Resveratrol-Enriched Rice Callus Extract Inhibits Oxidative and Cellular Melanogenic Activities in Melan-A Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Treatments
2.2. ABTS Radical Scavenging Ability
2.3. Vitamin C Equivalent Antioxidant Capacity and IC50
2.4. Melan-A Cell Viability
2.5. Melanin Content
2.6. L-DOPA Staining
2.7. Cellular Tyrosinase Activity
2.8. Morphological Appearance
2.9. Gene Expression Quantification
2.10. Western Blot
2.11. Statistical Analysis
3. Results
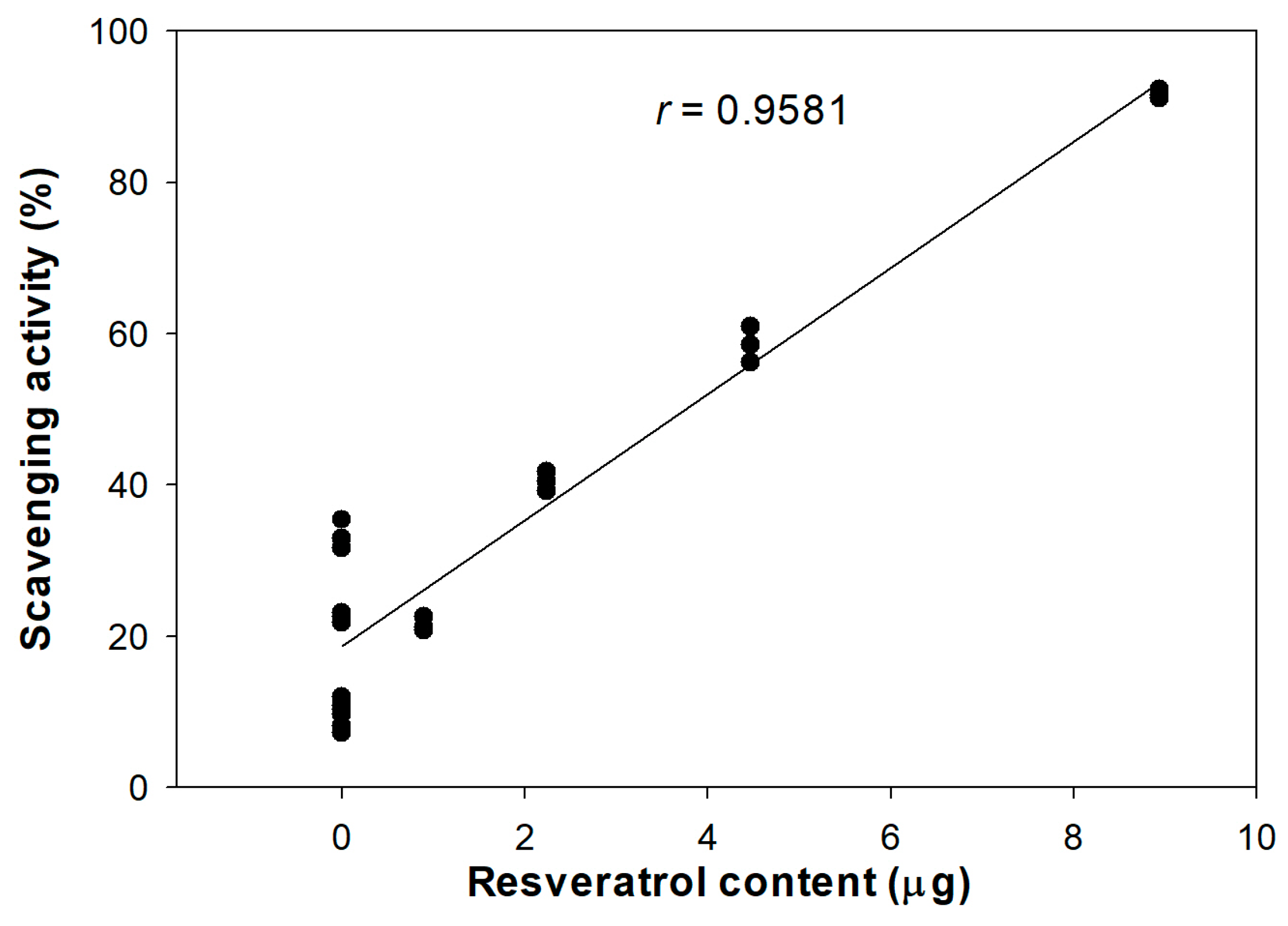
3.1. Antioxidant Activity of DJ526 Rice Callus Extract
3.2. Melan-A Cell Viability Effect of DJ526 Rice Callus Extract
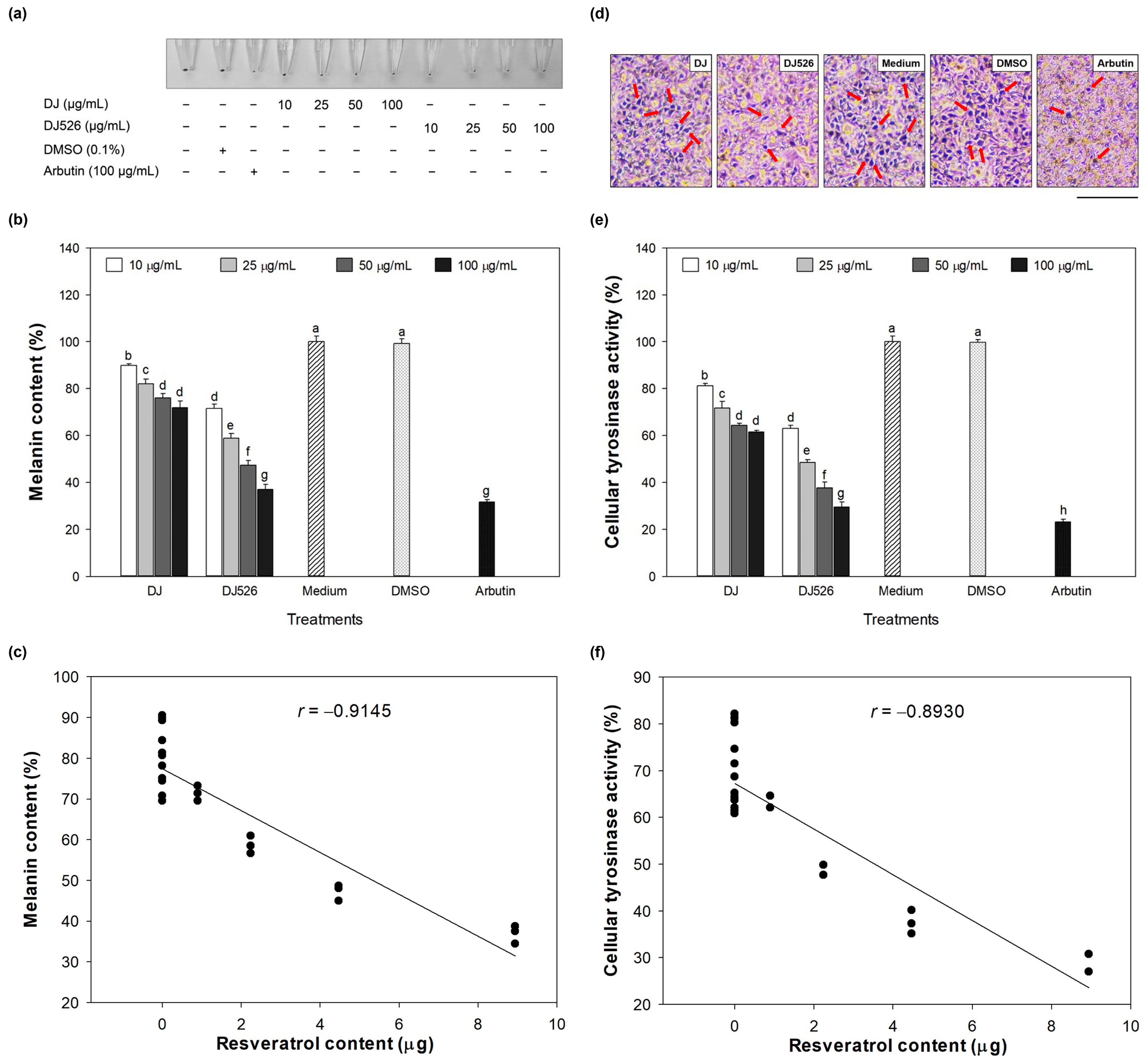
3.3. Melanin Content and Cellular Tyrosinase Activity Effect of DJ526 Rice Callus Extract
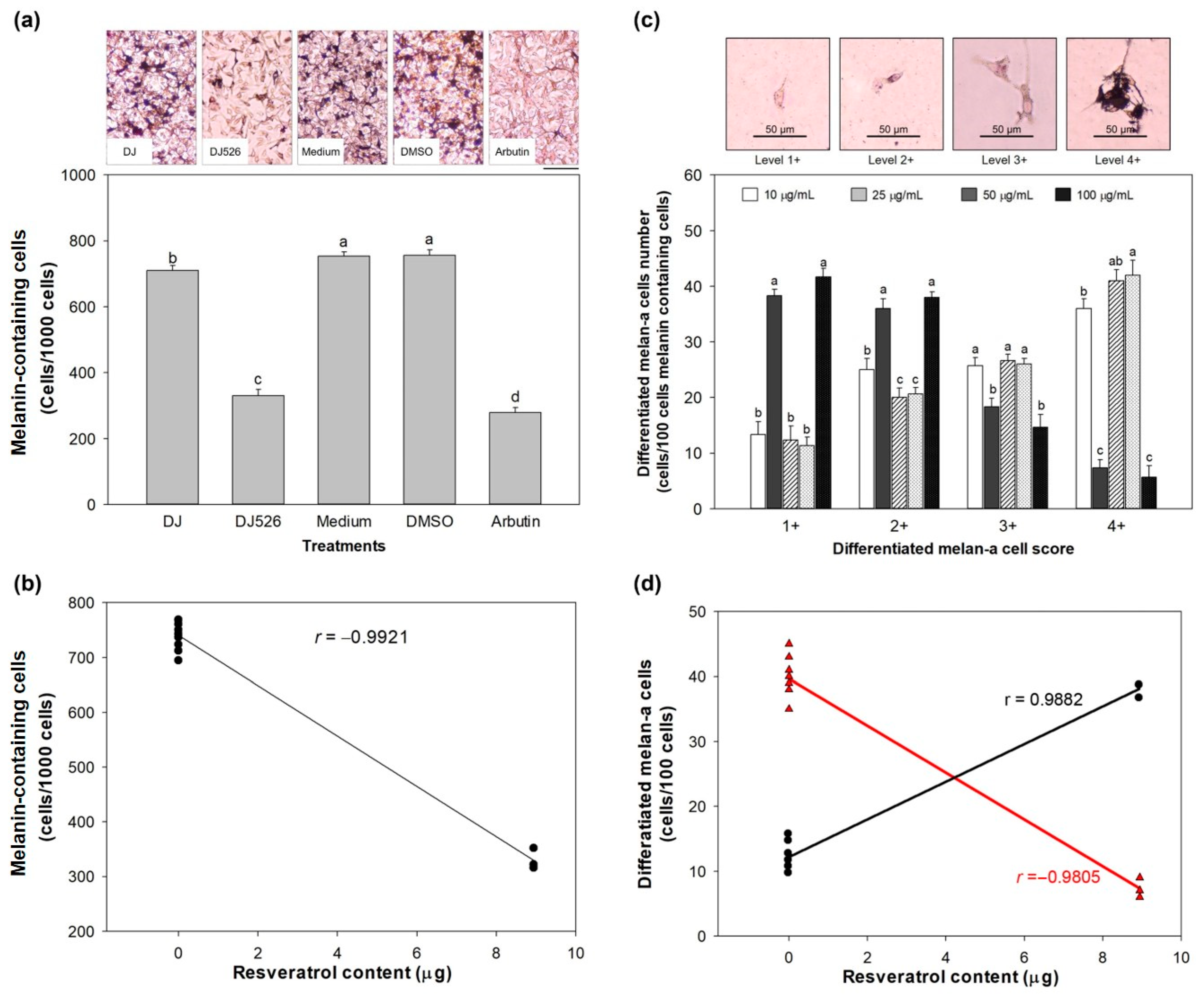
3.4. Effect of DJ526 Rice Callus Extract on Melanin Content and Morphological Appearance of Melan-A Cells
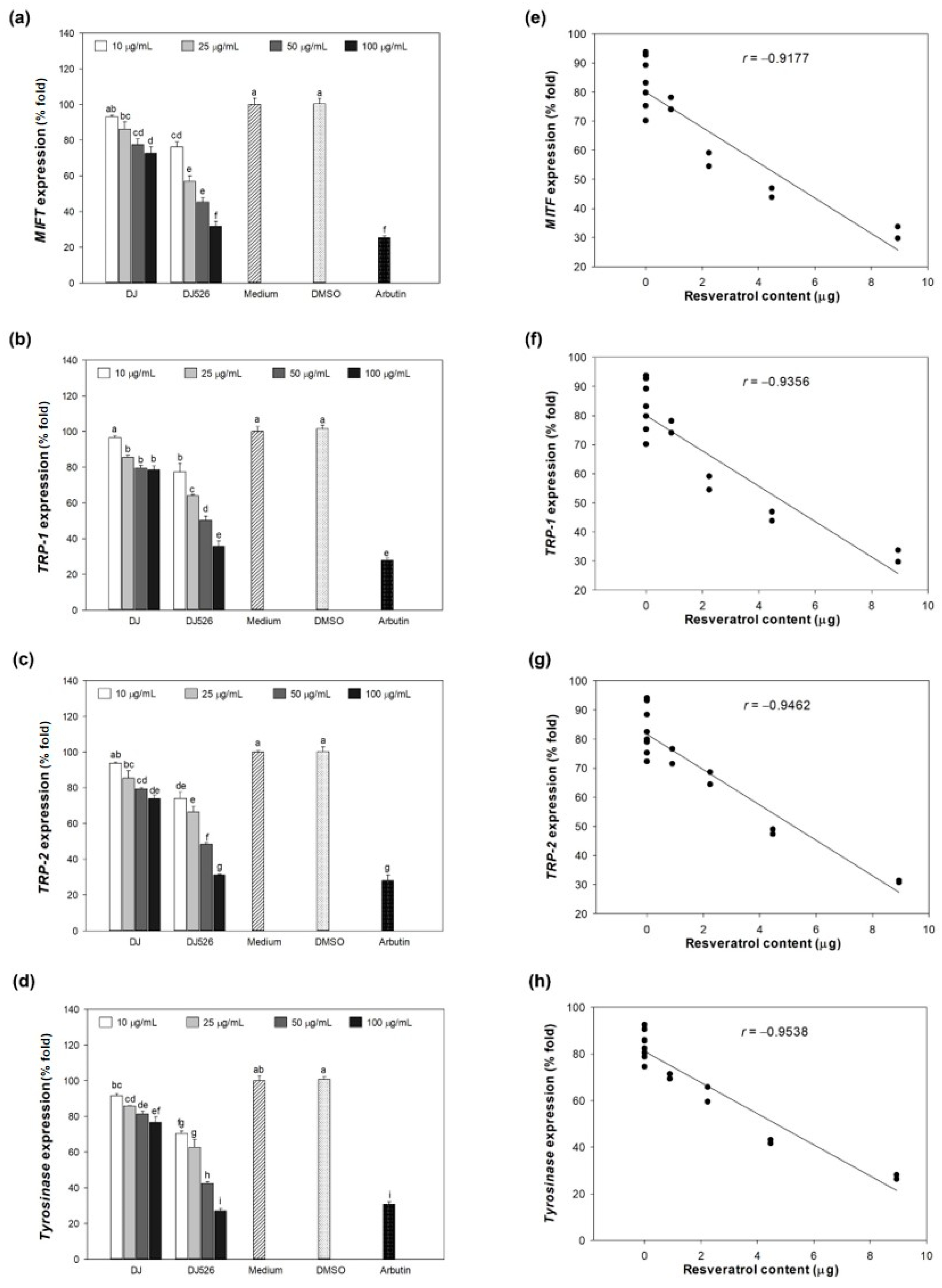
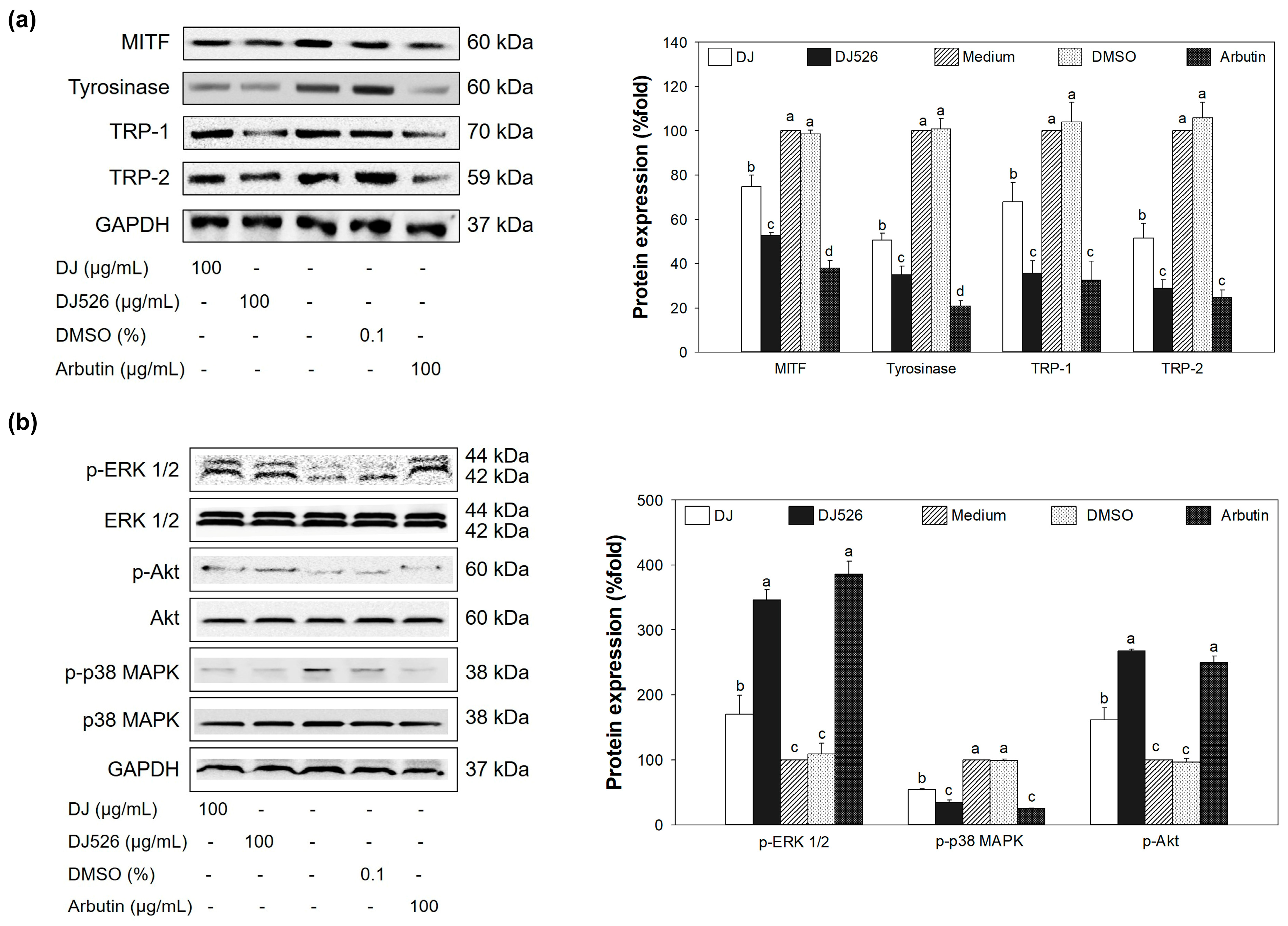
3.5. Effect of DJ526 Rice Callus Extract on Melanogenesis-Associated Gene Expression
3.6. Effect of DJ526 Rice Callus Extract on Protein Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- D’Mello, S.A.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Signaling pathways in melanogenesis. Int. J. Mol. Sci. 2016, 17, 1144. [Google Scholar] [CrossRef]
- Lee, S.-G.; Karadeniz, F.; Seo, Y.; Kong, C.-S. Anti-melanogenic effects of flavonoid glycosides from Limonium tetragonum (Thunb.) bullock via inhibition of tyrosinase and tyrosinase-related proteins. Molecules 2017, 22, 1480. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.C.; Lee, J.N.; Kim, B.S.; Hyun, C.-G. Anti-melanogenic effects of Paederia foetida L. extract via MAPK signaling-mediated MITF downregulation. Cosmetics 2021, 8, 22. [Google Scholar] [CrossRef]
- Wang, Y.; Duan, T.; Hong, M.; Zhou, Y.; Huang, H.; Xiao, X.; Zheng, J.; Zhou, H.; Lu, Z. Quantitative proteomic analysis uncovers inhibition of melanin synthesis by silk fibroin via MITF/tyrosinase axis in B16 melanoma cells. Life Sci. 2021, 284, 119930. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Lu, Y.; Zhong, Z.; Qu, B.; Wang, M.; Yu, X.; Chen, J. MITF involved in innate immunity by activating tyrosinase-mediated melanin synthesis in Pteria penguin. Front. Immunol. 2021, 12, 626493. [Google Scholar] [CrossRef]
- Alam, M.B.; Park, N.H.; Song, B.-R.; Lee, S.-H. Antioxidant potential-rich betel leaves (Piper betle L.) exert depigmenting action by triggering autophagy and downregulating MITF/tyrosinase in vitro and in vivo. Antioxidants 2023, 12, 374. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Yoon, J.-H.; Youn, K.; Jun, M. Decursin prevents melanogenesis by suppressing MITF expression through the regulation of PKA/CREB, MAPKs, and PI3K/Akt/GSK-3β cascades. Biomed. Pharmacother. 2022, 147, 112651. [Google Scholar] [CrossRef]
