Abstract
Hydrogen sulfide (H2S) and nitric oxide (NO) are long-known inhibitors of terminal oxidases in the respiratory chain. Yet, they exert pivotal signaling roles in physiological processes, and in several bacterial pathogens have been reported to confer resistance against oxidative stress, host immune responses, and antibiotics. Pseudomonas aeruginosa, an opportunistic pathogen causing life-threatening infections that are difficult to eradicate, has a highly branched respiratory chain including four terminal oxidases of the haem-copper type (aa3, cbb3-1, cbb3-2, and bo3) and one oxidase of the bd-type (cyanide-insensitive oxidase, CIO). As Escherichia coli bd-type oxidases have been shown to be H2S-insensitive and to readily recover their activity from NO inhibition, here we tested the effect of H2S and NO on CIO by performing oxygraphic measurements on membrane preparations from P. aeruginosa PAO1 and isogenic mutants depleted of CIO only or all other terminal oxidases except CIO. We show that O2 consumption by CIO is unaltered even in the presence of high levels of H2S, and that CIO expression is enhanced and supports bacterial growth under such stressful conditions. In addition, we report that CIO is reversibly inhibited by NO, while activity recovery after NO exhaustion is full and fast, suggesting a protective role of CIO under NO stress conditions. As P. aeruginosa is exposed to H2S and NO during infection, the tolerance of CIO towards these stressors agrees with the proposed role of CIO in P. aeruginosa virulence.
1. Introduction
Hydrogen sulfide (H2S) and nitric oxide (NO), similar to carbon monoxide, are endogenously produced gaseous molecules that can exhibit beneficial signalling and regulatory effects in cells and organisms at low physiological concentrations as well as toxic effects at higher concentrations [1]. As a result of this ambivalent behaviour, they play a crucial role in both physiological and pathological processes [2,3,4,5,6,7,8]. Notably, in bacteria the production of these gaseous molecules, known as “gasotransmitters”, has been suggested to be important during infection [9,10,11,12,13]. Specifically, these molecules have been reported to confer resistance to oxidative stress and antibiotics by enhancing the detoxification of reactive oxygen species caused by several antimicrobials [14] or by chemically modifying protein targets [15]. Indeed, H2S signalling mainly occurs via post-translational persulfidation of protein cysteine residues [16], which in turn modulates protein activity and can promote microbial virulence [17]. Proteomic profiling of S. aureus in response to exposure to exogenous sulfide has revealed that protein persulfidation regulates many key metabolic enzymes and genes related to virulence, influencing both cell surface composition and biofilm formation [18] Despite these beneficial roles exerted at lower concentrations, H2S and NO are better known for their ability to negatively affect energy metabolism and cell viability at higher concentrations, acting as inhibitors of antioxidant enzymes and respiratory oxidases by targeting Fe-S-containing and haem proteins [8]. The host immune system exploits this feature to fight infections by producing elevated levels of NO through the inducible NO synthase.
Unlike mammalians, bacteria possess branched respiratory chains with multiple electron entry sites and alternative oxidants as terminal acceptors [19]. Moreover, their flexible electron transfer pathways are reconfigured in response to changing conditions in the habitat [20], allowing certain bacteria to tolerate relatively high concentrations of NO and H2S. A highly branched respiratory chain is present in Pseudomonas aeruginosa, an important pathogen causing acute nosocomial lung, soft tissue, and systemic infections as well as chronic infections in individuals with underlying inflammatory lung diseases such as cystic fibrosis (CF) [21,22]. To date, P. aeruginosa chronic lung infection is the main cause of morbidity and mortality in individuals with CF. During permanence in the CF lung, P. aeruginosa undergoes evolutionary changes and adapted clones emerge with mutations conferring antibiotic resistance. Effective therapeutic options to treat P. aeruginosa infections are limited, and no antibiotic can eradicate the chronic lung infection that this bacterium establishes in CF individuals [23]. As a growing public health threat, P. aeruginosa is listed by the WHO as among the top priority pathogens for which new antimicrobials are urgently needed [24].
Remarkably, in recent years bacteria-specific energy metabolism has been garnering increasing attention as a therapeutic target [20]. Most antibiotics target the biosynthesis of DNA, RNA, proteins, or peptidoglycan, and are active against growing bacteria. However, these antibiotics are not effective in eradicating persistent infections where most of the bacterial cells are in slow-growing or non-growing states. Because maintenance of cellular energy and redox homeostasis is a requirement for non-growing cells, inhibition of energy metabolism is expected to be effective in killing persister cells as well [23].
The aerobic electron transport chain of P. aeruginosa includes five terminal oxidases, expressed under different growth conditions: three cytochrome c oxidases, aa3, cbb3-1, and cbb3-2, and two quinol oxidases, the cytochrome bo3, which belongs to the haem-copper oxidase superfamily, including all known cytochrome c oxidases, and cyanide insensitive oxidase (CIO), which is part of the family of copper-free bd-type oxidases [25]. All of these oxidases generate the proton motive force used for ATP production by catalyzing the four-electron reduction of O2 to 2 H2O. CIO is encoded by an operon containing the cioA and cioB genes, which share high similarity with the corresponding cydA and cydB genes in E. coli that code for cytochrome bd-I [26]. E. coli possesses three terminal quinol oxidases: the haem-copper cytochrome bo3, and the bd-type cytochromes bd-I and bd-II [25]. Despite its high similarity to E. coli cytochrome bd-I, CIO is reported to have a different haem composition, in that while bd-type oxidases typically carry one low-spin haem (b558) and two high-spin haems (b595 and d), in CIO haem d appears to be replaced by a b-type haem [26,27]. As in E. coli bd oxidases, CIO is predicted to have high affinity for O2; indeed, a cco1/cco2 double mutant lacking cbb3-1 and cbb3-2 was able to grow under microaerobic conditions (2% O2), whereas a cco1/cco2/cio triple mutant lacking cbb3-1, cbb3-2, and CIO did not grow under such conditions [28]. However, the affinity of P. aeruginosa CIO for O2 was later measured in a mutant strain expressing only CIO and found to be low, with a Km value for O2 of 4.0 ± 2.1 μM, significantly higher than those of cbb3-1 and cbb3-2 and comparable to those of aa3 and bo3 reported by Arai et al. [29]. These authors suggested that the inability of the cco1/cco2/cio triple mutant to grow under microaerobic conditions is due to the sensitivity of bo3 and aa3 to cyanide, a potent respiratory inhibitor that is produced by P. aeruginosa under microaerobiosis and during infections as a virulence factor [30]. Given its tolerance to cyanide, CIO is indeed capable of sustaining aerobic respiration under cyanogenic conditions [31,32]. Although P. aeruginosa CIO can resist high concentrations (>1 mM) of cyanide, growth and survival experiments indicate that this oxidase is unnecessary for protection against endogenous cyanide under physiological conditions [32]. Consistently, the increase in cio gene expression in the stationary phase occurs in both the wild type and in the ∆hcnB mutant which is unable to produce cyanide. Intriguingly, endogenous cyanide boosts transcription of P. aeruginosa ccoN4, encoding an orphan cbb3 oxidase subunit, which makes the oxidase resistant to cyanide [33]. On the contrary, CIO contributes to cyanide resistance when this agent is exogenously supplied and under aerobiosis; in the paralytic model of Caenorhabditis elegans infection, in which cyanide is the mediating factor causing nematode death, CIO is required for full pathogenicity, suggesting a possible role for this oxidase in bacterial virulence [30].
