Role of Advanced Glycation End Products and Mitohormesis in Cancer Development and Progression
Abstract
1. Introduction
2. Role of Mitochondria in Tumorigenesis and Cancer Progression
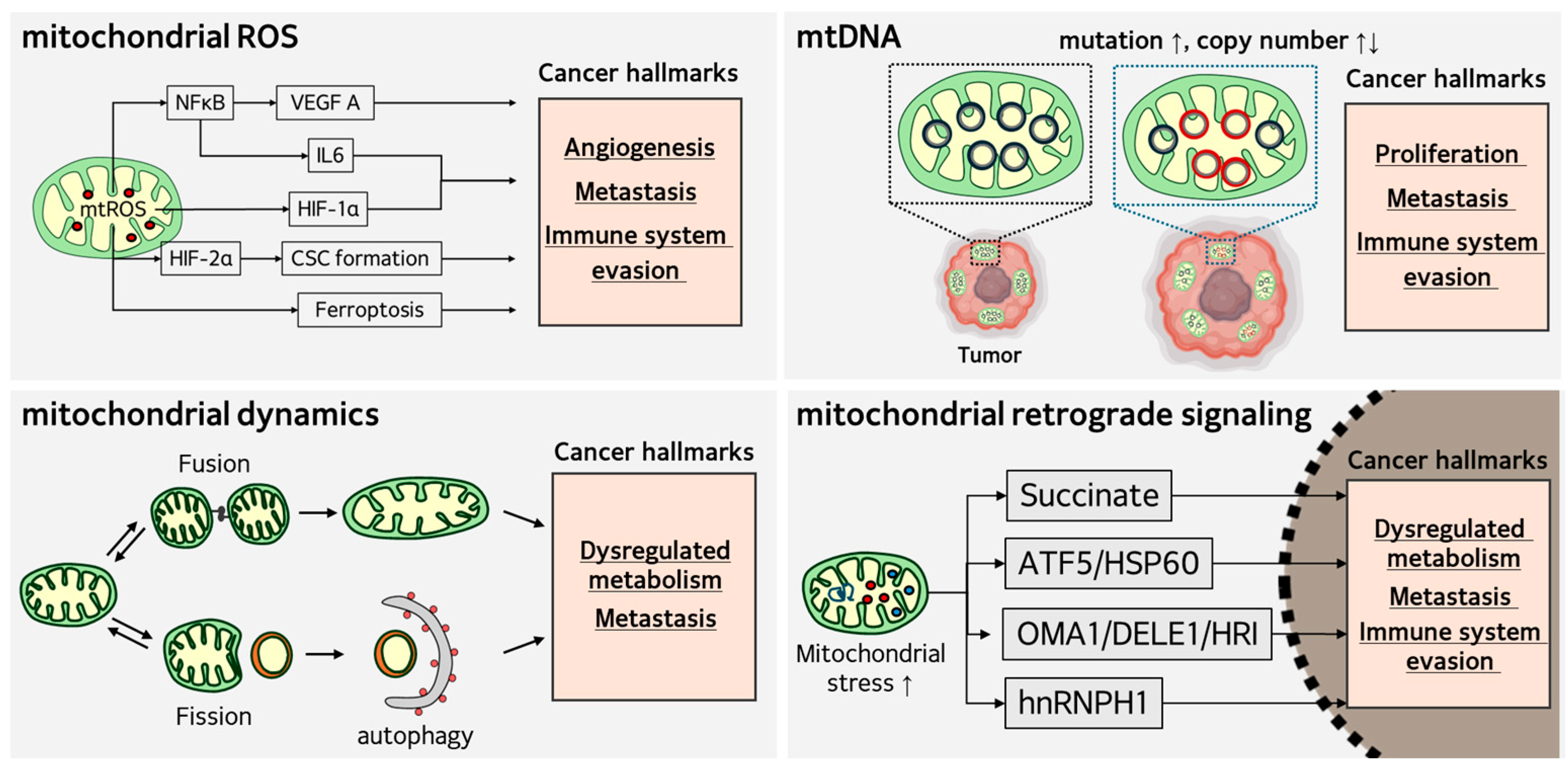
2.1. Mitochondrial Oxidative Stress in Cancer
2.2. Alteration of mtDNA in Cancer
2.3. Alteration of Mitochondrial Dynamics in Cancer
2.4. Mitochondrial Retrograde Signaling in Cancer
2.5. Mitochondrial Stress-Induced ISR in Cancer
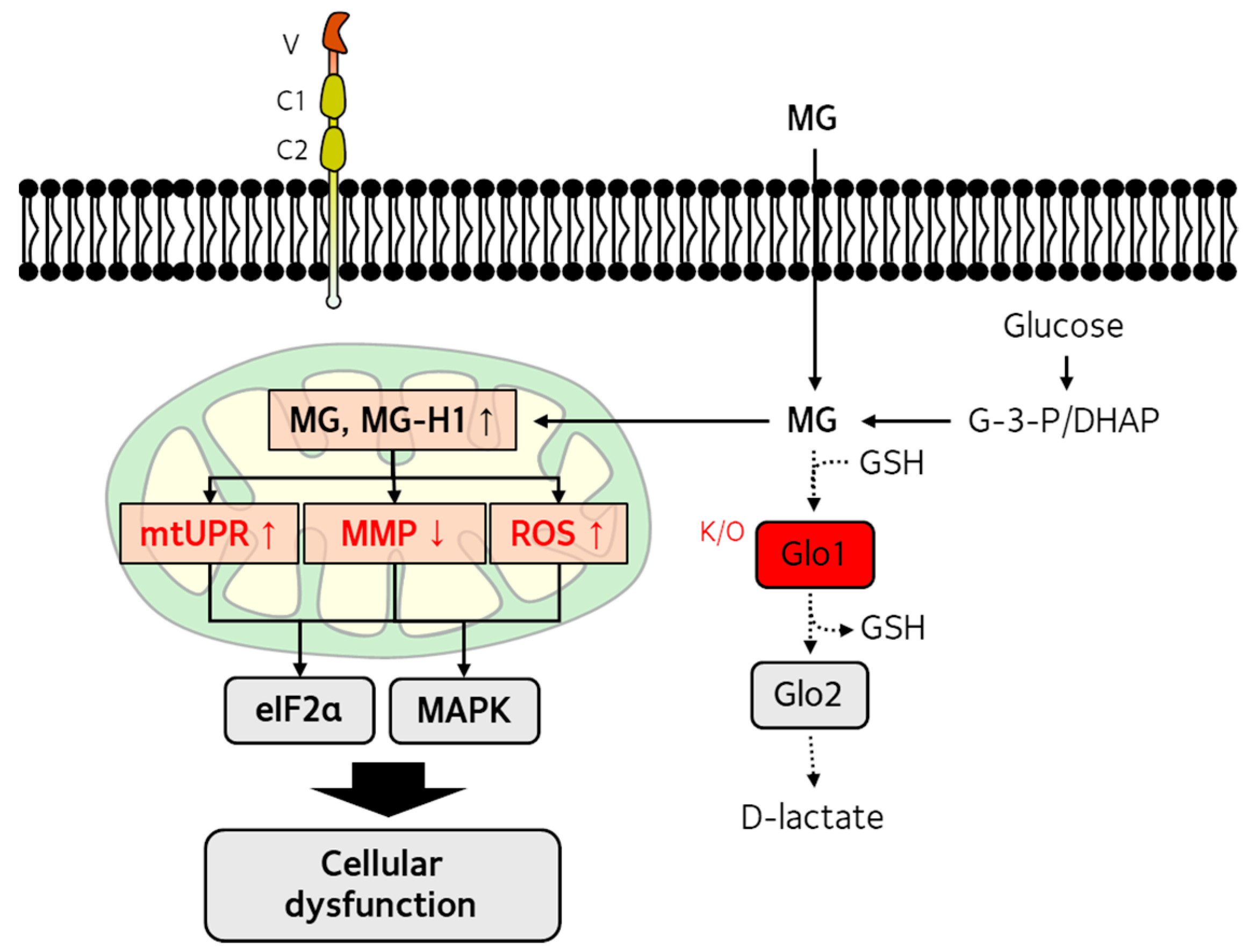
3. Dicarbonyl Stress and Mitochondrial Dysfunction
3.1. Production and Detoxification of Reactive Dicarbonyls
3.2. Effects of Dicarbonyl Stress on Mitochondrial Function
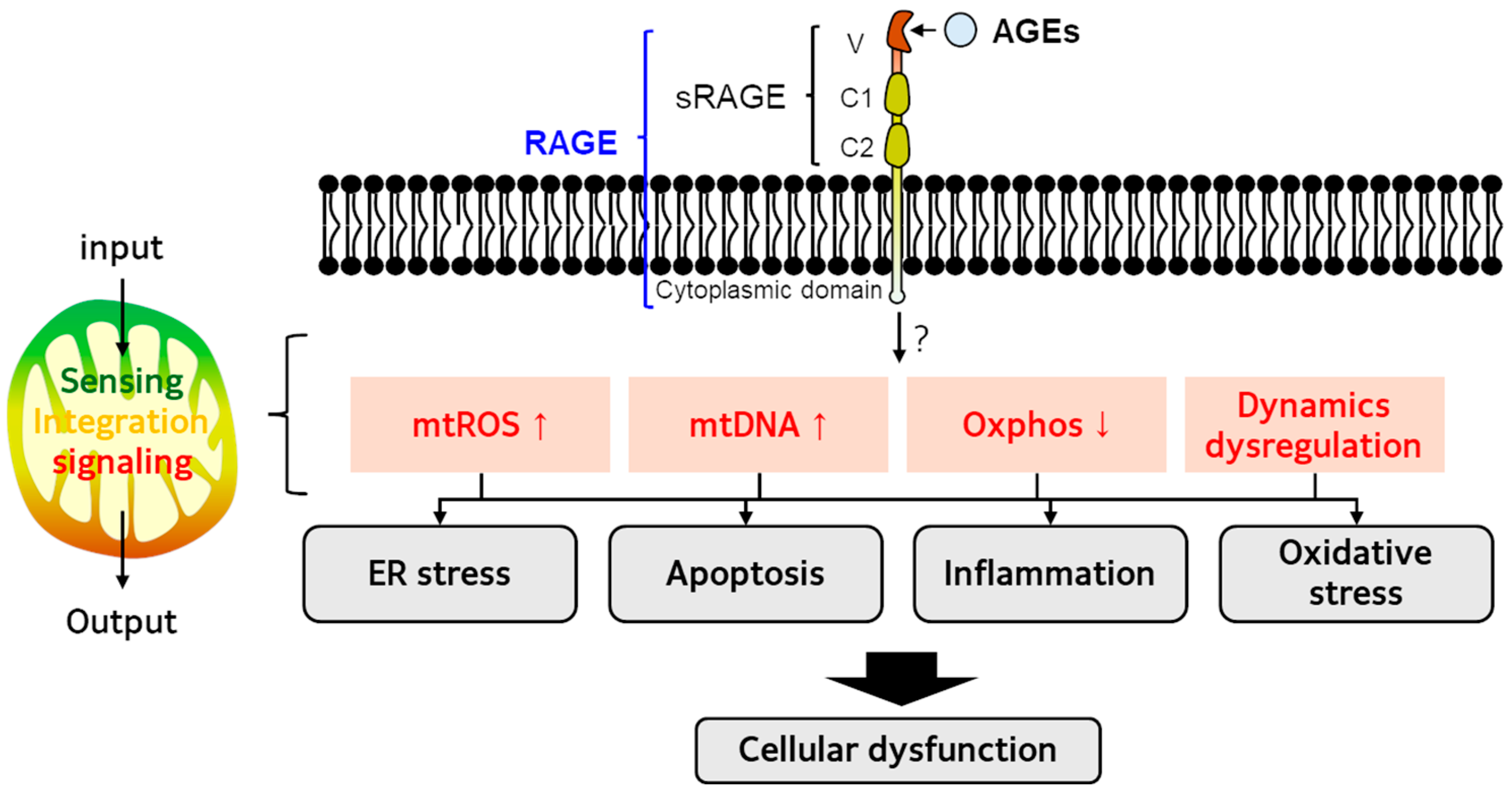
4. AGE-RAGE Axis and Mitochondrial Dysfunction
5. AGEs and Cancer
5.1. Epidemiology Studies
5.2. Animal Studies
5.3. In Vitro Studies
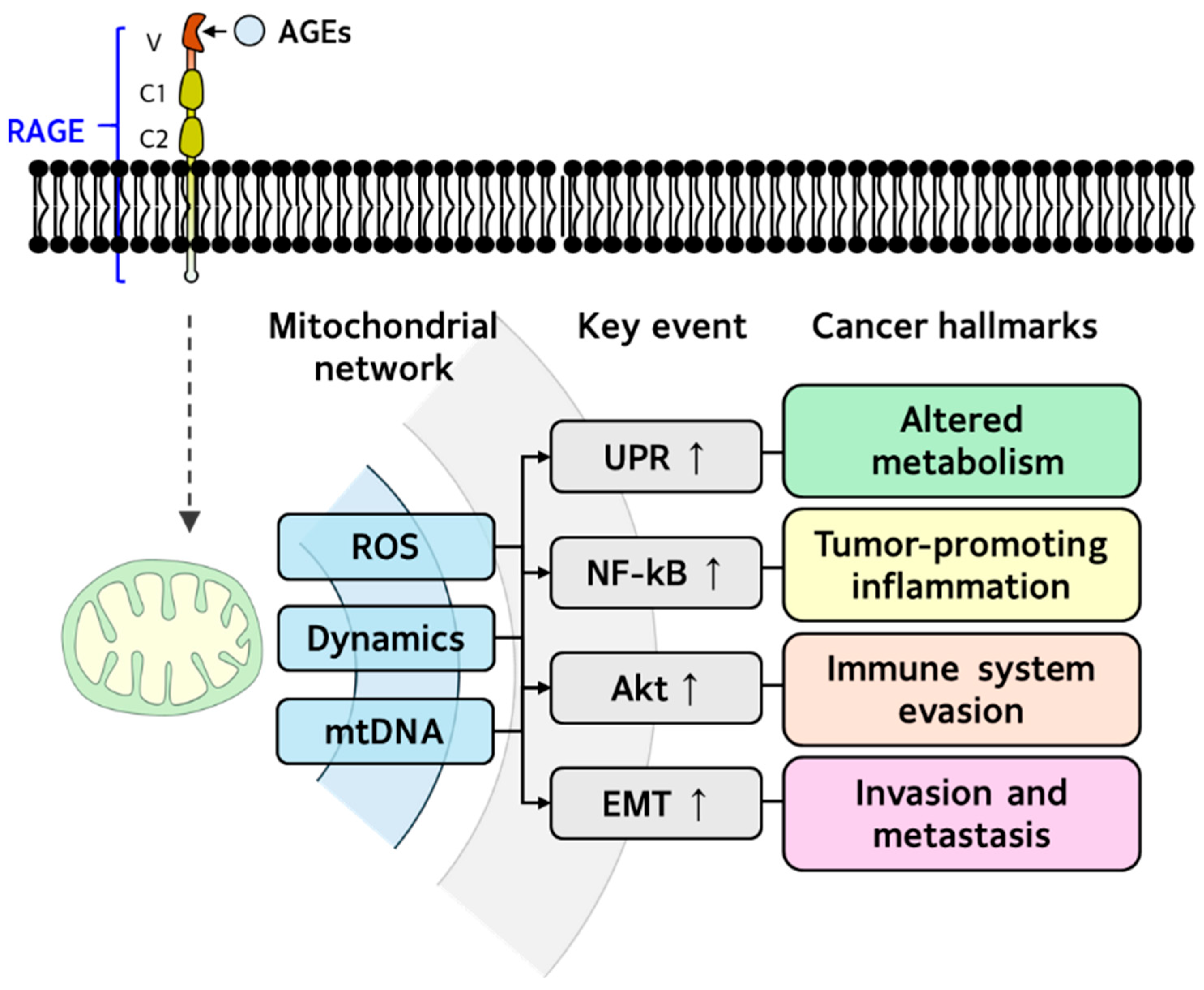
5.4. AGEs and Mitohormesis Dysregulation in Cancer
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| 3-DG | 3-deoxyglucosone |
| 5hmC | 5-hydroxymethylcytosine |
| AGEs | advanced glycation end products |
| CLL | chronic lymphocytic leukemia |
| CML | Nε-(carboxylmethyl)-l-lysine |
| CYTB | cytochrome B |
| DRP1 | dynamin-related protein 1 |
| EGPs | early glycation products |
| eIF2α | eukaryotic translation initiation factor 2A |
| EMT | Epithelial–mesenchymal transition |
| ERK | extracellular signal-regulated kinase |
| ERα | estrogen receptor alpha |
| ETC | electron transport chain |
| FH | fumarate hydratase |
| Glo1-KO hiPSCs | glyoxalase-1 knockout human induced pluripotent stem cells |
| HIF-1α | hypoxia-inducible factor 1α |
| hnRNP | H/F heterogeneous nuclear ribonucleoproteins H and F |
| HRI | Heme-regulated inhibitor |
| HSF1 | Heat Shock Factor 1 |
| HUVECs | human umbilical vein endothelial cells |
| IDH | isocitrate dehydrogenase |
| ISR | integrated stress response |
| JNK | c-Jun N-terminal kinase |
| MAPK | mitogen-activated protein kinase |
| MG | methylglyoxal |
| MG-H1 | Nδ-(5-hydro-5-methyl-4-imidazolon-2-yl)-l-ornithine |
| mtDNA | mitochondrial DNA |
| NOX | nicotinamide adenine dinucleotide phosphate oxidase |
| NSCLC | non-small cell lung cancer |
| PD-1 | programmed cell death protein 1 |
| ROS | reactive oxygen species |
| SDH | succinate dehydrogenase |
| SGs | stress granules |
| shRNA | short hairpin RNA |
| SIRT3 | Sirtuin 3 |
| SOD2 | superoxide dismutase 2 |
| TCA | tricarboxylic acid |
| TIL | tumor-infiltrating lymphocyte |
| UPRmt | mitochondrial unfolded protein response |
| VEGF | vascular endothelial growth factor |
| XBP1 | X-box Binding Protein 1 |
| XO | xanthine oxidase |
References
- Uceda, A.B.; Marino, L.; Casasnovas, R.; Adrover, M. An overview on glycation: Molecular mechanisms, impact on proteins, pathogenesis, and inhibition. Biophys. Rev. 2024, 16, 189–218. [Google Scholar] [CrossRef]
- Bolt, H.M.; Hengstler, J.G. Trends in research on advanced glycation end products (AGEs). Arch. Toxicol. 2024, 98, 3515–3517. [Google Scholar] [CrossRef] [PubMed]
- Rungratanawanich, W.; Qu, Y.; Wang, X.; Essa, M.M.; Song, B.J. Advanced glycation end products (AGEs) and other adducts in aging-related diseases and alcohol-mediated tissue injury. Exp. Mol. Med. 2021, 53, 168–188. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, Z.; Lv, C.; Li, M.; Wang, K.; Chen, Z. Advanced Glycation End Products and Health: A Systematic Review. Ann. Biomed. Eng. 2024, 52, 3145–3156. [Google Scholar] [CrossRef]
- Brown, J.S.; Amend, S.R.; Austin, R.H.; Gatenby, R.A.; Hammarlund, E.U.; Pienta, K.J. Updating the Definition of Cancer. Mol. Cancer Res. 2023, 21, 1142–1147. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Li, A.M.; Ye, J.B. Deciphering the Warburg Effect: Metabolic Reprogramming, Epigenetic Remodeling, and Cell Dedifferentiation. Annu. Rev. Cancer Biol. 2024, 8, 35–58. [Google Scholar] [CrossRef]
- Yang, J.; Shay, C.; Saba, N.F.; Teng, Y. Cancer metabolism and carcinogenesis. Exp. Hematol. Oncol. 2024, 13, 10. [Google Scholar] [CrossRef]
- Ghaddar, N.; Wang, S.; Woodvine, B.; Krishnamoorthy, J.; van Hoef, V.; Darini, C.; Kazimierczak, U.; Ah-son, N.; Popper, H.; Johnson, M.; et al. The integrated stress response is tumorigenic and constitutes a therapeutic liability in KRAS-driven lung cancer. Nat. Commun. 2021, 12, 4651. [Google Scholar] [CrossRef] [PubMed]
- Cerqua, M.; Foiani, M.; Boccaccio, C.; Comoglio, P.M.; Altintas, D.M. The integrated stress response drives MET oncogene overexpression in cancers. EMBO J. 2025, 44, 1107–1130. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J.; Osakabe, N.; Di Paola, R.; Siracusa, R.; Fusco, R.; D’Amico, R.; Impellizzeri, D.; Cuzzocrea, S.; Fritsch, T.; Abdelhameed, A.S.; et al. Hormesis defines the limits of lifespan. Ageing Res. Rev. 2023, 91, 102074. [Google Scholar] [CrossRef]
- Wan, Y.; Liu, J.; Mai, Y.; Hong, Y.; Jia, Z.; Tian, G.; Liu, Y.; Liang, H.; Liu, J. Current advances and future trends of hormesis in disease. NPJ Aging 2024, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Redding, A.; Grabocka, E. Stress granules and hormetic adaptation of cancer. Trends Cancer 2023, 9, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Yoshimasu, T.; Ohashi, T.; Oura, S.; Kokawa, Y.; Kawago, M.; Hirai, Y.; Miyasaka, M.; Nishiguchi, H.; Kawashima, S.; Yata, Y.; et al. A Theoretical Model for the Hormetic Dose-response Curve for Anticancer Agents. Anticancer. Res. 2015, 35, 5851–5855. [Google Scholar] [PubMed]
- Cheng, Y.W.; Liu, J.; Finkel, T. Mitohormesis. Cell Metab. 2023, 35, 1872–1886. [Google Scholar] [CrossRef]
- Yun, J.; Finkel, T. Mitohormesis. Cell Metab. 2014, 19, 757–766. [Google Scholar] [CrossRef]
- Inigo, J.R.; Chandra, D. The mitochondrial unfolded protein response (UPR(mt)): Shielding against toxicity to mitochondria in cancer. J. Hematol. Oncol. 2022, 15, 98. [Google Scholar] [CrossRef]
- Weber, D.; Thimm, J.; Seiz, T.; Kochlik, B.; Raupbach, J.; Burkle, A.; Grune, T.; Gruber, M.; Moreno-Villanueva, M. Association between redox biomarkers, DNA damage and aerobic capacity before and after physical stress in young men. Redox Biol. 2025, 85, 103764. [Google Scholar] [CrossRef]