- Chung, Y.C.; Kim, Y.B.; Kim, B.S.; Hyun, C.-G. Anti-melanogenic effects of bergamottin via mitogen-activated protein kinases and protein kinase B signaling pathways. Nat. Prod. Commun. 2019, 14, 1934578X19862105. [Google Scholar] [CrossRef]
- Lee, M.S.; Chung, Y.C.; Moon, S.-H.; Hyun, C.-G. Lincomycin induces melanogenesis through the activation of MITF via p38 MAPK, AKT, and PKA signaling pathways. J. Appl. Biol. Chem. 2021, 64, 323–331. [Google Scholar] [CrossRef]
- Siddiqui, M.F.; Jeon, S.; Kim, M.-M. CRISPR/Cas9 system mediated SIRT7 gene knockout promotes melanogenesis by MITF via MAPKS and BMP activation in melanoma cells. Appl. Biochem. Microbiol. 2023, 59, 122–131. [Google Scholar] [CrossRef]
- Uto, T.; Ohta, T.; Yamashita, A.; Fujii, S.; Shoyama, Y. Liquiritin and liquiritigenin induce melanogenesis via enhancement of p38 and PKA signaling pathways. Medicines 2019, 6, 68. [Google Scholar] [CrossRef]
- Otręba, M.; Buszman, E.; Miliński, M.; Wrześniok, D. Hypomelanoses transmitted from generation to generation. Postep. Hig. Med. Dosw. 2014, 68, 1081–1090. [Google Scholar] [CrossRef]
- Rathee, P.; Kumar, S.; Kumar, D.; Kumari, B.; Yadav, S.S. Skin hyperpigmentation and its treatment with herbs: An alternative method. Future J. Pharm. Sci. 2021, 7, 132. [Google Scholar] [CrossRef]
- Lee, M.; Park, H.Y.; Jung, K.H.; Kim, D.H.; Rho, H.S.; Choi, K. Anti-melanogenic effects of kojic acid and hydroxycinnamic acid derivatives. Biotechnol. Bioprocess Eng. 2020, 25, 190–196. [Google Scholar] [CrossRef]
- Zhao, X.; Fan, Z.; Zhu, C.; Zhang, W.; Qin, L. Melanin inspired microcapsules delivering immune metabolites for hepatic fibrosis management. Mater. Today Bio 2023, 21, 100711. [Google Scholar] [CrossRef] [PubMed]
- Yurpolsky, D.; Lagunov, A.; Golyshev, A. Development of device for analysis problem skin zones of the circumpolar region population. AIP Conf. Proc. 2019, 2174, 020267. [Google Scholar] [CrossRef]
- Athar, M.; Back, J.H.; Tang, X.; Kim, K.H.; Kopelovich, L.; Bickers, D.R.; Kim, A.L. Resveratrol: A review of preclinical studies for human cancer prevention. Toxicol. Appl. Pharmacol. 2007, 224, 274–283. [Google Scholar] [CrossRef]
- Szajdek, A.; Borowska, E.J. Bioactive compounds and health-promoting properties of berry fruits: A review. Plant Foods Hum. Nutr. 2008, 63, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.S.; Salve, A.R.; Chauhan, S. Peanuts as functional food: A review. J. Food Sci. Technol. 2016, 53, 31–41. [Google Scholar] [CrossRef]