A large body of evidence has demonstrated that bd-type oxidases, in addition to sustaining cell bioenergetics, can support virulence in several pathogens by providing protection against several stressors [25], including antibiotics [25,34,35], peroxynitrite [36,37], NO [38,39], hydrogen peroxide (H2O2) [40,41,42], reactive oxygen species produced by macrophages, and H2S [43]. Significant sulfate concentrations and high H2S production have recently been reported in CF individuals infected by P. aeruginosa both at the site of infection and in the expectorate [44,45]. This suggests a possible involvement of sulfide in the pathophysiology of this bacterium.
In light of these observations, and considering that resistance of aerobic respiration to reactive nitrogen species and sulfide is likely advantageous for P. aeruginosa during infection, we tested the effect of sulfide and NO on CIO activity by performing respirometric experiments on membrane fractions derived from P. aeruginosa PAO1 and on isogenic mutants expressing either only CIO or all other oxidases except CIO as their terminal oxidases.
2. Materials and Methods
2.1. Materials
All chemicals were from Merck unless otherwise specified. Sulfide stock solutions were prepared by dissolving Na2S or NaHS crystals in degassed ultra-pure water (Milli-Q®, Merck Millipore, Burlington, MA, USA), as reported in [46]. In aqueous solution, H2S is in equilibrium with hydrosulfide (HS−) and sulfide (S2−) according to the pKa1~7.0 (H2S/HS) and pKa2~19 (HS−/S2−) measured at 25 °C. For simplicity, unless otherwise stated, the terms ‘H2S’ and ‘sulfide’ are employed interchangeably here to collectively indicate the H2S/HS−/S2− species. The overall concentration of sulfide species in solution was quantified spectrophotometrically with DTNB according to [47], using the molar extinction coefficient ε412 = 14,150 M−1 cm−1 suggested by the producer. Stock solutions of NO were prepared at room temperature in a tonometer by equilibrating degassed ultra-pure water (Milli-Q®, Merck Millipore, Burlington, MA, USA) with pure NO gas (Air Liquide, Paris, France) at 1 atm. The concentration of NO in solution was obtained by titration with reduced beef heart cytochrome c oxidase according to [48].
2.2. Bacterial Strains, Growth Conditions, and Membrane Preparations
The P. aeruginosa PAO1 wild type strain used in this study was from the American Type Culture Collection (ATCC15692). For growth studies in the presence of sulfide, the PAO1 wild type strain and isogenic mutants with deletions in the cioAB locus (∆cio, [49,50], GeneBank accession numbers: cioA AAG07317.1 and cioB AAG07316.1) or in the cyo, cco-1, cco-2, and cox gene loci (∆cyo∆cco∆cox, [49]) were grown overnight in Lysogeny Broth (LB) complex medium at 37 °C with shaking (200 rpm). Following overnight growth, bacterial cells were diluted to obtain an optical density at 600 nm (OD600) of 0.1 in 10 mL of LB supplemented with 4 mM L-cysteine (LB-cys) in order to increase endogenous H2S levels [14] in the absence or presence of 200 µM NaHS as H2S donor. The resulting cultures were grown in 50 mL tubes at 37 °C with shaking (200 rpm). The OD600 of each culture was monitored every 2 h with a Lambda EZ 201 spectrophotometer (Perkin-Elmer, Waltham, MA, USA).
P. aeruginosa membranes were prepared as described by Cunningham and Williams [31]. P. aeruginosa cultures were grown in LB medium until an OD600 of about 0.9 was achieved. Cells were then harvested by centrifugation at 5000× g and washed twice in 0.05 M phosphate buffer pH 7.0. The cell pellets were resuspended in 0.01 M phosphate buffer pH 7.0 containing 5 mM MgCl2, 0.1 mg/mL lysozyme, and 50 µg/mL DNAse. The cells were then lysed by sonication and debris were removed by centrifugation at 5000× g. The membrane fraction was collected by centrifugation for 2 h at 100,000× g at 4 °C and resuspended in 0.5 mL of 0.01 M phosphate buffer pH 7.0 containing 5 mM MgCl2. The protein content was determined by the Bradford method using Bradford reagent with bovine serum albumin as the standard.
2.3. Oxygraphic Measurements
Oxygraphic measurements were performed using high-resolution respirometers (Oxygraph-2k and NextGen-O2k all-in-one, Oroboros Instruments GmbH, Innsbruck, Austria) equipped with a 1.5 mL chamber and Clark-type oxygen electrodes. The assays were carried out at 25 °C in 50 mM K/phosphate buffer pH 7.0 supplemented with: (i) 0.5 mM NADH to test the activity of all terminal oxidases; (ii) 10 mM dithiothreitol (DTT) and 0.25 mM coenzyme Q1 to selectively reduce quinol oxidases; or (iii) 2 mM ascorbate and 0.2 mM tetramethylene-p-phenylenediamine (TMPD) to preferentially reduce cytochrome c oxidases. Where indicated, 1 mM NaCN was added as a control on the cyanide resistance of CIO. Na2S was used as a sulfide donor. The high-resolution respirometer was employed to perform simultaneous measurements of O2 and NO. To assess changes in the levels of NO in solution, an NO-selective electrode (World Precision Instruments, Sarasota, FL, USA) was used. The electrode was calibrated with subsequent additions of NO from a stock solution prepared as described previously. Oxygraphic assays were conducted in the dark, as the NO-ferrous haem adduct is light sensitive [51]. The half-maximal inhibition concentration value (apparent IC50) for NO, defined as the NO concentration accounting for 50% inhibition of O2 reductase activity of membranes, was estimated by monitoring the activity in the recovery phase from NO inhibition.