- Bou-Teen, D.; Miro-Casas, E.; Ruiz-Meana, M. Dicarbonyl stress and mitochondrial dysfunction in the aged heart. Aging 2023, 15, 3223–3225. [Google Scholar] [CrossRef]
- Akhter, F.; Chen, D.; Akhter, A.; Yan, S.F.; Yan, S.S. Age-dependent accumulation of dicarbonyls and advanced glycation endproducts (AGEs) associates with mitochondrial stress. Free Radic. Biol. Med. 2021, 164, 429–438. [Google Scholar] [CrossRef]
- Monzel, A.S.; Enriquez, J.A.; Picard, M. Multifaceted mitochondria: Moving mitochondrial science beyond function and dysfunction. Nat. Metab. 2023, 5, 546–562. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.F.; Tseng, L.M.; Lee, H.C. Role of mitochondrial alterations in human cancer progression and cancer immunity. J. Biomed. Sci. 2023, 30, 61. [Google Scholar] [CrossRef]
- Li, X.; Fang, P.; Mai, J.; Choi, E.T.; Wang, H.; Yang, X.F. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J. Hematol. Oncol. 2013, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qi, H.; Liu, Y.; Duan, C.; Liu, X.; Xia, T.; Chen, D.; Piao, H.L.; Liu, H.X. The double-edged roles of ROS in cancer prevention and therapy. Theranostics 2021, 11, 4839–4857. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yu, K.; Hu, H.; Zhang, X.; Zeng, S.; Li, J.; Dong, X.; Deng, X.; Zhang, J.; Zhang, Y. METTL17 coordinates ferroptosis and tumorigenesis by regulating mitochondrial translation in colorectal cancer. Redox Biol. 2024, 71, 103087. [Google Scholar] [CrossRef]
- Kuo, C.L.; Ponneri Babuharisankar, A.; Lin, Y.C.; Lien, H.W.; Lo, Y.K.; Chou, H.Y.; Tangeda, V.; Cheng, L.C.; Cheng, A.N.; Lee, A.Y. Mitochondrial oxidative stress in the tumor microenvironment and cancer immunoescape: Foe or friend? J. Biomed. Sci. 2022, 29, 74. [Google Scholar] [CrossRef]
- Martins, C.; Rasbach, E.; Heppt, M.V.; Singh, P.; Kulcsar, Z.; Holzgruber, J.; Chakraborty, A.; Mucciarone, K.; Kleffel, S.; Brandenburg, A.; et al. Tumor cell-intrinsic PD-1 promotes Merkel cell carcinoma growth by activating downstream mTOR-mitochondrial ROS signaling. Sci. Adv. 2024, 10, eadi2012. [Google Scholar] [CrossRef]
- Kuo, C.L.; Chou, H.Y.; Chiu, Y.C.; Cheng, A.N.; Fan, C.C.; Chang, Y.N.; Chen, C.H.; Jiang, S.S.; Chen, N.J.; Lee, A.Y. Mitochondrial oxidative stress by Lon-PYCR1 maintains an immunosuppressive tumor microenvironment that promotes cancer progression and metastasis. Cancer Lett. 2020, 474, 138–150. [Google Scholar] [CrossRef]
- Idelchik, M.; Begley, U.; Begley, T.J.; Melendez, J.A. Mitochondrial ROS control of cancer. Semin. Cancer Biol. 2017, 47, 57–66. [Google Scholar] [CrossRef]
- Chen, Y.; Fu, L.L.; Wen, X.; Wang, X.Y.; Liu, J.; Cheng, Y.; Huang, J. Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis. 2014, 5, e1047. [Google Scholar] [CrossRef]
- Zhang, J.; Xiang, H.; Liu, J.; Chen, Y.; He, R.R.; Liu, B. Mitochondrial Sirtuin 3: New emerging biological function and therapeutic target. Theranostics 2020, 10, 8315–8342. [Google Scholar] [CrossRef]
- Panda, B.; Tripathy, A.; Patra, S.; Kullu, B.; Tabrez, S.; Jena, M. Imperative connotation of SODs in cancer: Emerging targets and multifactorial role of action. IUBMB Life 2024, 76, 592–613. [Google Scholar] [CrossRef]
- Bell, E.L.; Emerling, B.M.; Ricoult, S.J.; Guarente, L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene 2011, 30, 2986–2996. [Google Scholar] [CrossRef]
- He, C.; Danes, J.M.; Hart, P.C.; Zhu, Y.; Huang, Y.; de Abreu, A.L.; O’Brien, J.; Mathison, A.J.; Tang, B.; Frasor, J.M.; et al. SOD2 acetylation on lysine 68 promotes stem cell reprogramming in breast cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 23534–23541. [Google Scholar] [CrossRef]
- Hjelmeland, A.B.; Patel, R.P. SOD2 acetylation and deacetylation: Another tale of Jekyll and Hyde in cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 23376–23378. [Google Scholar] [CrossRef] [PubMed]
- Maiti, G.P.; Sinha, S.; Mahmud, H.; Boysen, J.; Mendez, M.T.; Vesely, S.K.; Holter-Chakrabarty, J.; Kay, N.E.; Ghosh, A.K. SIRT3 overexpression and epigenetic silencing of catalase regulate ROS accumulation in CLL cells activating AXL signaling axis. Blood Cancer J. 2021, 11, 93. [Google Scholar] [CrossRef] [PubMed]
- Castellani, C.A.; Longchamps, R.J.; Sun, J.; Guallar, E.; Arking, D.E. Thinking outside the nucleus: Mitochondrial DNA copy number in health and disease. Mitochondrion 2020, 53, 214–223. [Google Scholar] [CrossRef]
- Hosgood, H.D., 3rd; Liu, C.S.; Rothman, N.; Weinstein, S.J.; Bonner, M.R.; Shen, M.; Lim, U.; Virtamo, J.; Cheng, W.L.; Albanes, D.; et al. Mitochondrial DNA copy number and lung cancer risk in a prospective cohort study. Carcinogenesis 2010, 31, 847–849. [Google Scholar] [CrossRef]
- Lemnrau, A.; Brook, M.N.; Fletcher, O.; Coulson, P.; Tomczyk, K.; Jones, M.; Ashworth, A.; Swerdlow, A.; Orr, N.; Garcia-Closas, M. Mitochondrial DNA Copy Number in Peripheral Blood Cells and Risk of Developing Breast Cancer. Cancer Res. 2015, 75, 2844–2850. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; You, Y.Q.; Li, M.Y.; Guan, X.; Fu, M.; Wang, C.M.; Xiao, Y.; He, M.A.; Guo, H. Mitochondrial DNA copy number mediated the associations between perfluoroalkyl substances and breast cancer incidence: A prospective case-cohort study. Sci. Total Environ. 2024, 941, 173767. [Google Scholar] [CrossRef]
- Weerts, M.J.; Sieuwerts, A.M.; Smid, M.; Look, M.P.; Foekens, J.A.; Sleijfer, S.; Martens, J.W. Mitochondrial DNA content in breast cancer: Impact on in vitro and in vivo phenotype and patient prognosis. Oncotarget 2016, 7, 29166–29176. [Google Scholar] [CrossRef]
- Sun, X.; Zhan, L.; Chen, Y.; Wang, G.; He, L.; Wang, Q.; Zhou, F.; Yang, F.; Wu, J.; Wu, Y.; et al. Increased mtDNA copy number promotes cancer progression by enhancing mitochondrial oxidative phosphorylation in microsatellite-stable colorectal cancer. Signal Transduct. Target. Ther. 2018, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sundquist, K.; Vats, S.; Hong, M.G.; Wang, X.; Chen, Y.; Hedelius, A.; Saal, L.H.; Sundquist, J.; Memon, A.A. Mitochondrial heteroplasmic shifts reveal a positive selection of breast cancer. J. Transl. Med. 2023, 21, 696. [Google Scholar] [CrossRef]