- Sebastià, N.; Montoro, A.; Mañes, J.; Soriano, J.M. A preliminary study of presence of resveratrol in skins and pulps of European and Japanese plum cultivars. J. Sci. Food Agric. 2012, 92, 3091–3094. [Google Scholar] [CrossRef]
- Nassiri-Asl, M.; Hosseinzadeh, H. Review of the pharmacological effects of Vitis vinifera (grape) and its bioactive compounds. Phytother. Res. 2009, 23, 1197–1204. [Google Scholar] [CrossRef] [PubMed]
- Koushki, M.; Amiri-Dashatan, N.; Ahmadi, N.; Abbaszadeh, H.-A.; Rezaei-Tavirani, M. Resveratrol: A miraculous natural compound for diseases treatment. Food Sci. Nutr. 2018, 6, 2473–2490. [Google Scholar] [CrossRef] [PubMed]
- Baxter, R.A. Anti-aging properties of resveratrol: Review and report of a potent new antioxidant skin care formulation. J. Cosmet. Dermatol. 2008, 7, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Farris, P.; Krutmann, J.; Li, Y.-H.; McDaniel, D.; Krol, Y. Resveratrol: A unique antioxidant offering a multi-mechanistic approach for treating aging skin. J. Drugs Dermatol. 2013, 12, 1389–1394. [Google Scholar] [PubMed]
- Na, J.-I.; Shin, J.-W.; Choi, H.-R.; Kwon, S.-H.; Park, K.-C. Resveratrol as a multifunctional topical hypopigmenting agent. Int. J. Mol. Sci. 2019, 20, 956. [Google Scholar] [CrossRef] [PubMed]
- Banez, M.J.; Geluz, M.I.; Chandra, A.; Hamdan, T.; Biswas, O.S.; Bryan, N.S.; Von Schwarz, E.R. A systemic review on the antioxidant and anti-inflammatory effects of resveratrol, curcumin, and dietary nitric oxide supplementation on human cardiovascular health. Nutr. Res. 2020, 78, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Sharifi-Rad, J.; Quispe, C.; Durazzo, A.; Lucarini, M.; Souto, E.B.; Santini, A.; Imran, M.; Moussa, A.Y.; Mostafa, N.M.; El-Shazly, M.; et al. Resveratrol’ biotechnological applications: Enlightening its antimicrobial and antioxidant properties. J. Herb. Med. 2022, 32, 100550. [Google Scholar] [CrossRef]
- Meng, T.; Xiao, D.; Muhammed, A.; Deng, J.; Chen, L.; He, J. Anti-inflammatory action and mechanisms of resveratrol. Molecules 2021, 26, 229. [Google Scholar] [CrossRef]
- Chen, L.Z.; Yao, L.; Jiao, M.M.; Shi, J.B.; Tan, Y.; Ruan, B.F.; Liu, X.H. Novel resveratrol-based flavonol derivatives: Synthesis and anti-inflammatory activity in vitro and in vivo. Eur. J. Med. Chem. 2019, 175, 114–128. [Google Scholar] [CrossRef]
- Hsu, Y.-A.; Chen, C.-S.; Wang, Y.-C.; Lin, E.-S.; Chang, C.-Y.; Chen, J.-J.; Wu, M.-Y.; Lin, H.-J.; Wan, L. Anti-inflammatory effects of resveratrol on human retinal pigment cells and a myopia animal model. Curr. Issues Mol. Biol. 2021, 43, 716–727. [Google Scholar] [CrossRef]