2.4. RNA Extraction and RT-qPCR Analyses
P. aeruginosa PAO1 cultures were grown overnight in LB at 37 °C with shaking (200 rpm). Following overnight growth, bacteria were placed in 50 mL tubes diluted to an OD600 of 0.1 in 10 mL of LB alone or supplemented with 4 mM L-cysteine (LB-cys) in the presence or absence of 200 µM NaHS. The resulting cultures were grown at 37 °C with shaking (200 rpm). Total RNA was extracted as previously described [52] from 1 mL of each culture at an OD600 of 1.2 (the late exponential/early stationary phase of growth). Briefly, cells were mixed with 2 mL RNA Protect Bacteria Reagent (Qiagen, Hilden, Germany)) and RNA was purified using an RNeasy Mini Kit (Qiagen), including the on-column DNase I digestion step. The isolated RNA was incubated for 1 h at 37 °C with TURBO DNase (0.2 U per mg of RNA; Ambion Austin, TX, USA) and with SUPERase-In (0.4 U per mg of RNA; Ambion). Total RNA was purified with the RNeasy Column Purification Kit (Qiagen). Three different pools of RNA were extracted for each sample in independent experiments. Purified RNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The absence of DNA was verified by PCR using the oligonucleotides FWpqsB 5′-CCGCTCGAGCGACCAGGGCTATCGCA-3′ and RVpqsB 5′-CCGGAATTCCTTATGCATGAGCTTCTCC-3′. For RT-qPCR analyses, the iScript Reverse Transcription Supermix for RT-qPCR kit (Bio-Rad Laboratories, Hercules, CA, USA) was used to synthesize cDNA from 1 µg of purified RNA. Real-time PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories) and the AriaMX Real Time PCR system (Agilent Technologies, Santa Clara, CA, USA). 16S ribosomal RNA was used as the internal control to normalize the RT-qPCR data in every single run and to calculate the relative FC in gene expression using the 2−ΔΔCt method. cioA-specific primers (FWcioART 5′-GCCTCTGGCTGAAAACGAAC-3′; RVcioART 5′-GTGAGCACCTCGTAGGTCAG-3′) and 16S-specific primers (FW16SRT 5′-GAGAGTTTGATCCTGGCTCAG-3′; RV16SRT 5′-CTACGGCTACCTTGTTACGA-3′) were designed using Primer-BLAST.
3. Results
3.1. Effect of Reducing Systems on Respiration of P. aeruginosa Membranes
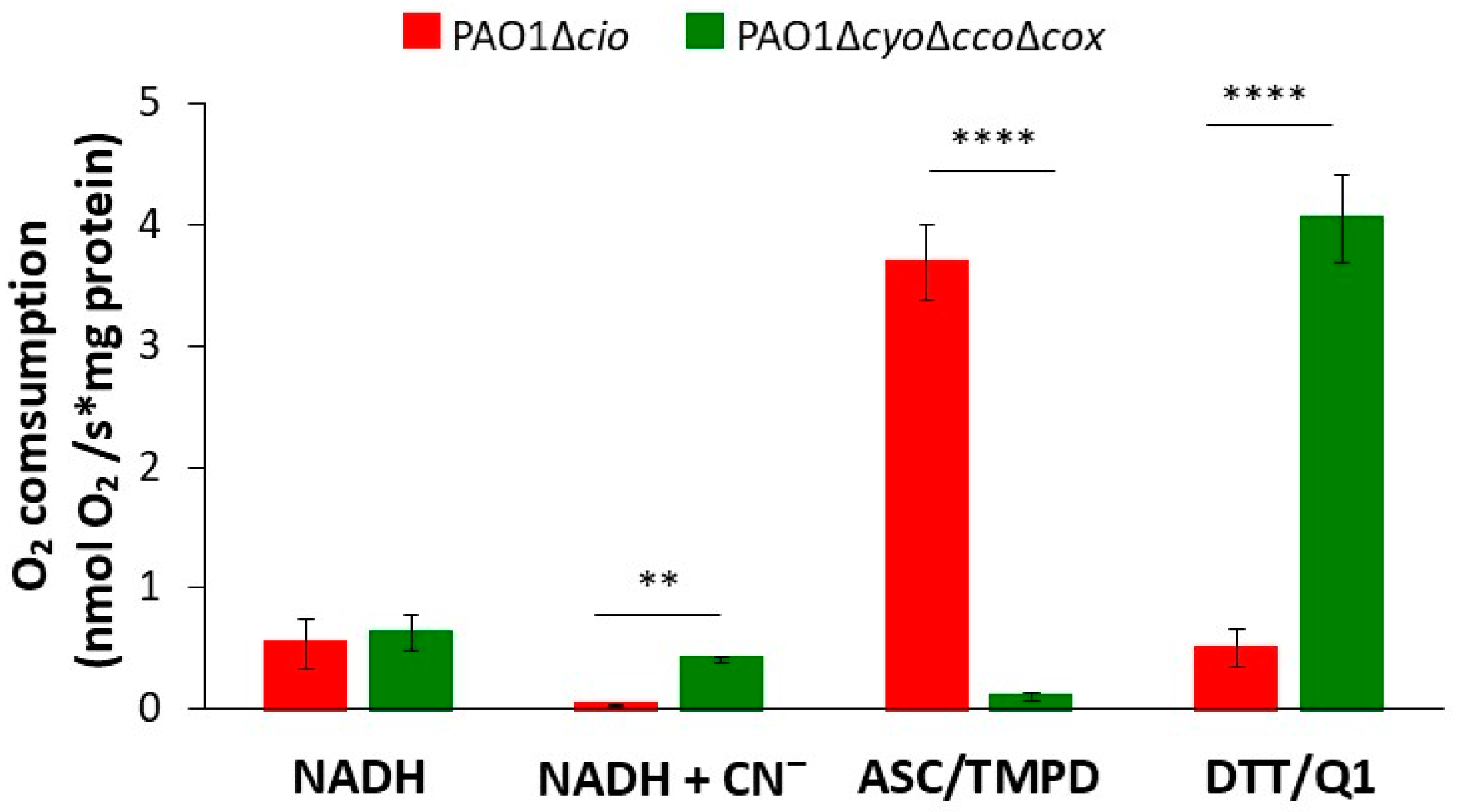
The respiratory activity of PAO1-derived mutants deleted in the cyo, cco-1, cco-2, and cox gene loci (∆cyo∆cco∆cox, [49]) or the cioAB locus (∆cio, [49]), respectively containing either only CIO or all other terminal oxidases except CIO, was initially investigated by testing the ability of membrane preparations from each strain to consume O2 in the presence of three different reducing systems: NADH (feeding the respiratory chain at the level of NADH-dehydrogenases, with donated electrons eventually reducing all terminal oxidases), DTT/Q1 (selectively reducing directly quinol oxidases), or ascorbate/TMPD (preferentially reducing directly cytochrome c oxidases). The results of these experiments are shown in Figure 1. When the physiological substrate NADH was used as the electron donor, the O2 consumption activity of both strains was similar (0.54 ± 0.21 and 0.63 ± 0.14 nmol O2/s*mg protein for Δcio and ΔcyoΔccoΔcox, respectively). As expected, 1 mM cyanide, a potent inhibitor of haem-copper oxidases, completely suppressed NADH-mediated O2 consumption in the Δcio strain only, indicating that CIO is the sole terminal oxidase able to sustain cyanide-insensitive respiration under the tested conditions. In the presence of ascorbate/TMPD, the Δcio mutant exhibited high O2 consumption activity (3.7 ± 0.31 nmol O2/s*mg protein), while the ΔcyoΔccoΔcox mutant (with the quinol oxidizing CIO as the only terminal oxidase) displayed almost undetectable respiratory activity (0.1 ± 0.3 nmol O2/s*mg protein). In contrast, in the presence of DTT/Q1, O2 consumption activity by the ΔcyoΔccoΔcox mutant was much higher (4.0 ± 0.4 nmol O2/s*mg protein) than observed with the Δcio mutant (0.5 ± 0.2 nmol O2/s*mg protein), indicating low expression of the bo3 quinol oxidase in the latter mutant.