- Kulawiec, M.; Owens, K.M.; Singh, K.K. Cancer cell mitochondria confer apoptosis resistance and promote metastasis. Cancer Biol. Ther. 2009, 8, 1378–1385. [Google Scholar] [CrossRef]
- Petros, J.A.; Baumann, A.K.; Ruiz-Pesini, E.; Amin, M.B.; Sun, C.Q.; Hall, J.; Lim, S.; Issa, M.M.; Flanders, W.D.; Hosseini, S.H.; et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 719–724. [Google Scholar] [CrossRef]
- Sun, Q.; Arnold, R.S.; Sun, C.Q.; Petros, J.A. A mitochondrial DNA mutation influences the apoptotic effect of statins on prostate cancer. Prostate 2015, 75, 1916–1925. [Google Scholar] [CrossRef] [PubMed]
- Arnold, R.S.; Sun, C.Q.; Richards, J.C.; Grigoriev, G.; Coleman, I.M.; Nelson, P.S.; Hsieh, C.L.; Lee, J.K.; Xu, Z.; Rogatko, A.; et al. Mitochondrial DNA mutation stimulates prostate cancer growth in bone stromal environment. Prostate 2009, 69, 1–11. [Google Scholar] [CrossRef]
- Dasgupta, S.; Hoque, M.O.; Upadhyay, S.; Sidransky, D. Mitochondrial cytochrome B gene mutation promotes tumor growth in bladder cancer. Cancer Res. 2008, 68, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, H.; Kawase, K.; Nishi, T.; Watanabe, T.; Takenaga, K.; Inozume, T.; Ishino, T.; Aki, S.; Lin, J.S.; Kawashima, S.; et al. Immune evasion through mitochondrial transfer in the tumour microenvironment. Nature 2025, 638, 225–236. [Google Scholar] [CrossRef]
- Mahmood, M.; Liu, E.M.; Shergold, A.L.; Tolla, E.; Tait-Mulder, J.; Huerta-Uribe, A.; Shokry, E.; Young, A.L.; Lilla, S.; Kim, M.; et al. Mitochondrial DNA mutations drive aerobic glycolysis to enhance checkpoint blockade response in melanoma. Nat. Cancer 2024, 5, 659–672. [Google Scholar] [CrossRef]
- Tabara, L.C.; Segawa, M.; Prudent, J. Molecular mechanisms of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2025, 26, 123–146. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhao, H.; Li, Y. Mitochondrial dynamics in health and disease: Mechanisms and potential targets. Signal Transduct. Target. Ther. 2023, 8, 333. [Google Scholar] [CrossRef] [PubMed]
- Trotta, A.P.; Chipuk, J.E. Mitochondrial dynamics as regulators of cancer biology. Cell Mol. Life Sci. 2017, 74, 1999–2017. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yang, H.T.; Zhang, B.; Phillips, J.W.; Cheng, D.; Rigo, F.; Witte, O.N.; Xing, Y.; Black, D.L. The RNA-binding proteins hnRNP H and F regulate splicing of a MYC-dependent HRAS exon in prostate cancer cells. Proc. Natl. Acad. Sci. USA 2023, 120, e2220190120. [Google Scholar] [CrossRef]
- Zhao, L.; Zou, X.; Deng, J.; Sun, B.; Li, Y.; Zhao, L.; Zhao, H.; Zhang, X.; Yuan, X.; Zhao, X.; et al. hnRNPH1 maintains mitochondrial homeostasis by establishing NRF1/DRP1 retrograde signaling under mitochondrial stress. Cell Death Differ. 2025, 32, 118–133. [Google Scholar] [CrossRef]
- Ghosh, D.; Pakhira, S.; Das Ghosh, D.; Roychoudhury, S.; Roy, S.S. Ets1 facilitates EMT/invasion through Drp1-mediated mitochondrial fragmentation in ovarian cancer. Iscience 2023, 26, 107537. [Google Scholar] [CrossRef]
- Greier, M.C.; Runge, A.; Dudas, J.; Pider, V.; Skvortsova, I.I.; Savic, D.; Riechelmann, H. Mitochondrial dysfunction and epithelial to mesenchymal transition in head neck cancer cell lines. Sci. Rep. 2022, 12, 13255. [Google Scholar] [CrossRef]
- Vellinga, T.T.; Borovski, T.; de Boer, V.C.; Fatrai, S.; van Schelven, S.; Trumpi, K.; Verheem, A.; Snoeren, N.; Emmink, B.L.; Koster, J.; et al. SIRT1/PGC1alpha-Dependent Increase in Oxidative Phosphorylation Supports Chemotherapy Resistance of Colon Cancer. Clin. Cancer Res. 2015, 21, 2870–2879. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, J.; Yu, M.; Xie, Y.; Huang, Y.; Wolff, D.W.; Abel, P.W.; Tu, Y. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene 2013, 32, 4814–4824. [Google Scholar] [CrossRef]
- Artusa, V.; De Luca, L.; Clerici, M.; Trabattoni, D. Connecting the dots: Mitochondrial transfer in immunity, inflammation, and cancer. Immunol. Lett. 2025, 274, 106992. [Google Scholar] [CrossRef]
- Saha, T.; Dash, C.; Jayabalan, R.; Khiste, S.; Kulkarni, A.; Kurmi, K.; Mondal, J.; Majumder, P.K.; Bardia, A.; Jang, H.L.; et al. Intercellular nanotubes mediate mitochondrial trafficking between cancer and immune cells. Nat. Nanotechnol. 2022, 17, 98–106. [Google Scholar] [CrossRef]
- Hoover, G.; Gilbert, S.; Curley, O.; Obellianne, C.; Lin, M.T.; Hixson, W.; Pierce, T.W.; Andrews, J.F.; Alexeyev, M.F.; Ding, Y.; et al. Nerve-to-cancer transfer of mitochondria during cancer metastasis. Nature 2025, 644, 252–262. [Google Scholar] [CrossRef]
- Quiros, P.M.; Mottis, A.; Auwerx, J. Mitonuclear communication in homeostasis and stress. Nat. Rev. Mol. Cell Biol. 2016, 17, 213–226. [Google Scholar] [CrossRef]
- Jazwinski, S.M. The retrograde response: When mitochondrial quality control is not enough. Biochim. Biophys. Acta 2013, 1833, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Picard, M.; Shirihai, O.S. Mitochondrial signal transduction. Cell Metab. 2022, 34, 1620–1653. [Google Scholar] [CrossRef] [PubMed]
- Guha, M.; Avadhani, N.G. Mitochondrial retrograde signaling at the crossroads of tumor bioenergetics, genetics and epigenetics. Mitochondrion 2013, 13, 577–591. [Google Scholar] [CrossRef]
- Eniafe, J.; Jiang, S. The functional roles of TCA cycle metabolites in cancer. Oncogene 2021, 40, 3351–3363. [Google Scholar] [CrossRef]
- Kuo, C.C.; Wu, J.Y.; Wu, K.K. Cancer-derived extracellular succinate: A driver of cancer metastasis. J. Biomed. Sci. 2022, 29, 93. [Google Scholar] [CrossRef]
- Morin, A.; Goncalves, J.; Moog, S.; Castro-Vega, L.J.; Job, S.; Buffet, A.; Fontenille, M.J.; Woszczyk, J.; Gimenez-Roqueplo, A.P.; Letouze, E.; et al. TET-Mediated Hypermethylation Primes SDH-Deficient Cells for HIF2alpha-Driven Mesenchymal Transition. Cell Rep. 2020, 30, 4551–4566.e4557. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.; Qi, Y.; Xiong, G.; Li, J.; Scott, T.L.; Chen, J.; He, D.; Li, L.; Wang, C.; Lane, A.N.; et al. The PLOD2/succinate axis regulates the epithelial-mesenchymal plasticity and cancer cell stemness. Proc. Natl. Acad. Sci. USA 2023, 120, e2214942120. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Ong, L.T.; Jiang, Z.; Lee, W.C.; Lee, P.L.; Yusuf, M.; Ditzel, H.J.; Wang, Y.; Chen, Q.; Wang, W.; et al. Targeting P4HA1 promotes CD8(+) T cell progenitor expansion toward immune memory and systemic anti-tumor immunity. Cancer Cell 2025, 43, 213–231.e219. [Google Scholar] [CrossRef]