- Angellotti, G.; Di Prima, G.; Belfiore, E.; Campisi, G.; De Caro, V. Chemopreventive and anticancer role of resveratrol against oral squamous cell carcinoma. Pharmaceutics 2023, 15, 275. [Google Scholar] [CrossRef]
- Ferraz da Costa, D.C.; Pereira Rangel, L.; Martins-Dinis, M.M.; Ferretti, G.D.; Ferreira, V.F.; Silva, J.L. Anticancer potential of resveratrol, β-lapachone and their analogues. Molecules 2020, 25, 893. [Google Scholar] [CrossRef] [PubMed]
- Vestergaard, M.; Ingmer, H. Antibacterial and antifungal properties of resveratrol. Int. J. Antimicrob. Agents 2019, 53, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Oh, J.; Averilla, J.-N.; Kim, H.-J.; Kim, J.-S.; Kim, J.-S. Grape peel extract and resveratrol inhibit wrinkle formation in mice model through activation of Nrf2/HO-1 signaling pathway. J. Food Sci. 2019, 84, 1600–1608. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Yun, J.; Lee, C.K.; Lee, H.; Min, K.R.; Kim, Y. Oxyresveratrol and hydroxystilbene compounds. Inhibitory effect on tyrosinase and mechanism of action. J. Biol. Chem. 2002, 277, 16340–16344. [Google Scholar] [CrossRef]
- Shin, N.H.; Ryu, S.Y.; Choi, E.J.; Kang, S.H.; Chang, I.M.; Min, K.R.; Kim, Y. Oxyresveratrol as the potent inhibitor on dopa oxidase activity of mushroom tyrosinase. Biochem. Biophys. Res. Commun. 1998, 243, 801–803. [Google Scholar] [CrossRef]
- Zheng, Z.-P.; Tan, H.-Y.; Wang, M. Tyrosinase inhibition constituents from the roots of Morus australis. Fitoterapia 2012, 83, 1008–1013. [Google Scholar] [CrossRef]
- Yokozawa, T.; Kim, Y.J. Piceatannol inhibits melanogenesis by its antioxidative actions. Biol. Pharm. Bull. 2007, 30, 2007–2011. [Google Scholar] [CrossRef]
- Baek, S.-H.; Shin, W.-C.; Ryu, H.-S.; Lee, D.-W.; Moon, E.; Seo, C.-S.; Hwang, E.; Lee, H.-S.; Ahn, M.-H.; Jeon, Y.; et al. Creation of resveratrol-enriched rice for the treatment of metabolic syndrome and related diseases. PLoS ONE 2013, 8, e57930. [Google Scholar] [CrossRef]
- Monmai, C.; Kim, J.-S.; Baek, S.-H. Use of germination to enhance resveratrol content and its anti-inflammatory activity in lipopolysaccharide-stimulated RAW264.7 cells. Molecules 2023, 28, 4898. [Google Scholar] [CrossRef]
- Monmai, C.; Kim, J.-S.; Baek, S.-H. Anti-inflammatory efficacy of resveratrol-enriched rice callus extract on lipopolysaccharide-stimulated RAW264.7 macrophages. Immuno 2024, 4, 131–146. [Google Scholar] [CrossRef]
- Monmai, C.; Kim, J.-S.; Promyot, K.; Baek, S.-H. Protopanaxadiol-enriched rice extracts suppressed oxidative and melanogenic activities in melan-a cells. Antioxidants 2023, 12, 166. [Google Scholar] [CrossRef] [PubMed]
- Boo, Y.C. Arbutin as a skin depigmenting agent with antimelanogenic and antioxidant properties. Antioxidants 2021, 10, 1129. [Google Scholar] [CrossRef] [PubMed]