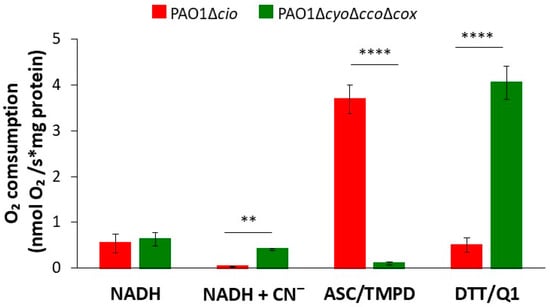
Figure 1.
O2 consumption by membrane preparations from P. aeruginosa mutants sustained with different reducing systems. O2 consumption activity of membranes prepared from the Δcio and ΔcyoΔccoΔcox mutants, measured at 25 °C in the presence of NADH (0.5 mM), ascorbate/TMPD (2 mM/0.2 mM), or DTT/Q1 (10 mM/0.25 mM). The average of at least three different experiments is reported together with the SD. For each condition, significant differences between the Δcio and ΔcyoΔccoΔcox mutants are indicated with asterisks (**, p < 0.01; ****, p < 0.0001).
3.2. Effect of Sulfide on Respiration of P. aeruginosa Membrane Preparations
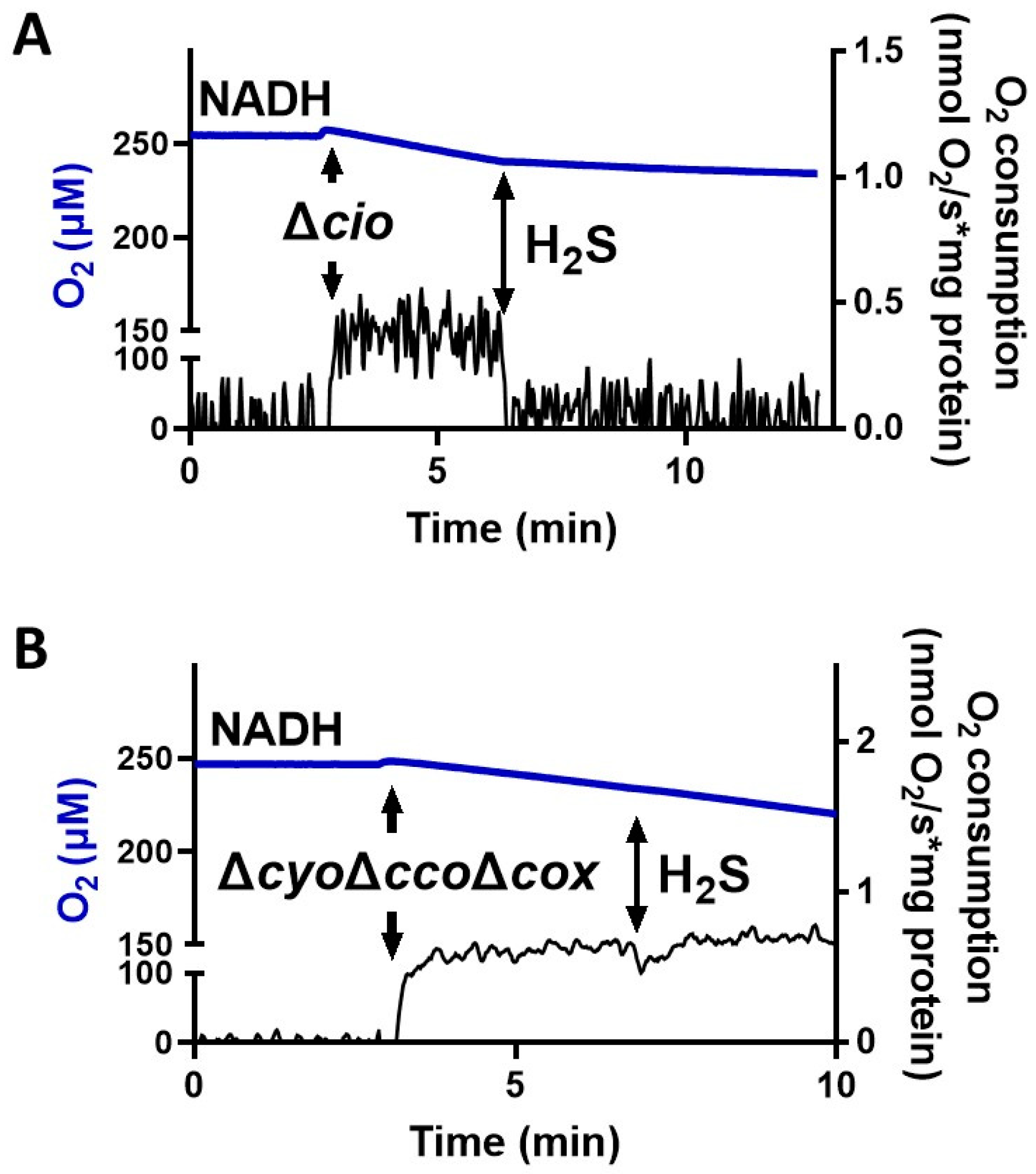
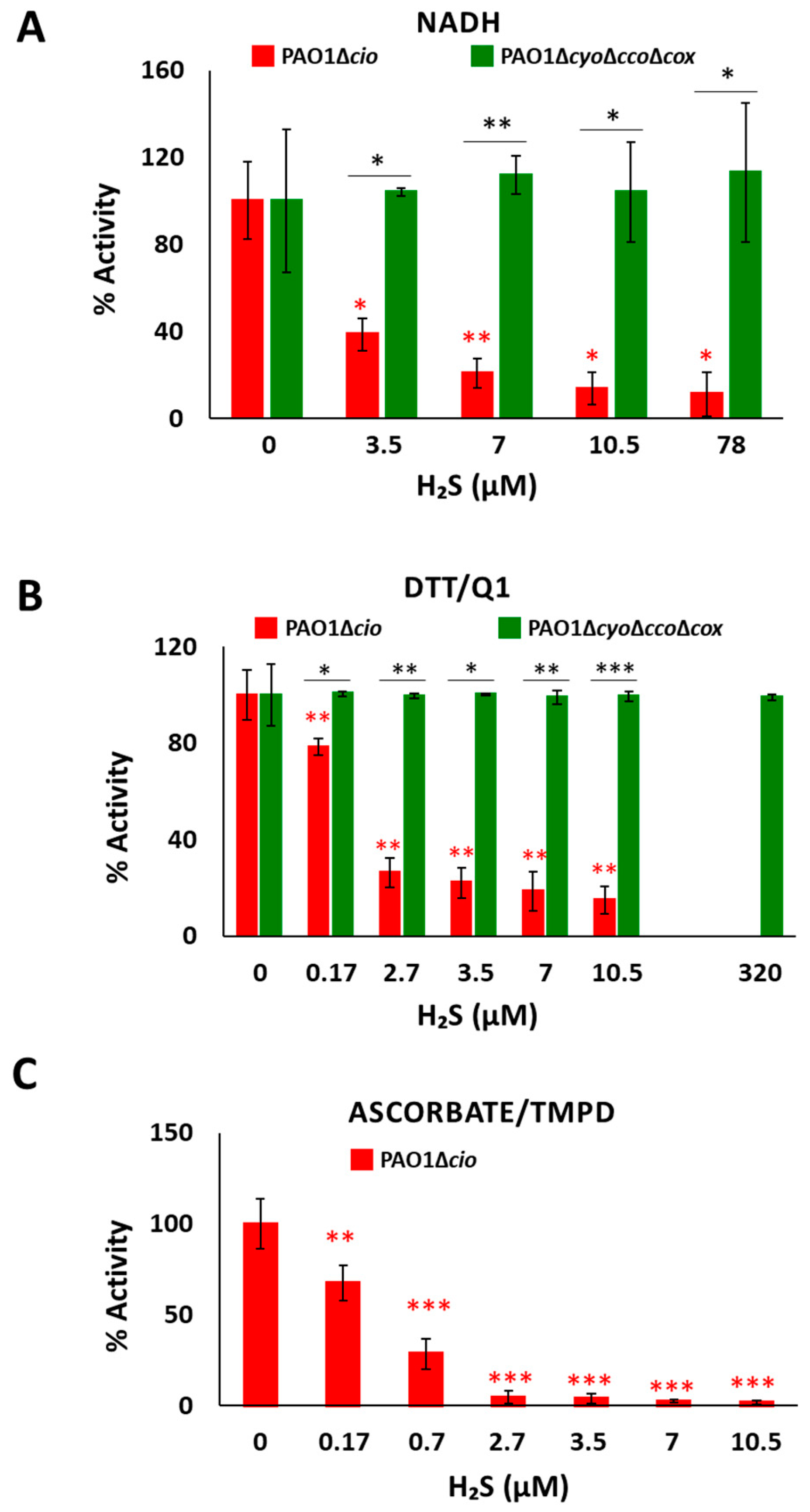
To test the effect of sulfide on P. aeruginosa CIO, we performed oxygraphic measurements on membrane preparations from the ΔcyoΔccoΔcox and Δcio mutants. As shown in Figure 2, the addition of 78 µM sulfide completely suppressed the NADH-mediated O2 consumption activity of Δcio (panel A) but did not affect that of the ΔcyoΔccoΔcox strain (panel B), demonstrating that CIO is insensitive to sulfide. As shown in Figure 3A, in the presence of NADH the respiratory activity of Δcio mutant was drastically affected at even lower sulfide concentrations (3.5, 7, or 10.5 µM), with the O2 consumption activity decreasing by