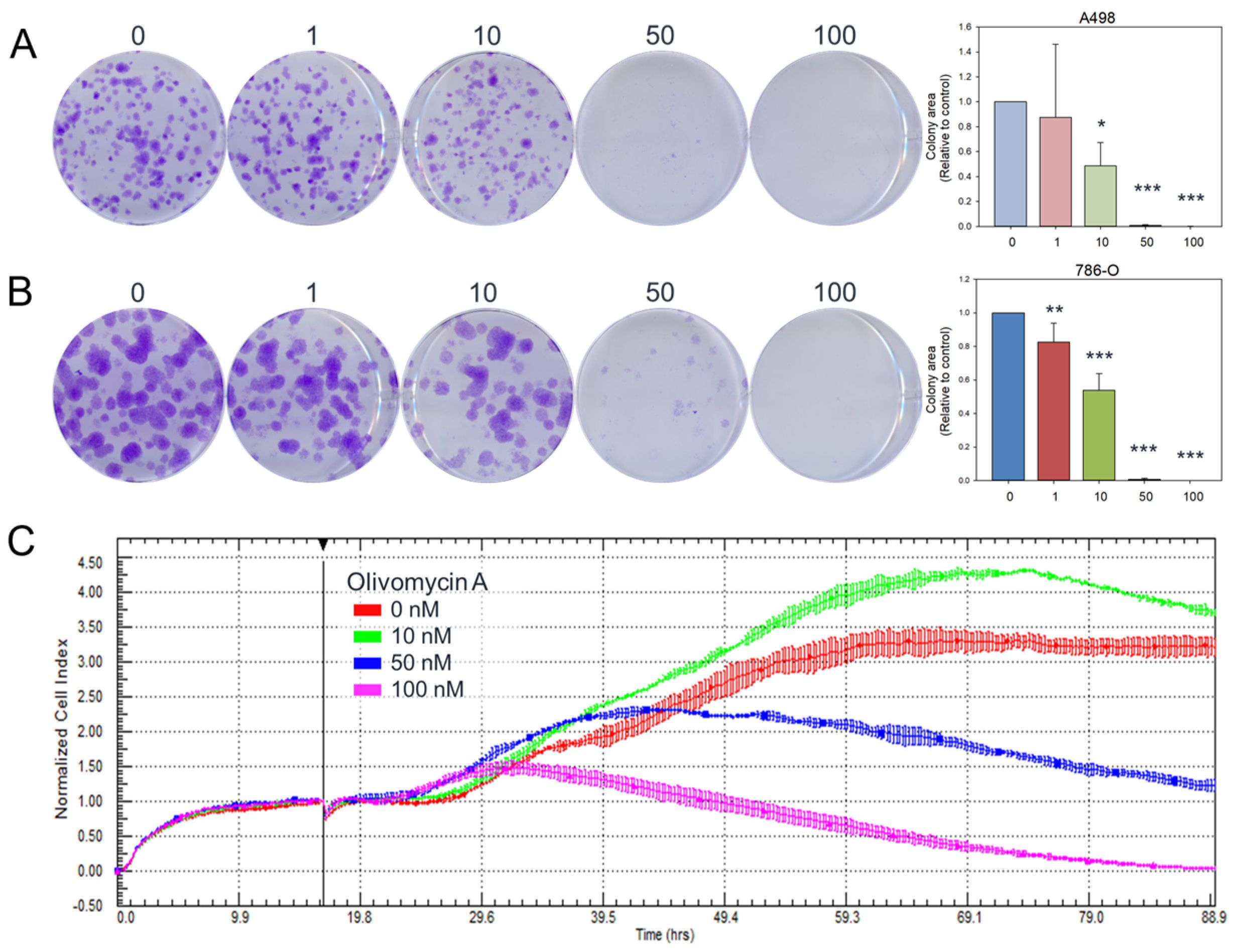
2.6. Colony-Forming Assay
2.7. Apoptosis Determination
2.8. Measurement of Cellular Reactive Oxygen Species (ROS)
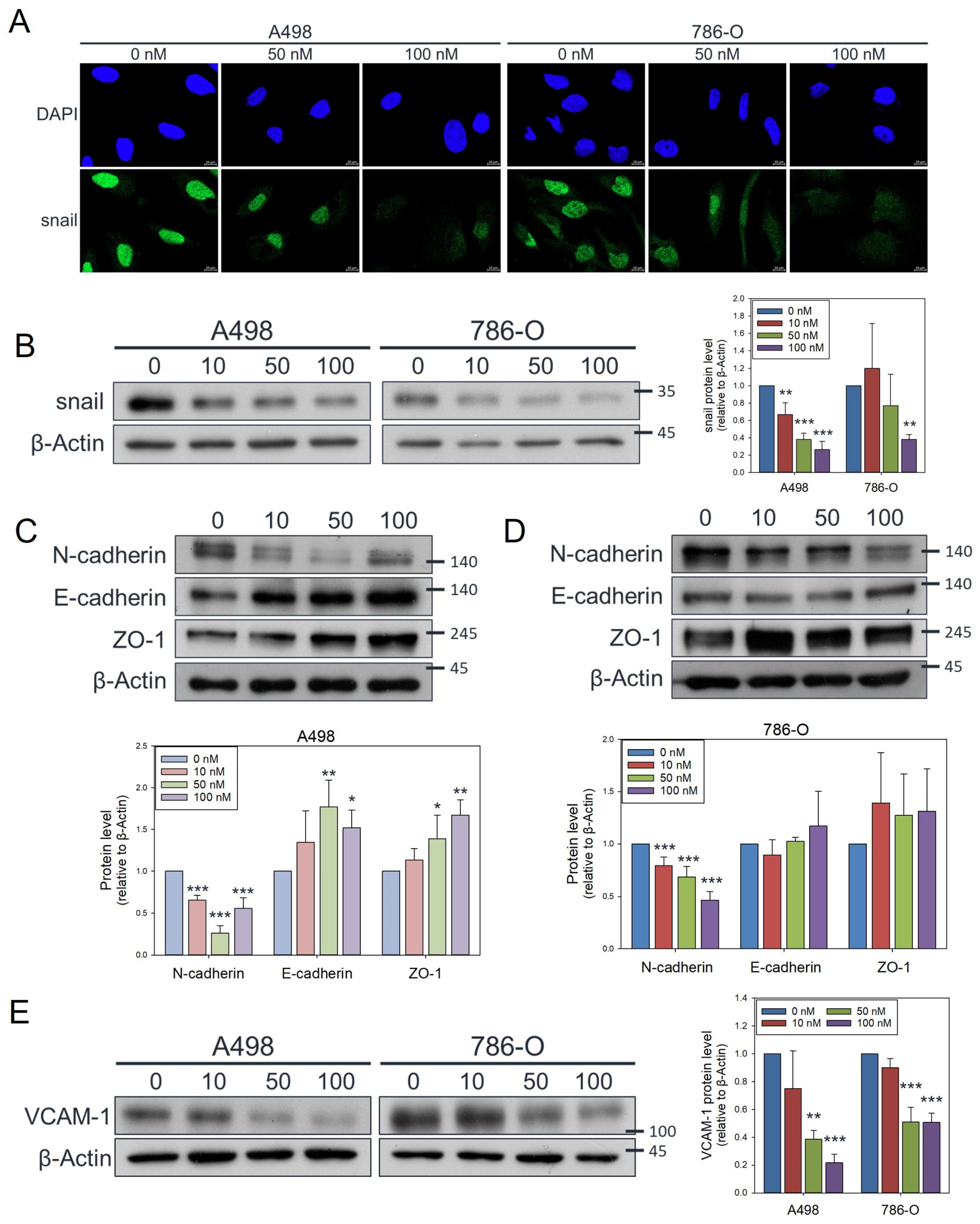
2.9. Immunofluorescence Staining
2.10. Western Blot Analysis
2.11. Statistics
3. Results
3.1. Olivomycin a Induces Antiproliferative Effects and Cellular Senescence in Renal Cancer Cells
3.2. Olivomycin a Suppresses Renal Cancer Cell Migration by Modulating EMT Markers
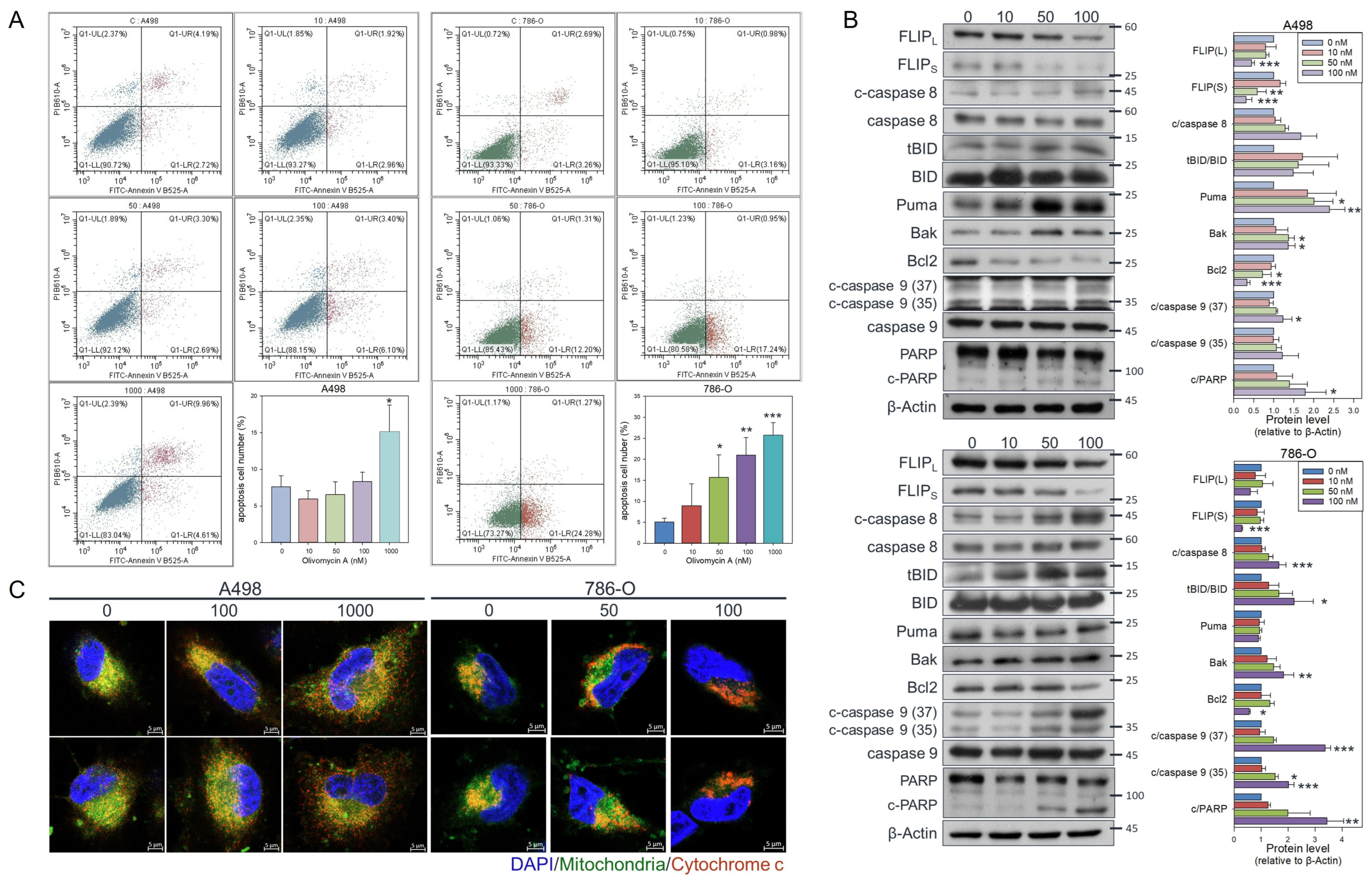
3.3. Olivomycin a Induces Cell Line-Specific Apoptotic Pathways in Renal Cancer Cells
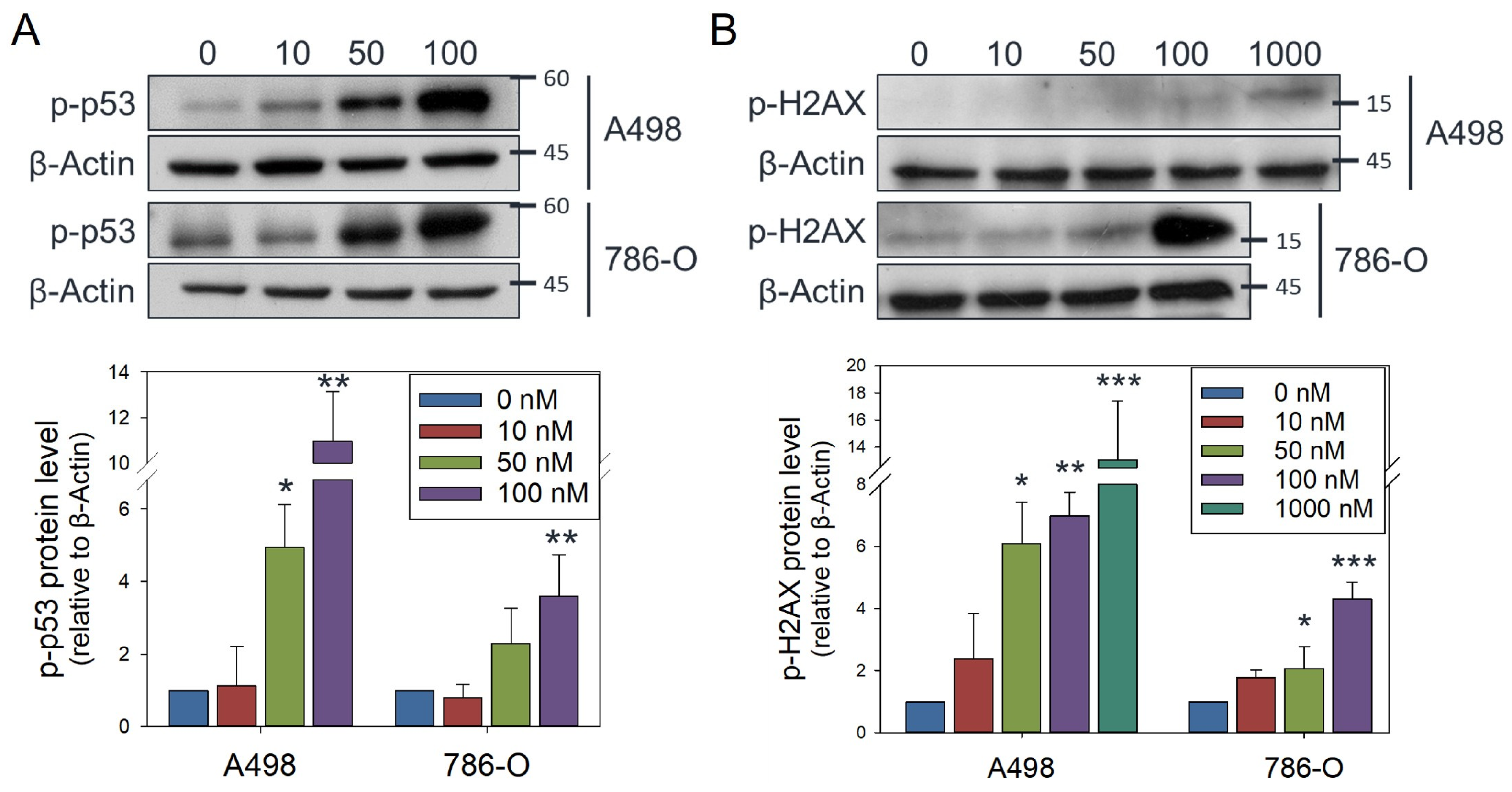
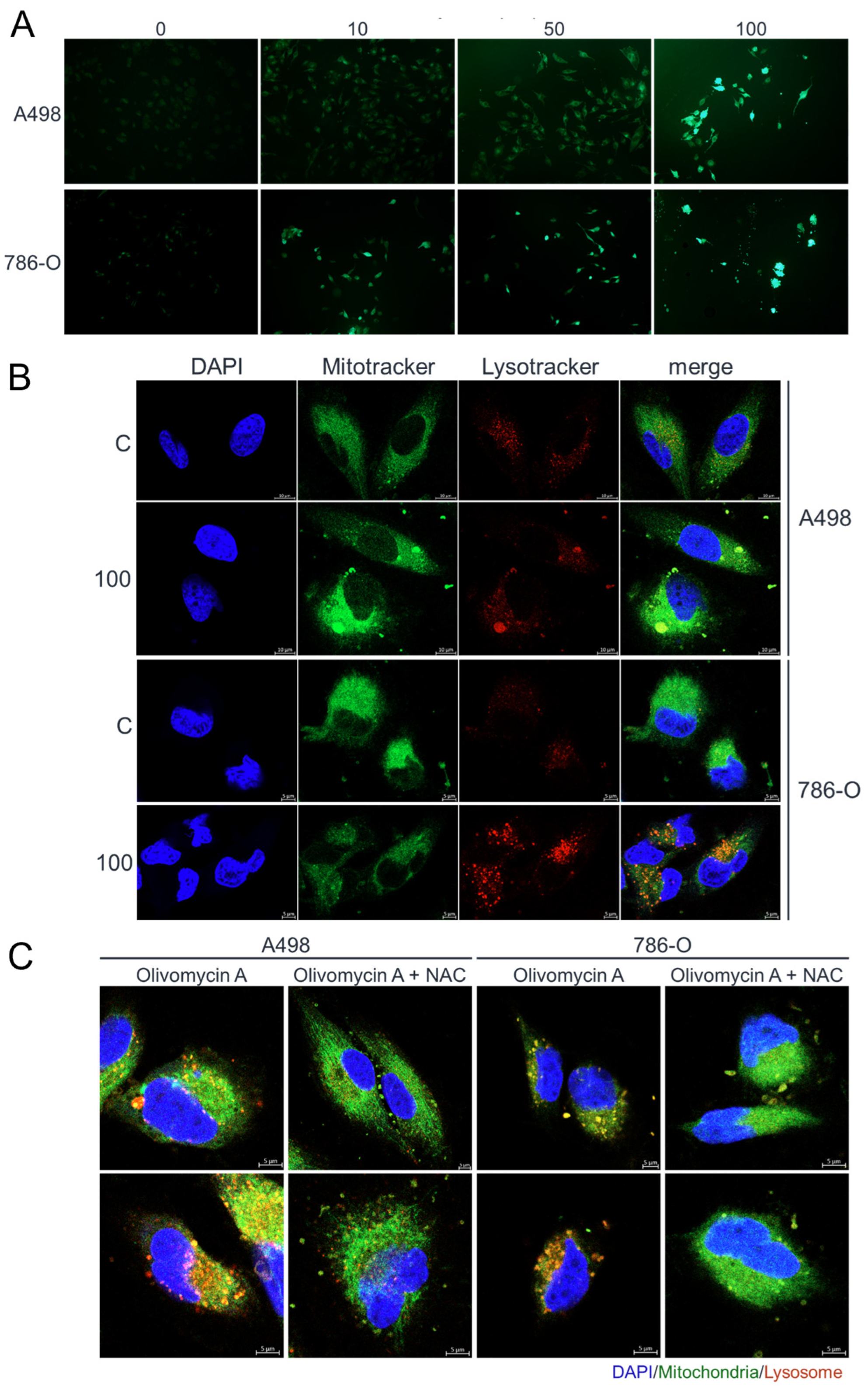
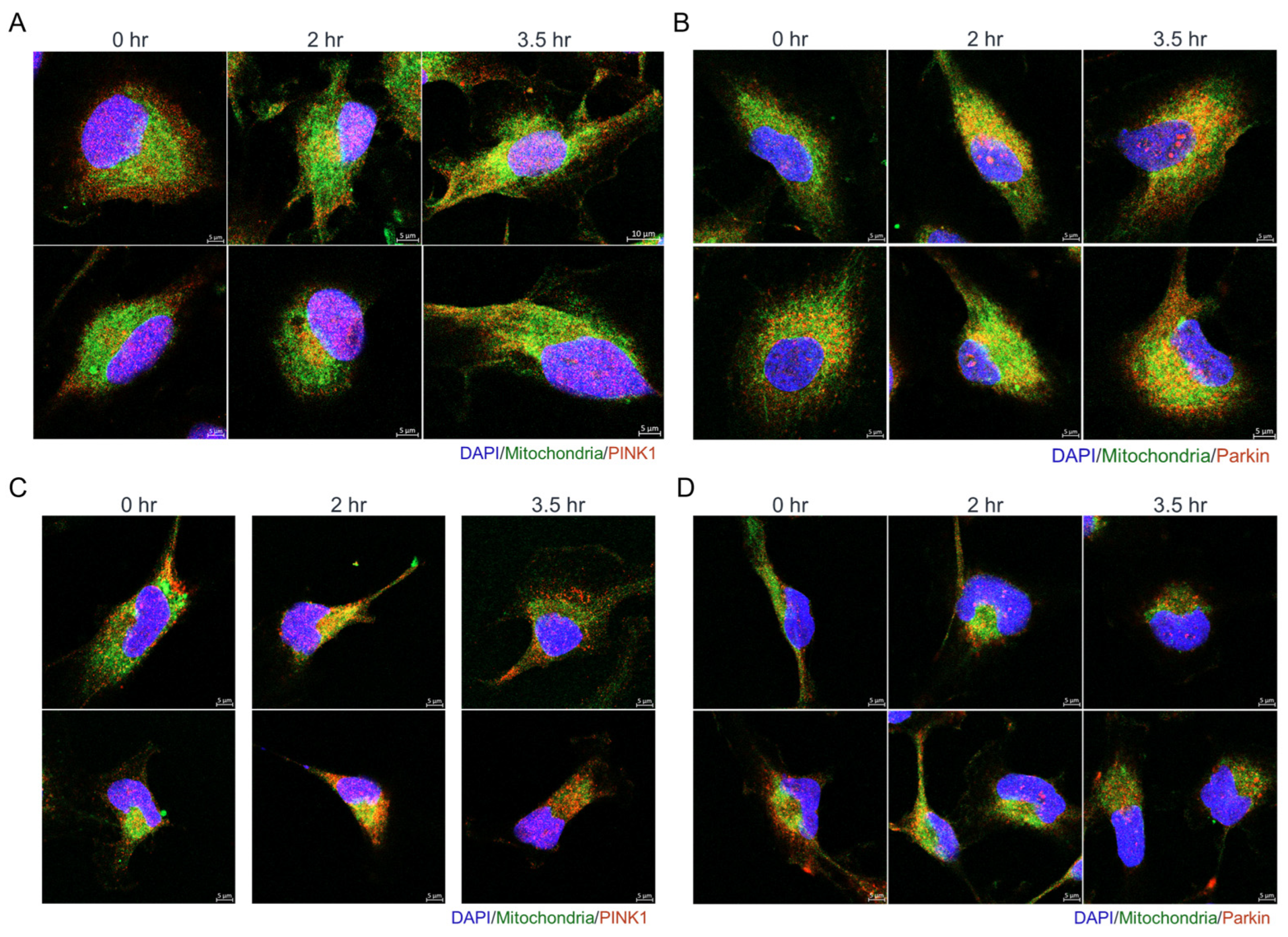
3.4. Olivomycin a Induces Mitochondrial Stress and Mitophagy Through PINK1 Signaling in Renal Cancer Cells with Mutant P53
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| EMT | Epithelial–mesenchymal transition |
| NAC | N-acetylcysteine |
| PINK1 | PTEN-induced kinase 1 |
| PTEN | Phosphatase and tensin homolog |
| ROS | Reactive oxygen species |
| RCC | Renal cell carcinoma |
| VHL | Von Hippel-Lindau tumor suppressor |
References
- Berlin, Y.A.; Kiseleva, O.A.; Kolosov, M.N.; Shemyakin, M.M.; Soifer, V.S.; Vasina, I.V.; Yartseva, I.V. Aureolic Acid Group of Anti-Tumour Antibiotics. Nature 1968, 218, 193–194. [Google Scholar] [CrossRef]
- Kumar, V.; Remers, W.A.; Bradner, W.T. Preparation and antitumor activity of olivomycin A analogues. J. Med. Chem. 1980, 23, 376–379. [Google Scholar] [CrossRef]