- Ebadollahi, S.H.; Pouramir, M.; Zabihi, E.; Golpour, M.; Aghajanpour-Mir, M. The effect of arbutin on the expression of tumor suppressor P53, BAX/BCL-2 ratio and oxidative stress induced by tert-butyl hydroperoxide in fibroblast and LNcap cell lines. Cell J. 2021, 22, 532–541. [Google Scholar] [CrossRef]
- Safari, H.; Zabihi, E.; Pouramir, M.; Morakabati, P.; Abedian, Z.; Karkhah, A.; Nouri, H.R. Decrease of intracellular ROS by arbutin is associated with apoptosis induction and downregulation of IL-1β and TNF-α in LNCaP; prostate cancer. J. Food Biochem. 2020, 44, e13360. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Liu, W.; Zhu, D.; Cao, Y.; Tang, A.; Gong, G.; Su, H. Natural skin-whitening compounds for the treatment of melanogenesis (Review). Exp. Ther. Med. 2020, 20, 173–185. [Google Scholar] [CrossRef] [PubMed]
- Manap, A.S.A.; Lum, Y.K.; Ong, L.H.; Tang, Y.-Q.; Gew, L.T.; Chia, A.Y.Y. Perspective approaches on melanogenesis inhibition. Dermatol. Sin. 2021, 39, 1–12. [Google Scholar] [CrossRef]
- Park, J.-Y.; Song, M.W.; Kim, K.-T.; Paik, H.-D. Improved antioxidative, anti-inflammatory, and antimelanogenic effects of fermented hydroponic ginseng with Bacillus Strains. Antioxidants 2022, 11, 1848. [Google Scholar] [CrossRef] [PubMed]
- Rodboon, T.; Okada, S.; Suwannalert, P. Germinated riceberry rice enhanced protocatechuic acid and vanillic acid to suppress melanogenesis through cellular oxidant-related tyrosinase activity in B16 cells. Antioxidants 2020, 9, 247. [Google Scholar] [CrossRef]
- Martínez-López, C.; Vázquez-Carrada, M.; Flores-Herrera, O.; Pardo, J.P.; Olicón-Hernández, D.R.; Guerra-Sánchez, G. Ustilago maydis yeast mutant produces cytosolic melanin by tyrosine-tyrosinase activity with stain biosorption capability. Appl. Sci. 2023, 13, 11288. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Lyu, J.-L.; Kuo, Y.-H.; Chiu, C.-Y.; Wen, K.-C.; Chiang, H.-M. The anti-melanogenesis effect of 3,4-dihydroxybenzalacetone through downregulation of melanosome maturation and transportation in B16F10 and human epidermal melanocytes. Int. J. Mol. Sci. 2021, 22, 2823. [Google Scholar] [CrossRef]
- Li, Z.; Chen, X.; Liu, G.; Li, J.; Zhang, J.; Cao, Y.; Miao, J. Antioxidant activity and mechanism of resveratrol and polydatin isolated from mulberry (Morus alba L.). Molecules 2021, 26, 7574. [Google Scholar] [CrossRef]
- Lagoutari, E.; Liouni, M.; Lagoutari, M.; Detsi, A.; Zoumpoulakis, P.; Proestos, C. Measurement of antioxidants and phenolics in wines and tsipouro enriched with powerful antioxidants such as vitamins and resveratrol. Pharmakeftiki 2024, 36, 10–26. [Google Scholar]