ca. 60% at 3.5 µM sulfide; in contrast, no change in the respiratory activity of ΔcyoΔccoΔcox membranes was observed in response to sulfide addition at all tested concentrations. To compare the sensitivity to sulfide of P. aeruginosa quinol oxidases, bo3, and CIO, oxygraphic experiments on the ΔcyoΔccoΔcox and Δcio mutants were carried out with DTT/Q1 as the reducing system. Figure 3B shows that whereas CIO in the ΔcyoΔccoΔcox mutant is insensitive to up to 320 µM sulfide, the O2 consumption activity of bo3 in the Δcio mutant proved to be highly susceptible to sulfide, already being reduced by 20% at 0.17 µM sulfide. An even higher sensitivity to sulfide was observed for cytochrome c oxidases in the Δcio strain with ascorbate/TMPD as the reducing system (Figure 3C).
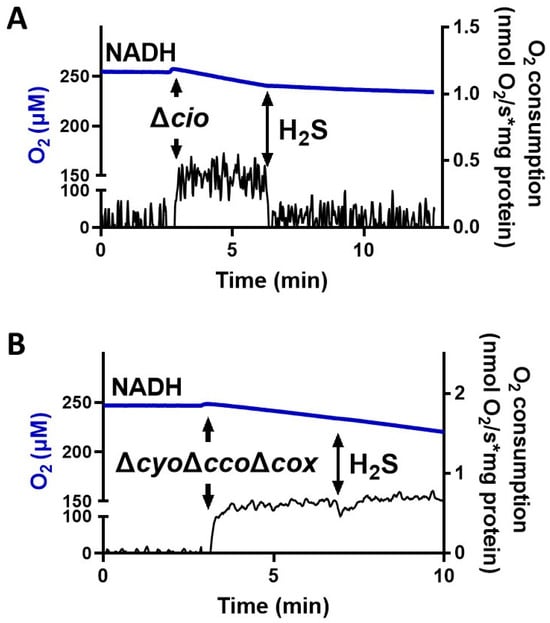
Figure 2.
Effect of sulfide (H2S) on NADH-mediated O2 consumption activity of membranes from P. aeruginosa Δcio and ΔcyoΔccoΔcox. Representative oxygraphic traces collected at 25 °C in the presence of 1 mM NADH before and after addition of 78 µM sulfide to membrane preparations of Δcio (A) or ΔcyoΔccoΔcox (B) mutants. Blue line: O2 concentration. Black line: O2 consumption rate. The protein concentrations of the Δcio and ΔcyoΔccoΔcox membranes were equal to 0.20 and 0.11 mg/mL, respectively.
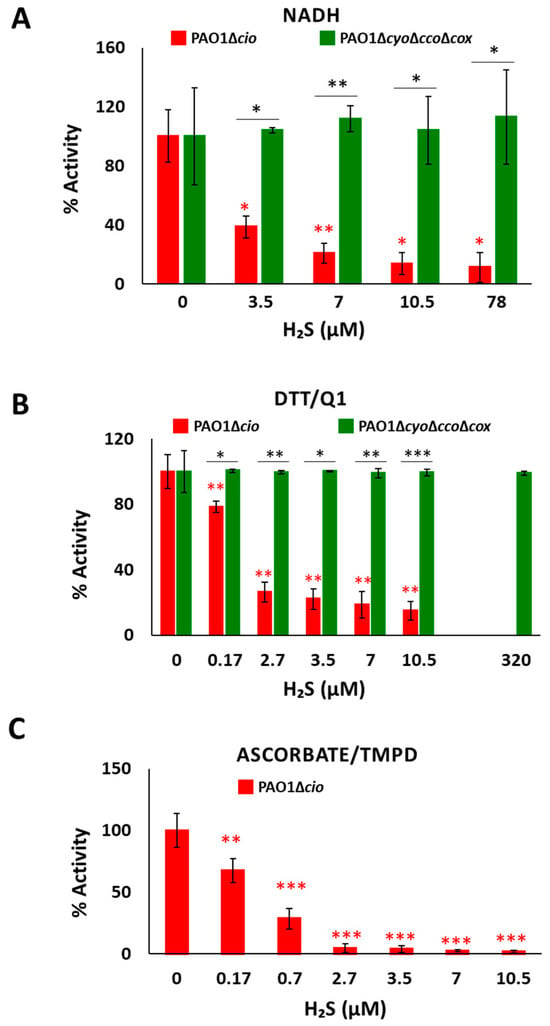
Figure 3.