- Radzievskaia, V.V.; Terent’eva, T.G.; Sokolov, A.B. Action of aureolic acid and dactinomycin group antibiotics on human brain tumors in culture. Antibiotiki 1978, 23, 103–109. [Google Scholar]
- Brikenshtein, V.; Pitina, L.R.; Barenboim, G.M.; Gurskii, G.V. Stereochemistry and kinetics of interaction with DNA of the antineoplastic antibiotic olivomycin. Mol. Biol. 1984, 18, 1606–1616. [Google Scholar]
- Fox, K.R.; Howarth, N.R. Investigations into the sequence-selective binding of mithramycin and related ligands to DNA. Nucleic Acids Res. 1985, 13, 8695–8714. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.X.; Gresh, N.; Hui, X.; Pullman, B.; Zakrzewska, K. Modelling basic features of specificity in DNA-aureolic acid-derived antibiotic interactions. FEBS Lett. 1989, 245, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Beniaminov, A.D.; Chashchina, G.V.; Livshits, M.A.; Kechko, O.I.; Mitkevich, V.A.; Mamaeva, O.K.; Tevyashova, A.N.; Shtil, A.A.; Shchyolkina, A.K.; Kaluzhny, D.N. Discrimination between G/C Binding Sites by Olivomycin A Is Determined by Kinetics of the Drug-DNA Interaction. Int. J. Mol. Sci. 2020, 21, 5299. [Google Scholar] [CrossRef]
- Kudla, G.; Lipinski, L.; Caffin, F.; Helwak, A.; Zylicz, M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006, 4, e180. [Google Scholar] [CrossRef]
- Nanjunda, R.; Wilson, W.D. Binding to the DNA minor groove by heterocyclic dications: From AT-specific monomers to GC recognition with dimers. Curr. Protoc. Nucleic Acid Chem. 2012, 51, 8.8.1–8.8.20. [Google Scholar] [CrossRef]