- Ren, J.G.; Seth, P.; Ye, H.; Guo, K.; Hanai, J.I.; Husain, Z.; Sukhatme, V.P. Citrate Suppresses Tumor Growth in Multiple Models through Inhibition of Glycolysis, the Tricarboxylic Acid Cycle and the IGF-1R Pathway. Sci. Rep. 2017, 7, 4537. [Google Scholar] [CrossRef]
- Gonsalves, W.I.; Jang, J.S.; Jessen, E.; Hitosugi, T.; Evans, L.A.; Jevremovic, D.; Pettersson, X.M.; Bush, A.G.; Gransee, J.; Anderson, E.I.; et al. In vivo assessment of glutamine anaplerosis into the TCA cycle in human pre-malignant and malignant clonal plasma cells. Cancer Metab. 2020, 8, 29. [Google Scholar] [CrossRef]
- San-Millan, I.; Brooks, G.A. Reexamining cancer metabolism: Lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 2017, 38, 119–133. [Google Scholar] [CrossRef]
- Shpilka, T.; Haynes, C.M. The mitochondrial UPR: Mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 2018, 19, 109–120. [Google Scholar] [CrossRef]
- Zhang, X.; Fan, Y.; Tan, K. A bird’s eye view of mitochondrial unfolded protein response in cancer: Mechanisms, progression and further applications. Cell Death Dis. 2024, 15, 667. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, M.; Jenkins, E.C.; Lechuga-Vieco, A.V.; Nie, K.; Fiel, M.I.; Rialdi, A.; Guccione, E.; Enriquez, J.A.; Sia, D.; Lujambio, A.; et al. The portrait of liver cancer is shaped by mitochondrial genetics. Cell Rep. 2022, 38, 110254. [Google Scholar] [CrossRef] [PubMed]
- Nwosu, G.O.; Powell, J.A.; Pitson, S.M. Targeting the integrated stress response in hematologic malignancies. Exp. Hematol. Oncol. 2022, 11, 94. [Google Scholar] [CrossRef]
- Deng, P.; Haynes, C.M. Mitochondrial dysfunction in cancer: Potential roles of ATF5 and the mitochondrial UPR. Semin. Cancer Biol. 2017, 47, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Kenny, T.C.; Craig, A.J.; Villanueva, A.; Germain, D. Mitohormesis Primes Tumor Invasion and Metastasis. Cell Rep. 2019, 27, 2292–2303.e2296. [Google Scholar] [CrossRef]
- Ishihara, S.; Enomoto, A.; Sakai, A.; Iida, T.; Tange, S.; Kioka, N.; Nukuda, A.; Nagasato, A.I.; Yasuda, M.; Tokino, T.; et al. Stiff extracellular matrix activates the transcription factor ATF5 to promote the proliferation of cancer cells. Iscience 2025, 28, 112057. [Google Scholar] [CrossRef] [PubMed]
- Woytash, J.A.; Kumar, R.; Chaudhary, A.K.; Donnelly, C.; Wojtulski, A.; Bethu, M.; Wang, J.; Spernyak, J.; Bross, P.; Yadav, N.; et al. Mitochondrial unfolded protein response-dependent beta-catenin signaling promotes neuroendocrine prostate cancer. Oncogene 2025, 44, 820–834. [Google Scholar] [CrossRef]
- Kim, Y.S.; Gupta Vallur, P.; Phaeton, R.; Mythreye, K.; Hempel, N. Insights into the Dichotomous Regulation of SOD2 in Cancer. Antioxidants 2017, 6, 86. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, H.D. The integrated stress response in metabolic adaptation. J. Biol. Chem. 2024, 300, 107151. [Google Scholar] [CrossRef]
- Chen, J.J. HRI protein kinase in cytoplasmic heme sensing and mitochondrial stress response: Relevance to hematological and mitochondrial diseases. J. Biol. Chem. 2025, 301, 108494. [Google Scholar] [CrossRef]
- Walker, E.M.; Pearson, G.L.; Lawlor, N.; Stendahl, A.M.; Lietzke, A.; Sidarala, V.; Zhu, J.; Stromer, T.; Reck, E.C.; Li, J.; et al. Retrograde mitochondrial signaling governs the identity and maturity of metabolic tissues. Science 2025, 388, eadf2034. [Google Scholar] [CrossRef]
- Cen, Y.; Lou, G.; Qi, J.; Li, M.; Zheng, M.; Liu, Y. Adipose-Derived Mesenchymal Stem Cells Inhibit JNK-Mediated Mitochondrial Retrograde Pathway to Alleviate Acetaminophen-Induced Liver Injury. Antioxidants 2023, 12, 158. [Google Scholar] [CrossRef]
- Zaglia, T.; Campo, A.; Moro, N.; Di Mauro, V.; Borile, G.; Menabo, R.; Antonucci, S.; Poli, L.; Campesan, M.; Carullo, P.; et al. Enhancement of mitochondrial calcium uptake is cardioprotective against maladaptive hypertrophy by retrograde signaling uptuning Akt. Proc. Natl. Acad. Sci. USA 2025, 122, e2402639122. [Google Scholar] [CrossRef]
- Perez, M.J.; Ivanyuk, D.; Panagiotakopoulou, V.; Di Napoli, G.; Kalb, S.; Brunetti, D.; Al-Shaana, R.; Kaeser, S.A.; Fraschka, S.A.; Jucker, M.; et al. Loss of function of the mitochondrial peptidase PITRM1 induces proteotoxic stress and Alzheimer’s disease-like pathology in human cerebral organoids. Mol. Psychiatry 2021, 26, 5733–5750. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Du, R.; Reis Monteiro Dos Santos, G.R.; Yefidoff-Freedman, R.; Bohm, A.; Halperin, J.; Chorev, M.; Aktas, B.H. New activators of eIF2alpha Kinase Heme-Regulated Inhibitor (HRI) with improved biophysical properties. Eur. J. Med. Chem. 2020, 187, 111973. [Google Scholar] [CrossRef] [PubMed]
- Cervia, L.D.; Shibue, T.; Borah, A.A.; Gaeta, B.; He, L.; Leung, L.; Li, N.; Moyer, S.M.; Shim, B.H.; Dumont, N.; et al. A Ubiquitination Cascade Regulating the Integrated Stress Response and Survival in Carcinomas. Cancer Discov. 2023, 13, 766–795. [Google Scholar] [CrossRef]
- Guo, X.Y.; Aviles, G.; Liu, Y.; Tian, R.L.; Unger, B.A.; Lin, Y.H.T.; Wiita, A.P.; Xu, K.; Correia, M.A.; Kampmann, M. Mitochondrial stress is relayed to the cytosol by an OMA1-DELE1-HRI pathway. Nature 2020, 579, 427–432. [Google Scholar] [CrossRef]
- Kold-Christensen, R.; Johannsen, M. Methylglyoxal Metabolism and Aging-Related Disease: Moving from Correlation toward Causation. Trends Endocrinol. Metab. 2020, 31, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, N.; Thornalley, P.J. Measurement of methylglyoxal by stable isotopic dilution analysis LC-MS/MS with corroborative prediction in physiological samples. Nat. Protoc. 2014, 9, 1969–1979. [Google Scholar] [CrossRef]
- Rabbani, N.; Thornalley, P.J. Dicarbonyl stress in cell and tissue dysfunction contributing to ageing and disease. Biochem. Biophys. Res. Commun. 2015, 458, 221–226. [Google Scholar] [CrossRef] [PubMed]