- Simon, J.D.; Peles, D.; Wakamatsu, K.; Ito, S. Current challenges in understanding melanogenesis: Bridging chemistry, biological control, morphology, and function. Pigment Cell Melanoma Res. 2009, 22, 563–579. [Google Scholar] [CrossRef]
- Cichorek, M.; Wachulska, M.; Stasiewicz, A.; Tymińska, A. Skin melanocytes: Biology and development. Postępy Dermatol. Alergol. 2013, 30, 30–41. [Google Scholar] [CrossRef]
- Hirobe, T. How are proliferation and differentiation of melanocytes regulated? Pigment Cell Melanoma Res. 2011, 24, 462–478. [Google Scholar] [CrossRef]
- Song, W.; Liu, L.-L.; Ren, Y.-J.; Wei, S.-D.; Yang, H.-B. Inhibitory effects and molecular mechanism on mushroom tyrosinase by condensed tannins isolation from the fruit of Ziziphus jujuba Mill. var. spinosa (Bunge) Hu ex H.F. Chow. Int. J. Biol. Macromol. 2020, 165, 1813–1821. [Google Scholar] [CrossRef]
- Gouda, A.; Soavi, F.; Santato, C. Eumelanin electrodes in buffered aqueous media at different pH values. Electrochim. Acta 2020, 347, 136250. [Google Scholar] [CrossRef]
- Ito, S.; Del Bino, S.; Hirobe, T.; Wakamatsu, K. Improved HPLC conditions to determine eumelanin and pheomelanin contents in biological samples using an ion pair reagent. Int. J. Mol. Sci. 2020, 21, 5134. [Google Scholar] [CrossRef]
- Saha, B.; Singh, S.K.; Sarkar, C.; Bera, R.; Ratha, J.; Tobin, D.J.; Bhadra, R. Activation of the MITF promoter by lipid-stimulated activation of p38-stress signalling to CREB. Pigment Cell Res. 2006, 19, 595–605. [Google Scholar] [CrossRef]
- Hartman, M.L.; Czyz, M. MITF in melanoma: Mechanisms behind its expression and activity. Cell. Mol. Life Sci. 2015, 72, 1249–1260. [Google Scholar] [CrossRef]
- Jang, J.Y.; Lee, J.H.; Kang, B.W.; Chung, K.T.; Choi, Y.H.; Choi, B.T. Dichloromethane fraction of Cimicifuga heracleifolia decreases the level of melanin synthesis by activating the ERK or AKT signaling pathway in B16F10 cells. Exp. Dermatol. 2009, 18, 232–237. [Google Scholar] [CrossRef]
- Jin, M.-L.; Park, S.-Y.; Kim, Y.-H.; Park, G.; Son, H.-J.; Lee, S.-J. Suppression of α-MSH and IBMX-induced melanogenesis by cordycepin via inhibition of CREB and MITF, and activation of PI3K/Akt and ERK-dependent mechanisms. Int. J. Mol. Med. 2012, 29, 119–124. [Google Scholar] [CrossRef]
- Brenner, M.; Hearing, V.J. The protective role of melanin against UV damage in human skin. Photochem. Photobiol. 2008, 84, 539–549. [Google Scholar] [CrossRef]
- Mansky, K.C.; Sankar, U.; Han, J.; Ostrowski, M.C. Microphthalmia transcription factor is a target of the p38 MAPK pathway in response to receptor activator of NF-kappa B ligand signaling. J. Biol. Chem. 2002, 277, 11077–11083. [Google Scholar] [CrossRef] [PubMed]