- Versteeg, R.; van Schaik, B.D.; van Batenburg, M.F.; Roos, M.; Monajemi, R.; Caron, H.; Bussemaker, H.J.; van Kampen, A.H. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003, 13, 1998–2004. [Google Scholar] [CrossRef]
- Isagulieva, A.K.; Kaluzhny, D.N.; Beniaminov, A.D.; Soshnikova, N.V.; Shtil, A.A. Differential Impact of Random GC Tetrad Binding and Chromatin Events on Transcriptional Inhibition by Olivomycin A. Int. J. Mol. Sci. 2022, 23, 8871. [Google Scholar] [CrossRef]
- Isagulieva, A.K.; Soshnikova, N.V.; Shtil, A.A. Inhibition of the c-Myc Oncogene by the Aureolic Acid Group Antibiotics. Dokl. Biochem. Biophys. 2021, 500, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Cheglakov, I.B.; Tevyashova, A.N.; Kurbatov, L.K.; Tatarsky, V.V., Jr.; Samusenko, A.V.; Preobrazhenskaya, M.N.; Shtil, A.A. Altered transcription and replication are the mechanisms of cytotoxicity of antitumor antibiotic olivomycin A. Dokl. Biochem. Biophys. 2010, 435, 320–322. [Google Scholar] [CrossRef] [PubMed]
- Tevyashova, A.N.; Shtil, A.A.; Olsufyeva, E.N.; Luzikov, Y.N.; Reznikova, M.I.; Dezhenkova, L.G.; Isakova, E.B.; Bukhman, V.M.; Durandin, N.A.; Vinogradov, A.M.; et al. Modification of olivomycin A at the side chain of the aglycon yields the derivative with perspective antitumor characteristics. Bioorganic Med. Chem. 2011, 19, 7387–7393. [Google Scholar] [CrossRef] [PubMed]
- Tevyashova, A.N.; Durandin, N.A.; Vinogradov, A.M.; Zbarsky, V.B.; Reznikova, M.I.; Dezhenkova, L.G.; Bykov, E.E.; Olsufyeva, E.N.; Kuzmin, V.A.; Shtil, A.A.; et al. Role of the acyl groups in carbohydrate chains in cytotoxic properties of olivomycin A. J. Antibiot. 2013, 66, 523–530. [Google Scholar] [CrossRef]