- Vistoli, G.; De Maddis, D.; Cipak, A.; Zarkovic, N.; Carini, M.; Aldini, G. Advanced glycoxidation and lipoxidation end products (AGEs and ALEs): An overview of their mechanisms of formation. Free Radical Res. 2013, 47, 3–27. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhou, C.; Huang, M.; Tang, C.; Liu, X.; Yue, Y.; Diao, Q.; Zheng, Z.; Liu, D. Glyoxalase system: A systematic review of its biological activity, related-diseases, screening methods and small molecule regulators. Biomed. Pharmacother. 2020, 131, 110663. [Google Scholar] [CrossRef]
- Clemente-Suarez, V.J.; Martin-Rodriguez, A.; Redondo-Florez, L.; Ruisoto, P.; Navarro-Jimenez, E.; Ramos-Campo, D.J.; Tornero-Aguilera, J.F. Metabolic Health, Mitochondrial Fitness, Physical Activity, and Cancer. Cancers 2023, 15, 814. [Google Scholar] [CrossRef]
- Vaskova, J.; Kovacova, G.; Pudelsky, J.; Palencar, D.; Mickova, H. Methylglyoxal Formation-Metabolic Routes and Consequences. Antioxidants 2025, 14, 212. [Google Scholar] [CrossRef]
- Schalkwijk, C.G.; Stehouwer, C.D.A. Methylglyoxal, a Highly Reactive Dicarbonyl Compound, in Diabetes, Its Vascular Complications, and Other Age-Related Diseases. Physiol. Rev. 2020, 100, 407–461. [Google Scholar] [CrossRef]
- de Bari, L.; Scirè, A.; Minnelli, C.; Cianfruglia, L.; Kalapos, M.P.; Armeni, T. Interplay among Oxidative Stress, Methylglyoxal Pathway and S-Glutathionylation. Antioxidants 2021, 10, 19. [Google Scholar] [CrossRef]
- Zhao, Y.; Yang, M.; Liang, X. The role of mitochondria in iron overload-induced damage. J. Transl. Med. 2024, 22, 1057. [Google Scholar] [CrossRef] [PubMed]
- Ito, G.; Tatara, Y.; Itoh, K.; Yamada, M.; Yamashita, T.; Sakamoto, K.; Nozaki, T.; Ishida, K.; Wake, Y.; Kaneko, T.; et al. Novel dicarbonyl metabolic pathway via mitochondrial ES1 possessing glyoxalase III activity. BBA Adv. 2023, 3, 100092. [Google Scholar] [CrossRef]
- Smith, A.J.; Advani, J.; Brock, D.C.; Nellissery, J.; Gumerson, J.; Dong, L.; Aravind, L.; Kennedy, B.; Swaroop, A. GATD3A, a mitochondrial deglycase with evolutionary origins from gammaproteobacteria, restricts the formation of advanced glycation end products. BMC Biol. 2022, 20, 68. [Google Scholar] [CrossRef]
- Masania, J.; Wijten, P.; Keipert, S.; Ost, M.; Klaus, S.; Rabbani, N.; Thornalley, P.J. Decreased methylglyoxal-mediated protein glycation in the healthy aging mouse model of ectopic expression of UCP1 in skeletal muscle. Redox Biol. 2023, 59, 102574. [Google Scholar] [CrossRef]
- Ceriello, A.; Ihnat, M.A.; Thorpe, J.E. Clinical review 2: The “metabolic memory”: Is more than just tight glucose control necessary to prevent diabetic complications? J. Clin. Endocrinol. Metab. 2009, 94, 410–415. [Google Scholar] [CrossRef]
- Pun, P.B.; Murphy, M.P. Pathological significance of mitochondrial glycation. Int. J. Cell Biol. 2012, 2012, 843505. [Google Scholar] [CrossRef]
- Hara, T.; Toyoshima, M.; Hisano, Y.; Balan, S.; Iwayama, Y.; Aono, H.; Futamura, Y.; Osada, H.; Owada, Y.; Yoshikawa, T. Glyoxalase I disruption and external carbonyl stress impair mitochondrial function in human induced pluripotent stem cells and derived neurons. Transl. Psychiatry 2021, 11, 275. [Google Scholar] [CrossRef] [PubMed]
- Pun, P.B.; Logan, A.; Darley-Usmar, V.; Chacko, B.; Johnson, M.S.; Huang, G.W.; Rogatti, S.; Prime, T.A.; Methner, C.; Krieg, T.; et al. A mitochondria-targeted mass spectrometry probe to detect glyoxals: Implications for diabetes. Free Radic. Biol. Med. 2014, 67, 437–450. [Google Scholar] [CrossRef]
- Samanta, S.; Akhter, F.; Xue, R.; Sosunov, A.A.; Wu, L.; Chen, D.; Arancio, O.; Yan, S.F.; Yan, S.S. Synaptic mitochondria glycation contributes to mitochondrial stress and cognitive dysfunction. Brain 2025, 148, 262–275. [Google Scholar] [CrossRef]
- Park, M.; Nishimura, T.; Baeza-Garza, C.D.; Caldwell, S.T.; Pun, P.B.L.; Prag, H.A.; Young, T.; Sauchanka, O.; Logan, A.; Forkink, M.; et al. Confirmation of the Cardioprotective Effect of MitoGamide in the Diabetic Heart. Cardiovasc. Drugs Ther. 2020, 34, 823–834. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.M.; Lindblom, R.S.J.; Ziemann, M.; Laskowski, A.; Granata, C.; Snelson, M.; Thallas-Bonke, V.; El-Osta, A.; Baeza-Garza, C.D.; Caldwell, S.T.; et al. Targeting Methylglyoxal in Diabetic Kidney Disease Using the Mitochondria-Targeted Compound MitoGamide. Nutrients 2021, 13, 1457. [Google Scholar] [CrossRef]
- Rosca, M.G.; Mustata, T.G.; Kinter, M.T.; Ozdemir, A.M.; Kern, T.S.; Szweda, L.I.; Brownlee, M.; Monnier, V.M.; Weiss, M.F. Glycation of mitochondrial proteins from diabetic rat kidney is associated with excess superoxide formation. Am. J. Physiol-Renal 2005, 289, F420–F430. [Google Scholar] [CrossRef]
- Hamelin, M.; Mary, J.; Vostry, M.; Friguet, B.; Bakala, H. Glycation damage targets glutamate dehydrogenase in the rat liver mitochondrial matrix during aging. FEBS J. 2007, 274, 5949–5961. [Google Scholar] [CrossRef]
- Prestes, A.S.; Dos Santos, M.M.; Kamdem, J.P.; Mancini, G.; Schuler da Silva, L.C.; de Bem, A.F.; Barbosa, N.V. Methylglyoxal disrupts the functionality of rat liver mitochondria. Chem. Biol. Interact. 2022, 351, 109677. [Google Scholar] [CrossRef]
- Kim, D.; Kim, K.A.; Kim, J.H.; Kim, E.H.; Bae, O.N. Methylglyoxal-Induced Dysfunction in Brain Endothelial Cells via the Suppression of Akt/HIF-1alpha Pathway and Activation of Mitophagy Associated with Increased Reactive Oxygen Species. Antioxidants 2020, 9, 820. [Google Scholar] [CrossRef]
- Chan, C.M.; Huang, D.Y.; Huang, Y.P.; Hsu, S.H.; Kang, L.Y.; Shen, C.M.; Lin, W.W. Methylglyoxal induces cell death through endoplasmic reticulum stress-associated ROS production and mitochondrial dysfunction. J. Cell Mol. Med. 2016, 20, 1749–1760. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.Z.; Guo, C.; Dong, J.Q.; Zhang, J.; Sun, K.T.; Lu, G.J.; Wang, L.; Bo, D.Y.; Jiao, L.Y.; Zhao, G.A. Glyoxal damages human aortic endothelial cells by perturbing the glutathione, mitochondrial membrane potential, and mitogen-activated protein kinase pathways. BMC Cardiovasc. Disord. 2021, 21, 603. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, S.; Carvalho, C.; Marinho, R.; Simoes, A.; Sena, C.M.; Matafome, P.; Santos, M.S.; Seica, R.M.; Moreira, P.I. Effects of methylglyoxal and pyridoxamine in rat brain mitochondria bioenergetics and oxidative status. J. Bioenerg. Biomembr. 2014, 46, 347–355. [Google Scholar] [CrossRef]