Effect of sulfide on O2 consumption activity of membranes from P. aeruginosa Δcio and ΔcyoΔccoΔcox. O2 consumption activity of membrane preparations of the Δcio and ΔcyoΔccoΔcox strains measured in the presence of (A) NADH (1 mM), (B) DTT/Q1 (10 mM/0.25 mM), or (C) ascorbate/TMPD (2 mM/0.2 mM) at increasing concentrations of sulfide. The average of three independent experiments is reported with the SD. Asterisks (*, p < 0.05; **, p < 0.01; ***, p < 0.001) denote significant differences between ΔcyoΔccoΔcox and ∆cio (panels (A,B), black asterisks) or with respect to the control not treated with sulfide (panels (A–C), red asterisks).
Collectively, these results show that CIO is the only sulfide-insensitive terminal oxidase in P. aeruginosa, and agree with data on the model organism E. coli showing that the quinol haem-copper bo3 oxidase is effectively inhibited by sulfide (IC50 = 1.1 ± 0.1 μM) while the cytochromes bd-I and bd-II are both insensitive to sulfide [43]. As proposed for cytochrome E. coli bd-I, the sulfide insensitivity of CIO may arise from the lack of copper, which instead is present in the active site of haem-copper oxidases and has been proposed to be implicated in sulfide inhibition of mammalian cytochrome c oxidase [53]. H2S inhibition of mitochondrial cytochrome c oxidase is indeed suggested to involve the binding of the gaseous ligand at CuB in the enzyme in turnover, followed by intramolecular transfer of H2S to ferric haem a3 [53].
3.3. Effect of Sulfide on P. aeruginosa Cell Growth
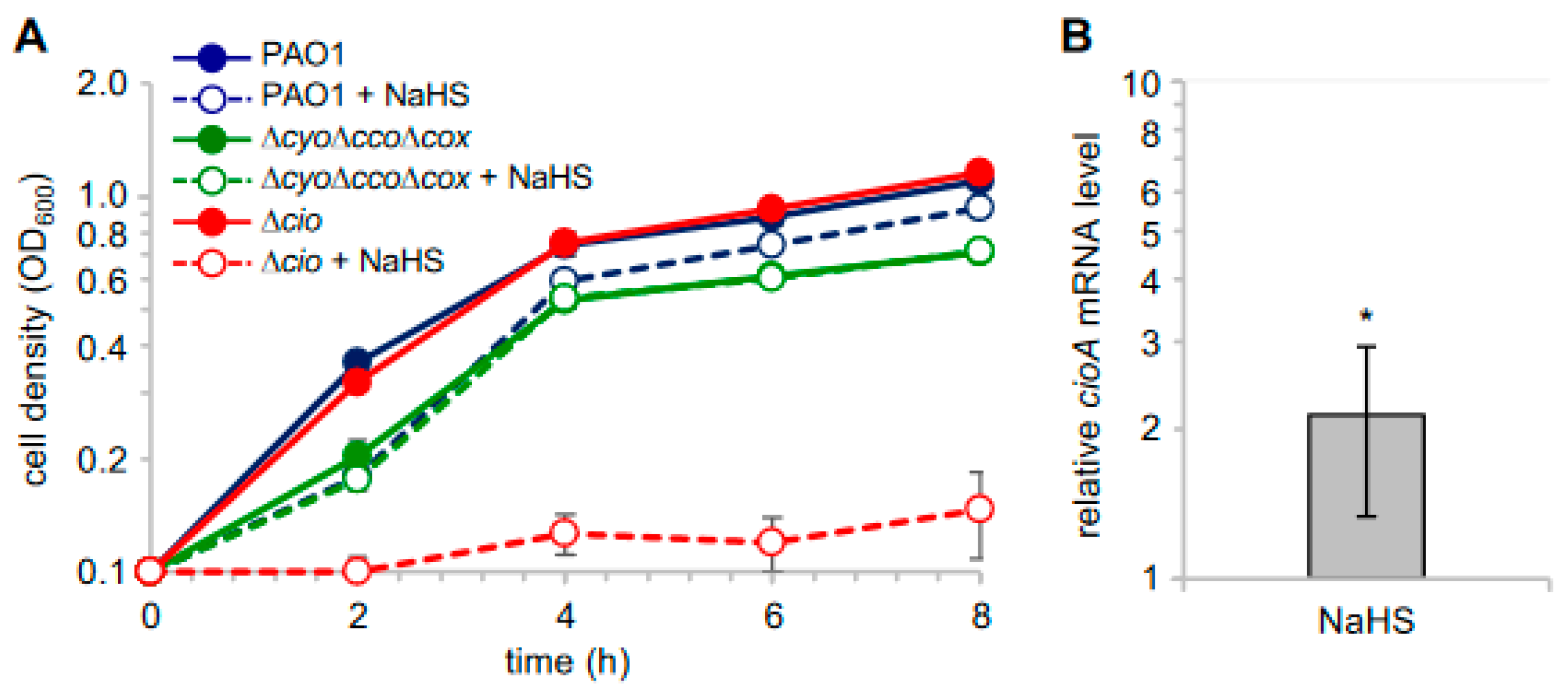
In light of the observed sulfide insensitivity of CIO, we investigated the effect of sulfide on P. aeruginosa cell growth by adding NaHS to cultures of P. aeruginosa PAO1 and its derived mutants ∆cio and ∆cyo∆cco∆cox [49]. As shown in Figure 4A, NaHS, while slightly decreasing the growth yield of PAO1, did not affect the growth of the ∆cyo∆cco∆cox mutant with CIO as the only terminal oxidase. Conversely, the growth of the ∆cio mutant was almost completely abrogated in the presence of NaHS, demonstrating that sulfide-insensitive CIO plays a pivotal role in aerobic respiration conditions in the presence of sulfide. In line with this evidence, our reverse transcriptase quantitative real time PCR (RT-qPCR) analyses revealed that cioA expression is upregulated by ca. two-fold in PAO1 cultures grown in the presence NaHS (Figure 4B).
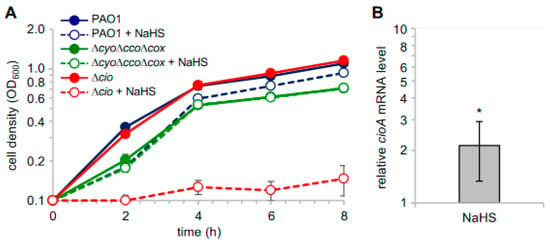
Figure 4.
Effect of NaHS on P. aeruginosa cell growth. (A) Growth curves of the indicated strains incubated at 37 °C with shaking (200 rpm) in Lysogeny Broth supplemented with 4 mM L-cysteine (LB-cys) in the absence (solid lines, full circles; untreated samples) or presence of 200 µM NaHS (dashed lines, empty circles; treated samples). The average of three independent experiments is reported with the SD. Differences between treated and untreated cultures of both PAO1 and ∆cio were statistically significant after 2 h incubation (p < 0.001). (B) Fold change in cioA mRNA level in PAO1 grown in LB-cys supplemented with 200 µM NaHS relative to the same strain grown in LB-cys alone. The average of three independent experiments is reported with the SD (*, p < 0.05).