- Sergeev, A.V.; Tevyashova, A.N.; Vorobyov, A.P.; Gromova, E.S. The Effect of Antitumor Antibiotic Olivomycin A and Its New Semi-synthetic Derivative Olivamide on the Activity of Murine DNA Methyltransferase Dnmt3a. Biochemistry 2019, 84, 62–70. [Google Scholar] [CrossRef]
- Simonova, V.S.; Samusenko, A.V.; Filippova, N.A.; Tevyashova, A.N.; Lyniv, L.S.; Kulik, G.I.; Chekhun, V.F.; Shtil, A.A. Olivomycin induces tumor cell apoptosis and suppresses p53-induced transcription. Bull. Exp. Biol. Med. 2005, 139, 455–459. [Google Scholar] [CrossRef]
- Leroy, B.; Girard, L.; Hollestelle, A.; Minna, J.D.; Gazdar, A.F.; Soussi, T. Analysis of TP53 mutation status in human cancer cell lines: A reassessment. Hum. Mutat. 2014, 35, 756–765. [Google Scholar] [CrossRef]
- Wang, M.; Luan, S.; Fan, X.; Wang, J.; Huang, J.; Gao, X.; Han, D. The emerging multifaceted role of PINK1 in cancer biology. Cancer Sci. 2022, 113, 4037–4047. [Google Scholar] [CrossRef]
- Linehan, W.M.; Schmidt, L.S.; Crooks, D.R.; Wei, D.; Srinivasan, R.; Lang, M.; Ricketts, C.J. The Metabolic Basis of Kidney Cancer. Cancer Discov. 2019, 9, 1006–1021. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Powles, T.; Albiges, L.; Bex, A.; Comperat, E.; Grunwald, V.; Kanesvaran, R.; Kitamura, H.; McKay, R.; Porta, C.; Procopio, G.; et al. Renal cell carcinoma: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2024, 35, 692–706. [Google Scholar] [CrossRef]