- Egawa, T.; Ogawa, T.; Yokokawa, T.; Kido, K.; Goto, K.; Hayashi, T. Methylglyoxal reduces molecular responsiveness to 4 weeks of endurance exercise in mouse plantaris muscle. J. Appl. Physiol. 2022, 132, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, N. Methylglyoxal and glyoxalase 1-a metabolic stress pathway-linking hyperglycemia to the unfolded protein response and vascular complications of diabetes. Clin. Sci. 2022, 136, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, N.; Xue, M.; Thornalley, P.J. Dicarbonyl stress, protein glycation and the unfolded protein response. Glycoconj. J. 2021, 38, 331–340. [Google Scholar] [CrossRef]
- Xue, M.; Irshad, Z.; Rabbani, N.; Thornalley, P.J. Increased cellular protein modification by methylglyoxal activates endoplasmic reticulum-based sensors of the unfolded protein response. Redox Biol. 2024, 69, 103025. [Google Scholar] [CrossRef]
- Ahmad, S.; Khan, H.; Siddiqui, Z.; Khan, M.Y.; Rehman, S.; Shahab, U.; Godovikova, T.; Silnikov, V.; Moinuddin. AGEs, RAGEs and s-RAGE; friend or foe for cancer. Semin. Cancer Biol. 2018, 49, 44–55. [Google Scholar] [CrossRef]
- Zhou, M.Z.; Zhang, Y.Y.; Shi, L.; Li, L.C.; Zhang, D.; Gong, Z.H.; Wu, Q. Activation and modulation of the AGEs-RAGE axis: Implications for inflammatory pathologies and therapeutic interventions—A review. Pharmacol. Res. 2024, 206, 107282. [Google Scholar] [CrossRef]
- Akhter, F.; Chen, D.; Akhter, A.; Sosunov, A.A.; Chen, A.; McKhann, G.M.; Yan, S.F.; Yan, S.S. High Dietary Advanced Glycation End Products Impair Mitochondrial and Cognitive Function. J. Alzheimers Dis. 2020, 76, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Velayoudom-Cephise, F.L.; Cano-Sanchez, M.; Bercion, S.; Tessier, F.; Yu, Y.; Boulanger, E.; Neviere, R. Receptor for advanced glycation end products modulates oxidative stress and mitochondrial function in the soleus muscle of mice fed a high-fat diet. Appl. Physiol. Nutr. Metab. 2020, 45, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Coughlan, M.T.; Thorburn, D.R.; Penfold, S.A.; Laskowski, A.; Harcourt, B.E.; Sourris, K.C.; Tan, A.L.; Fukami, K.; Thallas-Bonke, V.; Nawroth, P.P.; et al. RAGE-induced cytosolic ROS promote mitochondrial superoxide generation in diabetes. J. Am. Soc. Nephrol. 2009, 20, 742–752. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, L.; Delguste, F.; Durand, A.; Guilbaud, A.; Rousselin, C.; Schmidt, A.M.; Tessier, F.; Boulanger, E.; Neviere, R. Advanced glycation end products receptor RAGE controls myocardial dysfunction and oxidative stress in high-fat fed mice by sustaining mitochondrial dynamics and autophagy-lysosome pathway. Free Radic. Biol. Med. 2017, 112, 397–410. [Google Scholar] [CrossRef]
- Jeong, S.R.; Lee, K.W. Methylglyoxal-Derived Advanced Glycation End Product (AGE4)-Induced Apoptosis Leads to Mitochondrial Dysfunction and Endoplasmic Reticulum Stress through the RAGE/JNK Pathway in Kidney Cells. Int. J. Mol. Sci. 2021, 22, 6530. [Google Scholar] [CrossRef]
- Mao, Y.X.; Cai, W.J.; Sun, X.Y.; Dai, P.P.; Li, X.M.; Wang, Q.; Huang, X.L.; He, B.; Wang, P.P.; Wu, G.; et al. RAGE-dependent mitochondria pathway: A novel target of silibinin against apoptosis of osteoblastic cells induced by advanced glycation end products. Cell Death Dis. 2018, 9, 674. [Google Scholar] [CrossRef]
- Patel, S.H.; Yue, F.; Saw, S.K.; Foguth, R.; Cannon, J.R.; Shannahan, J.H.; Kuang, S.; Sabbaghi, A.; Carroll, C.C. Advanced Glycation End-Products Suppress Mitochondrial Function and Proliferative Capacity of Achilles Tendon-Derived Fibroblasts. Sci. Rep. 2019, 9, 12614. [Google Scholar] [CrossRef]
- Pearson-Stuttard, J.; Zhou, B.; Kontis, V.; Bentham, J.; Gunter, M.J.; Ezzati, M. Worldwide burden of cancer attributable to diabetes and high body-mass index: A comparative risk assessment. Lancet Diabetes Endocrinol. 2018, 6, e6–e15. [Google Scholar] [CrossRef]
- Lega, I.C.; Lipscombe, L.L. Review: Diabetes, Obesity, and Cancer-Pathophysiology and Clinical Implications. Endocr. Rev. 2020, 41, 33–52. [Google Scholar] [CrossRef] [PubMed]
- Mayen, A.L.; Aglago, E.K.; Knaze, V.; Cordova, R.; Schalkwijk, C.G.; Wagner, K.H.; Aleksandrova, K.; Fedirko, V.; Keski-Rahkonen, P.; Leitzmann, M.F.; et al. Dietary intake of advanced glycation endproducts and risk of hepatobiliary cancers: A multinational cohort study. Int. J. Cancer 2021, 149, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Stolzenberg-Solomon, R.; Zimmerman, T.P.; Duan, Z.; Chen, L.; Kahle, L.; Risch, A.; Subar, A.F.; Cross, A.J.; Hollenbeck, A.; et al. Dietary consumption of advanced glycation end products and pancreatic cancer in the prospective NIH-AARP Diet and Health Study. Am. J. Clin. Nutr. 2015, 101, 126–134. [Google Scholar] [CrossRef]
- Omofuma, O.O.; Turner, D.P.; Peterson, L.L.; Merchant, A.T.; Zhang, J.; Steck, S.E. Dietary Advanced Glycation End-products (AGE) and Risk of Breast Cancer in the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO). Cancer Prev. Res. 2020, 13, 601–610. [Google Scholar] [CrossRef]
- Peterson, L.L.; Park, S.; Park, Y.; Colditz, G.A.; Anbardar, N.; Turner, D.P. Dietary advanced glycation end products and the risk of postmenopausal breast cancer in the National Institutes of Health-AARP Diet and Health Study. Cancer 2020, 126, 2648–2657. [Google Scholar] [CrossRef] [PubMed]
- Córdova, R.; Mayén, A.L.; Knaze, V.; Aglago, E.K.; Schalkwijk, C.; Wagner, K.H.; Overvad, K.; Tjonneland, A.; Kyro, C.; Katzke, V.A.; et al. Dietary intake of advanced glycation endproducts (AGEs) and cancer risk across more than 20 anatomical sites: A multinational cohort study. Cancer Commun. 2022, 42, 1041–1045. [Google Scholar] [CrossRef]
- Deng, R.Y.; Mo, F.B.; Chang, B.W.; Zhang, Q.; Ran, H.; Yang, S.H.; Zhu, Z.Q.; Hu, L.; Su, Q. Glucose-derived AGEs enhance human gastric cancer metastasis through RAGE/ERK/Sp1/MMP2 cascade. Oncotarget 2017, 8, 104216–104226. [Google Scholar] [CrossRef]