- Tagashira, H.; Miyamoto, A.; Kitamura, S.-I.; Tsubata, M.; Yamaguchi, K.; Takagaki, K.; Imokawa, G. UVB stimulates the expression of endothelin B receptor in human melanocytes via a sequential activation of the p38/MSK1/CREB/MITF pathway which can be interrupted by a French maritime pine bark extract through a direct inactivation of MSK1. PLoS ONE 2015, 10, e0128678. [Google Scholar] [CrossRef]
- Bhat, A.M.; Haroon, R.; Naikoo, S.; Sharma, R.R.; Archoo, S.; Tasduq, S.A. (2-Methylbutyryl)shikonin naturally occurring shikonin derivative ameliorates the α-MSH-induced melanogenesis via ERK1/2 and p38 MAP kinase-mediated down-regulation of the MITF transcription factor. Chem. Res. Toxicol. 2024, 37, 274–284. [Google Scholar] [CrossRef]
- Kim, T.; Hyun, C.-G. Imperatorin positively regulates melanogenesis through signaling pathways involving PKA/CREB, ERK, AKT, and GSK3β/β-catenin. Molecules 2022, 27, 6512. [Google Scholar] [CrossRef]
- Huang, H.-C.; Chang, S.-J.; Wu, C.-Y.; Ke, H.-J.; Chang, T.-M. [6]-Shogaol inhibits α-MSH-induced melanogenesis through the acceleration of ERK and PI3K/Akt-mediated MITF degradation. Biomed. Res. Int. 2014, 2014, 842569. [Google Scholar] [CrossRef]
- Lee, T.-H.; Seo, J.-O.; Baek, S.-H.; Kim, S.-Y. Inhibitory effects of resveratrol on melanin synthesis in ultraviolet B-induced pigmentation in guinea pig skin. Biomol. Ther. 2014, 22, 35–40. [Google Scholar] [CrossRef]
- Zhao, H.; Han, L.; Jian, Y.; Ma, Y.; Yan, W.; Chen, X.; Xu, H.; Li, L. Resveratrol induces apoptosis in human melanoma cell through negatively regulating Erk/PKM2/Bcl-2 axis. OncoTargets Ther. 2018, 11, 8995–9006. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, B.; E, C.; Liu, J.; Zhang, Q.; Liu, J.; Chen, N.; Chen, R.; Zhu, R. Resveratrol inhibits the proliferation of human melanoma cells by inducing G1/S cell cycle arrest and apoptosis. Mol. Med. Rep. 2015, 11, 400–404. [Google Scholar] [CrossRef]
- Chen, Y.-J.; Chen, Y.-Y.; Lin, Y.-F.; Hu, H.-Y.; Liao, H.-F. Resveratrol inhibits alpha-melanocyte-stimulating hormone signaling, viability, and invasiveness in melanoma cells. Evid.-Based Complement. Altern. Med. 2013, 2013, 632121. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Sun, Z.; Nie, S.; Zhang, T.; Lu, H. Effects of resveratrol on mouse B16 melanoma cell proliferation through the SHCBP1-ERK1/2 signaling pathway. Molecules 2023, 28, 7614. [Google Scholar] [CrossRef] [PubMed]

| Treatment | A260:A280 | A260:A230 | RNA Concentration (ng/µL) | ||
|---|---|---|---|---|---|
| Mean | SD | CV | |||
| Medium | 1.832 | 1.962 | 306.45 | 29.75 | 9.71 |
| DMSO (0.1%) | 1.947 | 1.831 | 465.17 | 34.16 | 7.34 |
| Arbutin (100 µg/mL) | 1.981 | 1.998 | 768.59 | 21.54 | 2.80 |