- Haas, N.B.; Nathanson, K.L. Hereditary kidney cancer syndromes. Adv. Chronic Kidney Dis. 2014, 21, 81–90. [Google Scholar] [CrossRef]
- Jin, J.; Xie, Y.; Zhang, J.S.; Wang, J.Q.; Dai, S.J.; He, W.F.; Li, S.Y.; Ashby, C.R., Jr.; Chen, Z.S.; He, Q. Sunitinib resistance in renal cell carcinoma: From molecular mechanisms to predictive biomarkers. Drug Resist. Updat. 2023, 67, 100929. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, W.; Xiong, Z.; Zhang, X. Molecular mechanisms of renal cell carcinoma metastasis and potential targets for therapy. Front. Cell Dev. Biol. 2025, 13, 1521151. [Google Scholar] [CrossRef] [PubMed]
- Fei, C.; Zhen, X.; Shiqiang, Z.; Jun, P. Frontier knowledge and future directions of programmed cell death in clear cell renal cell carcinoma. Cell Death Discov. 2024, 10, 113. [Google Scholar] [CrossRef] [PubMed]
- Amendolare, A.; Marzano, F.; Petruzzella, V.; Vacca, R.A.; Guerrini, L.; Pesole, G.; Sbisa, E.; Tullo, A. The Underestimated Role of the p53 Pathway in Renal Cancer. Cancers 2022, 14, 5733. [Google Scholar] [CrossRef]
- Swiatkowska, A. p53 and Its Isoforms in Renal Cell Carcinoma-Do They Matter? Biomedicines 2022, 10, 1330. [Google Scholar] [CrossRef]
- Xu, Z.; Liu, L.; Jiang, W.; Qiu, Y.; Zhang, B.; Cheng, J.; Luo, J.; Guo, J.; Xu, J. VHL missense mutation delineate aggressive clear cell renal cell carcinoma subtype with favorable immunotherapeutic response. J. Immunother. Cancer 2024, 12, e009963. [Google Scholar] [CrossRef]