3.4. Effect of NO on Respiration of P. aeruginosa Membranes
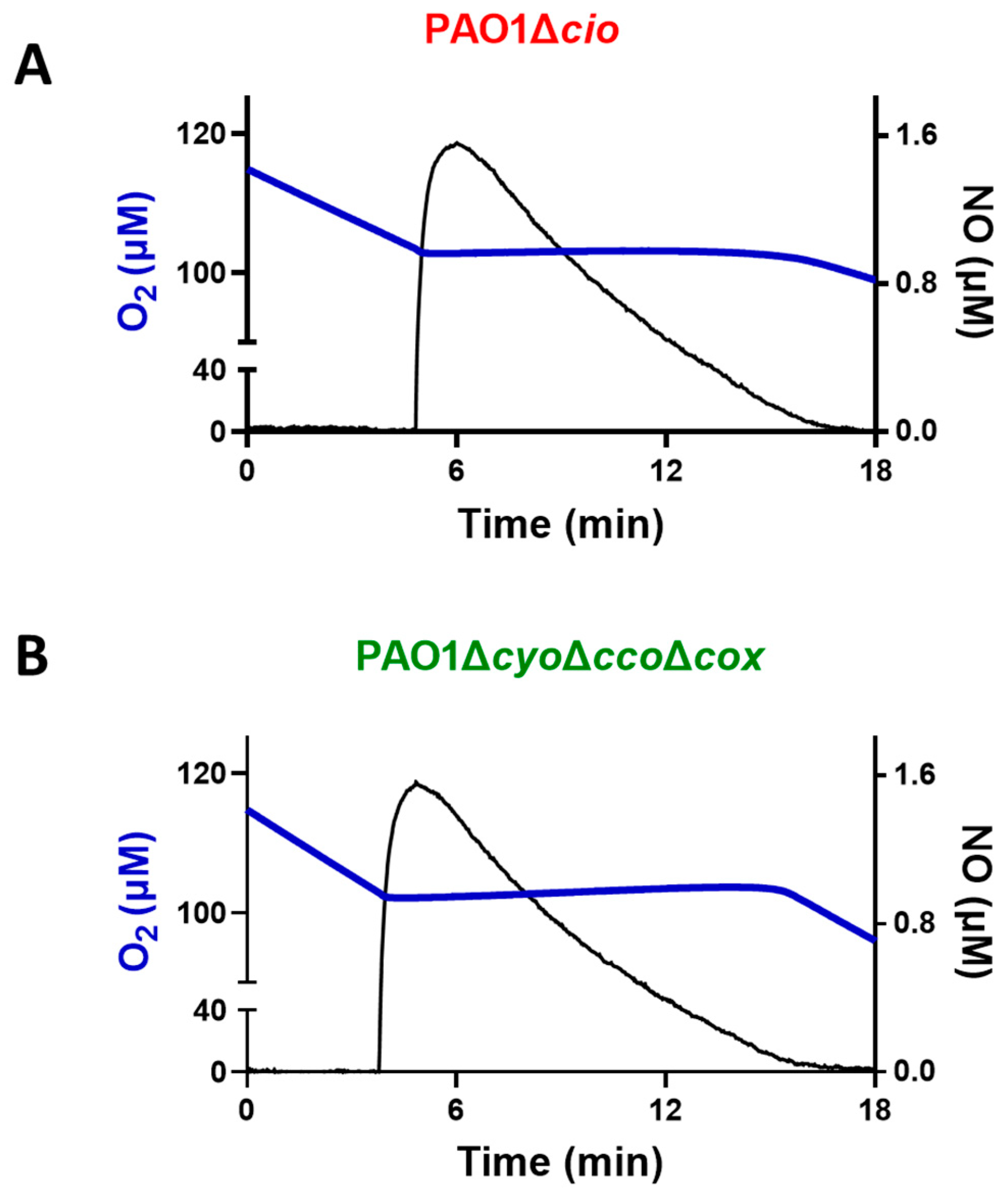
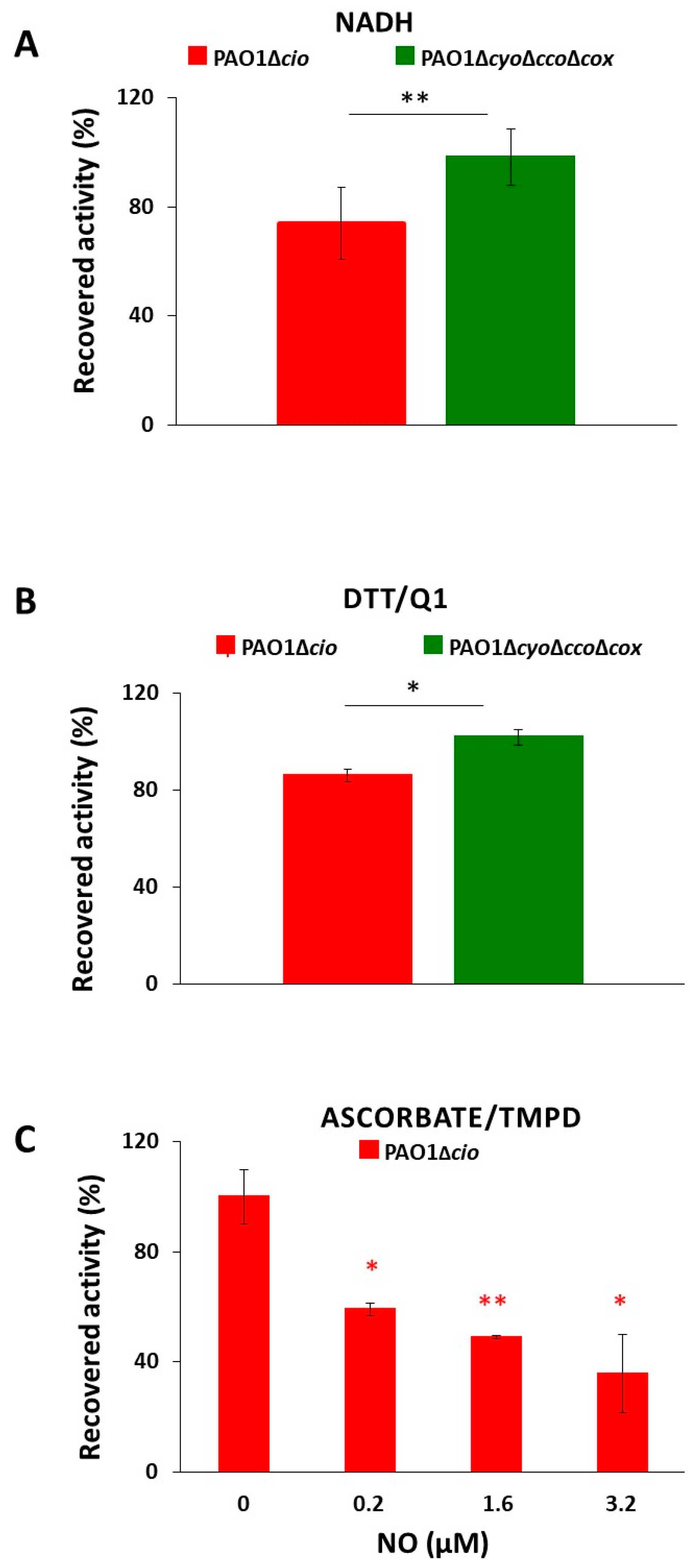
Triggered by the finding that the NO donor sodium nitroprusside increases cio gene expression [54], we investigated the effect of NO on the respiratory activity of membrane fractions from both the P. aeruginosa ΔcyoΔccoΔcox and Δcio mutant strains. Oxygraphic assays were conducted at 25 °C by simultaneously monitoring the O2 and NO concentrations in solution. As shown in Figure 5, in the presence of NADH as the electron donor, NO (1.6 μM) transiently and completely blocked the respiration of membrane preparations from both mutants, demonstrating inhibition by NO of all respiratory oxidases present in the membranes. When NO levels decayed upon reaction of NO with O2 in solution, this inhibition was relieved and respiration recovered. However, only the ΔcyoΔccoΔcox mutant regained full respiratory activity, whereas activity recovery was only partial (approximately 80% of the initial activity) in the Δcio strain (Figure 5 and Figure 6A). Because similar results were obtained with DTT/Q1 as the reducing system, which is selective for quinol oxidases (Figure 6B), we conclude that CIO is the only quinol oxidase from P. aeruginosa to fully recover its O2 reductase activity after NO inhibition. Interestingly, in the presence of ascorbate/TMPD, a remarkably lower percentage of maximal respiratory recovery (50%) was observed at the same NO concentration (1.6 μM) compared to that observed with DTT/Q1 (cfr. Figure 6C with Figure 6B). Surprisingly, in