- Pan, S.; Guan, Y.; Ma, Y.; Cui, Q.; Tang, Z.; Li, J.; Zu, C.; Zhang, Y.; Zhu, L.; Jiang, J.; et al. Advanced glycation end products correlate with breast cancer metastasis by activating RAGE/TLR4 signaling. BMJ Open Diabetes Res. Care 2022, 10, e002697. [Google Scholar] [CrossRef] [PubMed]
- Peterson, L.L.; Ligibel, J.A. Dietary and serum advanced glycation end-products and clinical outcomes in breast cancer. Biochim. Biophys. Acta Rev. Cancer 2024, 1879, 188995. [Google Scholar] [CrossRef] [PubMed]
- Krisanits, B.A.; Schuster, R.; Randise, J.; Nogueira, L.M.; Lane, J.T.; Panguluri, G.A.; Li, H.; Helke, K.; Cuitino, M.C.; Koivisto, C.; et al. Pubertal exposure to dietary advanced glycation end products disrupts ductal morphogenesis and induces atypical hyperplasia in the mammary gland. Breast Cancer Res. 2023, 25, 118. [Google Scholar] [CrossRef]
- Chen, Y.J.; Guo, T.L. Dietary Early Glycation Products Promote the Growth of Prostate Tumors More than Advanced Glycation End-Products through Modulation of Macrophage Polarization. Mol. Nutr. Food Res. 2019, 63, 1800885. [Google Scholar] [CrossRef]
- Menini, S.; Iacobini, C.; de Latouliere, L.; Manni, I.; Ionta, V.; Blasetti Fantauzzi, C.; Pesce, C.; Cappello, P.; Novelli, F.; Piaggio, G.; et al. The advanced glycation end-product N(ϵ) -carboxymethyllysine promotes progression of pancreatic cancer: Implications for diabetes-associated risk and its prevention. J. Pathol. 2018, 245, 197–208. [Google Scholar] [CrossRef]
- Chang, T.Y.; Lan, K.C.; Wu, C.H.; Sheu, M.L.; Yang, R.S.; Liu, S.H. Nε-(1-Carboxymethyl)-L-lysine, an advanced glycation end product, exerts malignancy on chondrosarcoma via the activation of cancer stemness. Arch. Toxicol. 2023, 97, 2231–2244. [Google Scholar] [CrossRef]
- Bartling, B.; Hofmann, H.S.; Sohst, A.; Hatzky, Y.; Somoza, V.; Silber, R.E.; Simm, A. Prognostic potential and tumor growth-inhibiting effect of plasma advanced glycation end products in non-small cell lung carcinoma. Mol. Med. 2011, 17, 980–989. [Google Scholar] [CrossRef]
- Pusterla, T.; Németh, J.; Stein, I.; Wiechert, L.; Knigin, D.; Marhenke, S.; Longerich, T.; Kumar, V.; Arnold, B.; Vogel, A.; et al. Receptor for Advanced Glycation Endproducts (RAGE) Is a Key Regulator of Oval Cell Activation and Inflammation-Associated Liver Carcinogenesis in Mice. Hepatology 2013, 58, 363–373. [Google Scholar] [CrossRef]
- Vernon, P.J.; Loux, T.J.; Schapiro, N.E.; Kang, R.; Muthuswamy, R.; Kalinski, P.; Tang, D.; Lotze, M.T.; Zeh, H.J., 3rd. The receptor for advanced glycation end products promotes pancreatic carcinogenesis and accumulation of myeloid-derived suppressor cells. J. Immunol. 2013, 190, 1372–1379. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Matsui, T.; Ishibashi, Y.; Sotokawauchi, A.; Fukami, K.; Higashimoto, Y.; Yamagishi, S.I. RAGE-aptamer Attenuates the Growth and Liver Metastasis of Malignant Melanoma in Nude Mice. Mol. Med. 2017, 23, 295–306. [Google Scholar] [CrossRef]
- Ojima, A.; Matsui, T.; Maeda, S.; Takeuchi, M.; Inoue, H.; Higashimoto, Y.; Yamagishi, S. DNA aptamer raised against advanced glycation end products inhibits melanoma growth in nude mice. Lab. Invest. 2014, 94, 422–429. [Google Scholar] [CrossRef]
- Elangovan, I.; Thirugnanam, S.; Chen, A.; Zheng, G.; Bosland, M.C.; Kajdacsy-Balla, A.; Gnanasekar, M. Targeting receptor for advanced glycation end products (RAGE) expression induces apoptosis and inhibits prostate tumor growth. Biochem. Biophys. Res. Commun. 2012, 417, 1133–1138. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.R.; Jensen, A.; Ng, S.; Yin, Z.R.; Li, A.M.; Misra, A.; Von Hoff, D.D.; Gruber, L.; Gruber, M.; Han, H.Y. Advanced glycation end product (AGE) targeting antibody SIWA318H is efficacious in preclinical models for pancreatic cancer. Sci. Rep-Uk 2023, 13, 16953. [Google Scholar] [CrossRef] [PubMed]
- Kwak, T.; Drews-Elger, K.; Ergonul, A.; Miller, P.C.; Braley, A.; Hwang, G.H.; Zhao, D.; Besser, A.; Yamamoto, Y.; Yamamoto, H.; et al. Targeting of RAGE-ligand signaling impairs breast cancer cell invasion and metastasis. Oncogene 2017, 36, 1559–1572. [Google Scholar] [CrossRef]
- Nasser, M.W.; Wani, N.A.; Ahirwar, D.K.; Powell, C.A.; Ravi, J.; Elbaz, M.; Zhao, H.; Padilla, L.; Zhang, X.; Shilo, K.; et al. RAGE mediates S100A7-induced breast cancer growth and metastasis by modulating the tumor microenvironment. Cancer Res. 2015, 75, 974–985. [Google Scholar] [CrossRef]
- Chen, M.C.; Chen, K.C.; Chang, G.C.; Lin, H.; Wu, C.C.; Kao, W.H.; Teng, C.J.; Hsu, S.L.; Yang, T.Y. RAGE acts as an oncogenic role and promotes the metastasis of human lung cancer. Cell Death Dis. 2020, 11, 265. [Google Scholar] [CrossRef] [PubMed]
- Takino, J.; Yamagishi, S.; Takeuchi, M. Cancer malignancy is enhanced by glyceraldehyde-derived advanced glycation end-products. J. Oncol. 2010, 2010, 739852. [Google Scholar] [CrossRef]
- Ko, S.Y.; Ko, H.A.; Shieh, T.M.; Chang, W.C.; Chen, H.I.; Chang, S.S.; Lin, I.H. Cell migration is regulated by AGE-RAGE interaction in human oral cancer cells in vitro. PLoS ONE 2014, 9, e110542. [Google Scholar] [CrossRef]
- Sharaf, H.; Matou-Nasri, S.; Wang, Q.; Rabhan, Z.; Al-Eidi, H.; Al Abdulrahman, A.; Ahmed, N. Advanced glycation endproducts increase proliferation, migration and invasion of the breast cancer cell line MDA-MB-231. Biochim. Biophys. Acta 2015, 1852, 429–441. [Google Scholar] [CrossRef]
- Kuniyasu, H.; Chihara, Y.; Kondo, H. Differential effects between amphoterin and advanced glycation end products on colon cancer cells. Int. J. Cancer 2003, 104, 722–727. [Google Scholar] [CrossRef]
- Goodson, W.H., 3rd; Lowe, L.; Carpenter, D.O.; Gilbertson, M.; Manaf Ali, A.; Lopez de Cerain Salsamendi, A.; Lasfar, A.; Carnero, A.; Azqueta, A.; Amedei, A.; et al. Assessing the carcinogenic potential of low-dose exposures to chemical mixtures in the environment: The challenge ahead. Carcinogenesis 2015, 36 (Suppl. S1), S254–S296. [Google Scholar] [CrossRef] [PubMed]
- Salaroglio, I.C.; Belisario, D.C.; Akman, M.; La Vecchia, S.; Godel, M.; Anobile, D.P.; Ortone, G.; Digiovanni, S.; Fontana, S.; Costamagna, C.; et al. Mitochondrial ROS drive resistance to chemotherapy and immune-killing in hypoxic non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2022, 41, 243. [Google Scholar] [CrossRef] [PubMed]