- Xu, X.; Tang, Y.Y.; Liang, X.; Luo, W.; Jiang, D.M.; Chen, J. PTEN suppresses renal cell carcinoma proliferation and migration via inhibition of the PI3K/AKT pathway. World J. Surg. Oncol. 2025, 23, 42. [Google Scholar] [CrossRef]
- Huang, X.; Zhang, D.; Zhang, D.; Guo, J.; Gu, G.; Wang, Y.; Wu, G.; Wang, C.; Fu, B.; Li, K. Decoding PTEN: From biological functions to signaling pathways in tumors. Mol. Biol. Rep. 2024, 51, 1089. [Google Scholar] [CrossRef]
- Nakanishi, A.; Kitagishi, Y.; Ogura, Y.; Matsuda, S. The tumor suppressor PTEN interacts with p53 in hereditary cancer (Review). Int. J. Oncol. 2014, 44, 1813–1819. [Google Scholar] [CrossRef]
- Lee, Y.R.; Chen, M.; Pandolfi, P.P. The functions and regulation of the PTEN tumour suppressor: New modes and prospects. Nat. Rev. Mol. Cell Biol. 2018, 19, 547–562. [Google Scholar] [CrossRef] [PubMed]
- Song, M.S.; Salmena, L.; Pandolfi, P.P. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012, 13, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.F.; Tang, M.Y.; Fon, E.A.; Durcan, T.M. Defending the mitochondria: The pathways of mitophagy and mitochondrial-derived vesicles. Int. J. Biochem. Cell Biol. 2016, 79, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Dan, X.L.; Babbar, M.; Moore, A.; Wechter, N.; Tian, J.Y.; Mohanty, J.G.; Croteau, D.L.; Bohr, V.A. DNA damage invokes mitophagy through a pathway involving Spata18. Nucleic Acids Res. 2020, 48, 6611–6623. [Google Scholar] [CrossRef]
- Feng, X.; Liu, X.; Zhang, W.; Xiao, W. p53 directly suppresses BNIP3 expression to protect against hypoxia-induced cell death. EMBO J. 2011, 30, 3397–3415. [Google Scholar] [CrossRef]