| DJ (10 µg/mL) | 1.894 | 1.890 | 268.20 | 19.72 | 7.35 |
| DJ (25 µg/mL) | 2.001 | 1.994 | 474.80 | 44.15 | 9.30 |
| DJ (50 µg/mL) | 1.989 | 1.891 | 647.25 | 53.72 | 8.30 |
| DJ (100 µg/mL) | 1.865 | 1.881 | 574.19 | 21.74 | 3.79 |
| DJ526 (10 µg/mL) | 1.975 | 1.938 | 640.72 | 10.19 | 1.59 |
| DJ526 (25 µg/mL) | 1.968 | 1.966 | 650.67 | 55.29 | 8.50 |
| DJ526 (50 µg/mL) | 1.942 | 1.947 | 143.60 | 2.70 | 1.88 |
| DJ526 (100 µg/mL) | 1.835 | 1.959 | 791.73 | 12.77 | 1.61 |
| Process | Temperature (°C) | Time (min:s) | Number of Cycles |
|---|---|---|---|
| Pre-denaturation | 95 | 10:00 | 1 |
| PCR cycle - Denaturation - Annealing - Extension | 95 60 72 | 0:20 0:20 0:30 | 40 |
| Final extension | 72 | 5:00 | 1 |
| Melt curve | 65–95 | Every 0:05 | 1 |
| Treatment | Concentration | Scavenging Activity (%) | VCEAC (mg/g) | IC50 (mg/mL) | ||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| Control (DW) | − | 0.00 | 0.61 | 0.000 | 0.002 | − |
| DMSO | 0.1% (v/v) | 0.10 | 0.09 | 0.000 | 0.000 | − |
| DJ | 10 mg/mL | 8.34 c | 1.21 | 0.004 c | 0.003 | Noncalculable |
| 25 mg/mL | 10.99 c | 0.83 | 0.012 c | 0.002 | ||
| 50 mg/mL | 22.48 c | 0.68 | 0.044 c | 0.002 | ||
| 100 mg/mL | 33.32 c | 1.91 | 0.074 c | 0.005 | ||
| DJ526 | 10 mg/mL | 21.48 b | 0.94 | 0.041 b | 0.003 | 42.24 ± 0.67 |
| 25 mg/mL | 40.46 b | 1.27 | 0.094 b | 0.004 | ||
| 50 mg/mL | 58.54 b | 2.32 | 0.145 b | 0.006 | ||
| 100 mg/mL | 91.66 a | 0.62 | 0.237 a | 0.002 | ||
| Arbutin | 10 mg/mL | 25.32 a | 0.62 | 0.052 a | 0.002 | 29.57 ± 0.55 |
| 25 mg/mL | 50.80 a | 0.74 | 0.123 a | 0.002 | ||
| 50 mg/mL | 77.77 a | 1.26 | 0.198 a | 0.004 | ||
| 100 mg/mL | 93.71 a | 0.40 | 0.243 a | 0.001 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monmai, C.; Kim, J.-S.; Baek, S.-H. Resveratrol-Enriched Rice Callus Extract Inhibits Oxidative and Cellular Melanogenic Activities in Melan-A Cells. Antioxidants 2024, 13, 625. https://doi.org/10.3390/antiox13060625
Monmai C, Kim J-S, Baek S-H. Resveratrol-Enriched Rice Callus Extract Inhibits Oxidative and Cellular Melanogenic Activities in Melan-A Cells. Antioxidants. 2024; 13(6):625. https://doi.org/10.3390/antiox13060625
Chicago/Turabian StyleMonmai, Chaiwat, Jin-Suk Kim, and So-Hyeon Baek. 2024. "Resveratrol-Enriched Rice Callus Extract Inhibits Oxidative and Cellular Melanogenic Activities in Melan-A Cells" Antioxidants 13, no. 6: 625. https://doi.org/10.3390/antiox13060625
APA StyleMonmai, C., Kim, J.-S., & Baek, S.-H. (2024). Resveratrol-Enriched Rice Callus Extract Inhibits Oxidative and Cellular Melanogenic Activities in Melan-A Cells. Antioxidants, 13(6), 625. https://doi.org/10.3390/antiox13060625