| Species (Sex) | Target Organ | Treatment | Adverse Outcome | Reference | |
|---|---|---|---|---|---|
| Mitochondria | Cell/Organ | ||||
| C57BL/6 mice (M, F) | Brain | * AGE diet (1000 mg/kg, 17 months) | ** Oxphos capacity ↓ ATP production ↓ | Cognitive impairment | [126] |
| C57BL/6N mice (M) | Kidney | * AGEs diet (800 mg/kg, p.o., 3 weeks) | - | ER stress ATF4/CHOP, GRP78 ↑ p-JNK/JNK ↑ | [130] |
| RAGE-deficient C57BL/6J mice (M) | Skeletal muscle | High-fat diet (4 months) | * Oxphos capacity ↓ | Inflammatory response (IL-1β) | [127] |
| RAGE-deficient C57BL/6J mice (M) | Heart | High-fat diet (4 months) | Mitochondria count ↓ Morphological change | Oxidative stress | [129] |
| SD rats (M) | Kidney | AGEs (20 mg/kg, i.p., 16 weeks) | * Oxphos capacity ↓ Mitochondrial NADH ↓ | - | [128] |
| Cell Type | Origin | AGE Treatment | Adverse Outcome | Reference | |
|---|---|---|---|---|---|
| Mitochondria | Cell | ||||
| Kidney proximal epithelial cell line (HK-2) | Human | 200 μg/mL, 24 h (MG-derived AGE) | ATP production ↓ MMP ↓ | ER stress ATF4/CHOP, GRP78 ↑ p-JNK/JNK ↑ | [130] |
| Osteoblastic cell line (MC3T3-E1) | Mouse | * 400 μg/mL, 24 h | ROS production ↑ ATP production ↓ MMP ↓, fission ↑ | Apoptosis | [131] |
| RAGE-overexpressed primary mesangial cells | Rat | 100 μg/mL, 48 h (Glucose-derived AGE) | ** Oxphos capacity ↓ Mitochondrial permeability transition ↑ | Oxidative stress | [128] |
| Primary rat fibroblast | Rat | 500 μg/mL, 0.5 h (Glycolaldehyde-derived AGE) | mtDNA count ↑ ROS production ↑ ATP production ↓ | Extracellular matrix remodeling | [132] |
| Cancer Type | Animals (Sex) | Treatment (Dosage, Route, Duration) | Adverse Outcome | Reference |
|---|---|---|---|---|
| Breast cancer | FVB/n mice (F) | 15–25 g AGE diet/week, p.o., 4–25 weeks |
| [143] |
| Chondrosarcoma | NOD/SCID mice (M) | 40 mg streptozotocin/kg body weight/day, 5 days |
| [146] |
| Lung cancer | NMRI nu/nu mice (F) | 6 g AGE diet/day, p.o., 14 days |
| [147] |
| Prostate cancer | C57BL/6 mice (M) | 600 mg EGPs/kg body weight/day, p.o., 4 weeks |
| [144] |
| Pancreatic cancer | KC mice (N/A) | 30 μg AGE/day, i.p., 6 weeks |
| [145] |
| Cancer Type | Animals (Sex) | Treatment (Dosage, Route, Duration) | Adverse Outcome | Reference |
|---|---|---|---|---|
| Breast cancer | C57BL6 mice (N/A) | RAGE knockout |
| [154] |
| C57B/6 mice (N/A) | RAGE knockout |
| [155] | |
| Liver cancer | Mdr2−/− C57Bl/6 mice (M) | RAGE knockout |
| [148] |
| Lung cancer | BALB/c nude mice (M) | Inoculating RAGE-overexpressed A549 |
| [156] |
| Prostate cancer | Nude mice (M) | shRAGE (100 μg, 5 times/week, i.p., 6 weeks) |
| [152] |
| Pancreatic cancer | Humanized CD34+ NSG mice (F) | AGE antibody (10 or 20 mg/kg BIW × 1 followed by 5 or 10 mg/kg BIW × 2) |
| [153] |
| Pdx1-Cre:KrasG12D/+ C57BL/6 mice (N/A) | RAGE knockout |
| [149] | |
| Skin cancer | Nude mice (F) | RAGE aptamer (38.4 pmol/day/g body weight, i.p., 42 days) |
| [150] |
| Athymic nude mice (F) | AGE aptamer (0.136 μg/day, i.p., 43 days) |
| [151] |
| Cell Type | Origin | Treatment | Adverse Outcome | Reference |
|---|---|---|---|---|
| Chondrosarcoma cell line (JJ012, SW1353) | Human | 25–100 μM CML, 24–72 h |
| [146] |
| Human umbilical vein endothelial cells (HUVECs) | Human | 100 μg/mL AGE-BSA, 24 h |
| [151] |
| 50 μg/mL AGE-BSA, 4 h |
| [150] | ||
| Melanoma cell line (G361) | Human | 1000 μg/mL AGE-BSA, 24 h |
| [151] |
| 1000 μg/mL AGE-BSA, 24 h |
| [150] | ||
| Prostate cancer cell line (LNCaP) /PMA-differentiated macrophages (d-U937) | Human | 2.5 mg/mL early glycation products (EGPs), 48 h |
| [144] |
| Pancreatic ductal adenocarcinoma cell line (PANC-1) | Human | 50 μg/mL CML, 24 h |
| [145] |
| Primary mammary fibroblast /Mammary epithelial cell-line (HC11)/mammary gland carcinoma (Met1) | Mouse | 50 μg/mL BSA-AGE, 24 h |
| [143] |
| Cancer Hallmarks | Key Mediators | Changes | Effect | Reference |
|---|---|---|---|---|
| Angiogenesis | CXCL10 | Decreased | Anticarcinogenic | [149] |
| Dysregulated metabolism | ROS | Increased | Procarcinogenic | [150] |
| Immune system evasion | Akt | Decreased | Anticarcinogenic | [146] |
| CXCL10 | Decreased | Anticarcinogenic | [149] | |
| Sustained proliferative signaling | Cyclin D1 | Increased | Procarcinogenic | [150] |
| Activating invasion and metastasis | EMT | Increased | Procarcinogenic | [146] |
| Tumor microenvironment | Oxidative stress | Increased | Procarcinogenic | [150] |
| IL-6 | Decreased | Anticarcinogenic | [149] | |
| Tumor-promoting inflammation | NFκB | Increased | Procarcinogenic | [145,146] |
| IL-6 | Decreased | Anticarcinogenic | [149] |
| Mitohormesis | Cancer Hallmarks | Effect | Reference | |
|---|---|---|---|---|
| Signaling | Response | |||
| mtROS | NF-κB ↑ | Tumor-promoting inflammation | Procarcinogenic | [131,132] |
| Akt ↑ | Immune system evasion | Procarcinogenic | ||
| * UPR ↑ | Dysregulated metabolism | Procarcinogenic | ||
| Dynamics | EMT ↑ | Invasion and metastasis | Procarcinogenic | [131] |
| * mtDNA | N/A | Invasion and metastasis | Procarcinogenic | [132] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.; Choi, K.-N.; Park, J.-I.; Kim, E.-H.; Majid, A.; Bae, O.-N. Role of Advanced Glycation End Products and Mitohormesis in Cancer Development and Progression. Antioxidants 2025, 14, 1165. https://doi.org/10.3390/antiox14101165
Kim D, Choi K-N, Park J-I, Kim E-H, Majid A, Bae O-N. Role of Advanced Glycation End Products and Mitohormesis in Cancer Development and Progression. Antioxidants. 2025; 14(10):1165. https://doi.org/10.3390/antiox14101165
Chicago/Turabian StyleKim, Donghyun, Kyung-Nam Choi, Jong-In Park, Eun-Hye Kim, Arshad Majid, and Ok-Nam Bae. 2025. "Role of Advanced Glycation End Products and Mitohormesis in Cancer Development and Progression" Antioxidants 14, no. 10: 1165. https://doi.org/10.3390/antiox14101165
APA StyleKim, D., Choi, K.-N., Park, J.-I., Kim, E.-H., Majid, A., & Bae, O.-N. (2025). Role of Advanced Glycation End Products and Mitohormesis in Cancer Development and Progression. Antioxidants, 14(10), 1165. https://doi.org/10.3390/antiox14101165