- Hoshino, A.; Ariyoshi, M.; Okawa, Y.; Kaimoto, S.; Uchihashi, M.; Fukai, K.; Iwai-Kanai, E.; Ikeda, K.; Ueyama, T.; Ogata, T.; et al. Inhibition of p53 preserves Parkin-mediated mitophagy and pancreatic beta-cell function in diabetes. Proc. Natl. Acad. Sci. USA 2014, 111, 3116–3121. [Google Scholar] [CrossRef]
- Palikaras, K.; Lionaki, E.; Tavernarakis, N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 2018, 20, 1013–1022. [Google Scholar] [CrossRef]
- Li, G.; Yang, J.; Yang, C.; Zhu, M.; Jin, Y.; McNutt, M.A.; Yin, Y. PTENalpha regulates mitophagy and maintains mitochondrial quality control. Autophagy 2018, 14, 1742–1760. [Google Scholar] [CrossRef]
- Wang, L.; Lu, G.; Shen, H.M. The Long and the Short of PTEN in the Regulation of Mitophagy. Front. Cell Dev. Biol. 2020, 8, 299. [Google Scholar] [CrossRef]
- Vousden, K.H.; Prives, C. Blinded by the Light: The Growing Complexity of p53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Lane, D.P. p53 in health and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Murray-Zmijewski, F.; Slee, E.A.; Lu, X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat. Rev. Mol. Cell Biol. 2008, 9, 702–712. [Google Scholar] [CrossRef] [PubMed]
- Sablina, A.A.; Budanov, A.V.; Ilyinskaya, G.V.; Agapova, L.S.; Kravchenko, J.E.; Chumakov, P.M. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005, 11, 1306–1313. [Google Scholar] [CrossRef]
- Liu, B.; Chen, Y.; St Clair, D.K. ROS and p53: A versatile partnership. Free Radic. Biol. Med. 2008, 44, 1529–1535. [Google Scholar] [CrossRef]
- Bristow, R.G.; Hill, R.P. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat. Rev. Cancer 2008, 8, 180–192. [Google Scholar] [CrossRef]
- Bartek, J.; Lukas, J. DNA damage checkpoints: From initiation to recovery or adaptation. Curr. Opin. Cell Biol. 2007, 19, 238–245. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsieh, C.-Y.; Liou, Y.-F.; Shih, Y.-T.; Tikhomirov, A.S.; Shchekotikhin, A.E.; Chueh, P.J. Olivomycin A Targets Epithelial–Mesenchymal Transition, Apoptosis, and Mitochondrial Quality Control in Renal Cancer Cells. Antioxidants 2025, 14, 1348. https://doi.org/10.3390/antiox14111348
Hsieh C-Y, Liou Y-F, Shih Y-T, Tikhomirov AS, Shchekotikhin AE, Chueh PJ. Olivomycin A Targets Epithelial–Mesenchymal Transition, Apoptosis, and Mitochondrial Quality Control in Renal Cancer Cells. Antioxidants. 2025; 14(11):1348. https://doi.org/10.3390/antiox14111348
Chicago/Turabian StyleHsieh, Ching-Yu, Yih-Farng Liou, Yu-Tung Shih, Alexander S. Tikhomirov, Andrey E. Shchekotikhin, and Pin Ju Chueh. 2025. "Olivomycin A Targets Epithelial–Mesenchymal Transition, Apoptosis, and Mitochondrial Quality Control in Renal Cancer Cells" Antioxidants 14, no. 11: 1348. https://doi.org/10.3390/antiox14111348
APA StyleHsieh, C.-Y., Liou, Y.-F., Shih, Y.-T., Tikhomirov, A. S., Shchekotikhin, A. E., & Chueh, P. J. (2025). Olivomycin A Targets Epithelial–Mesenchymal Transition, Apoptosis, and Mitochondrial Quality Control in Renal Cancer Cells. Antioxidants, 14(11), 1348. https://doi.org/10.3390/antiox14111348