the presence of ascorbate/TMPD, concentrations of NO as low as 0.2 μM significantly affected the ability to regain full oxidase activity (Figure 6C), implying a higher propensity of at least one cytochrome c oxidase to undergo NO damage compared to quinol oxidases.
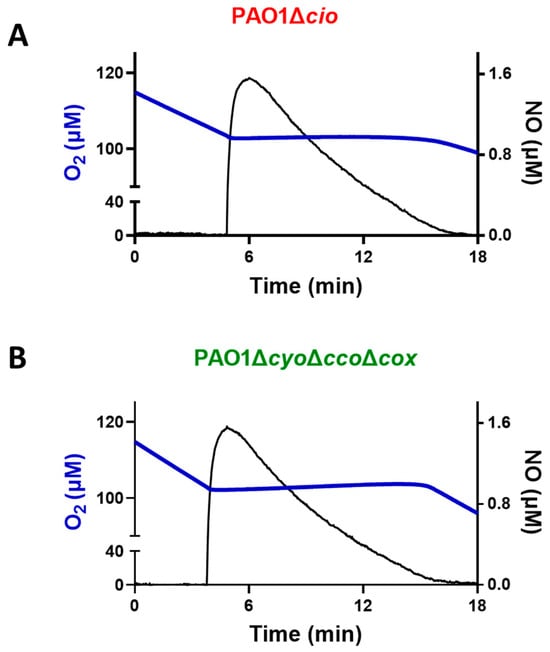
Figure 5.
Effect of nitric oxide (NO) on NADH-mediated O2 consumption activity of membranes of P. aeruginosa mutants Δcio and ΔcyoΔccoΔcox. Representative oxygraphic experiments performed at 25 °C in the presence of 1 mM NADH with membrane preparations of the Δcio (A) and ΔcyoΔccoΔcox (B) strains. [NO] = 1.6 µM. The protein concentration of the Δcio and ΔcyoΔccoΔcox membranes was equal to 0.27 and 0.23 mg/mL, respectively.
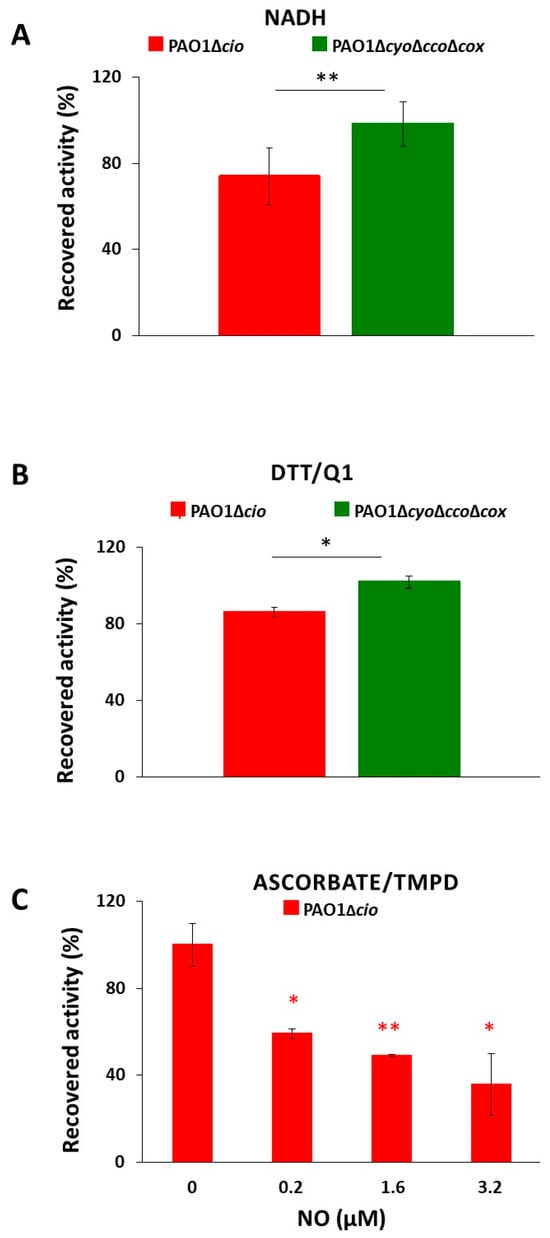
Figure 6.
O2 consumption activity recovered after NO inhibition of respiration of P. aeruginosa Δcio and ΔcyoΔccoΔcox membranes. O2 reductase activity recovered after NO inhibition of respiration of membranes prepared from the indicated mutants measured in the presence of (A) NADH (1 mM), (B) DTT/ Q1 (10 mM/0.25 mM), or (C) ascorbate/TMPD (2 mM/0.2 mM). The average of three independent experiments is reported with the SD. Asterisks (*, p < 0.05; **, p < 0.01) denote significant differences between ΔcyoΔccoΔcox and ∆cio (panels (A,B)) or with respect to the control untreated with NO (panel (C)).
Taken together, these results show that CIO is less vulnerable to irreversible NO damage compared to other terminal oxidases in P. aeruginosa, possibly contributing to the resistance of P. aeruginosa to NO stress.
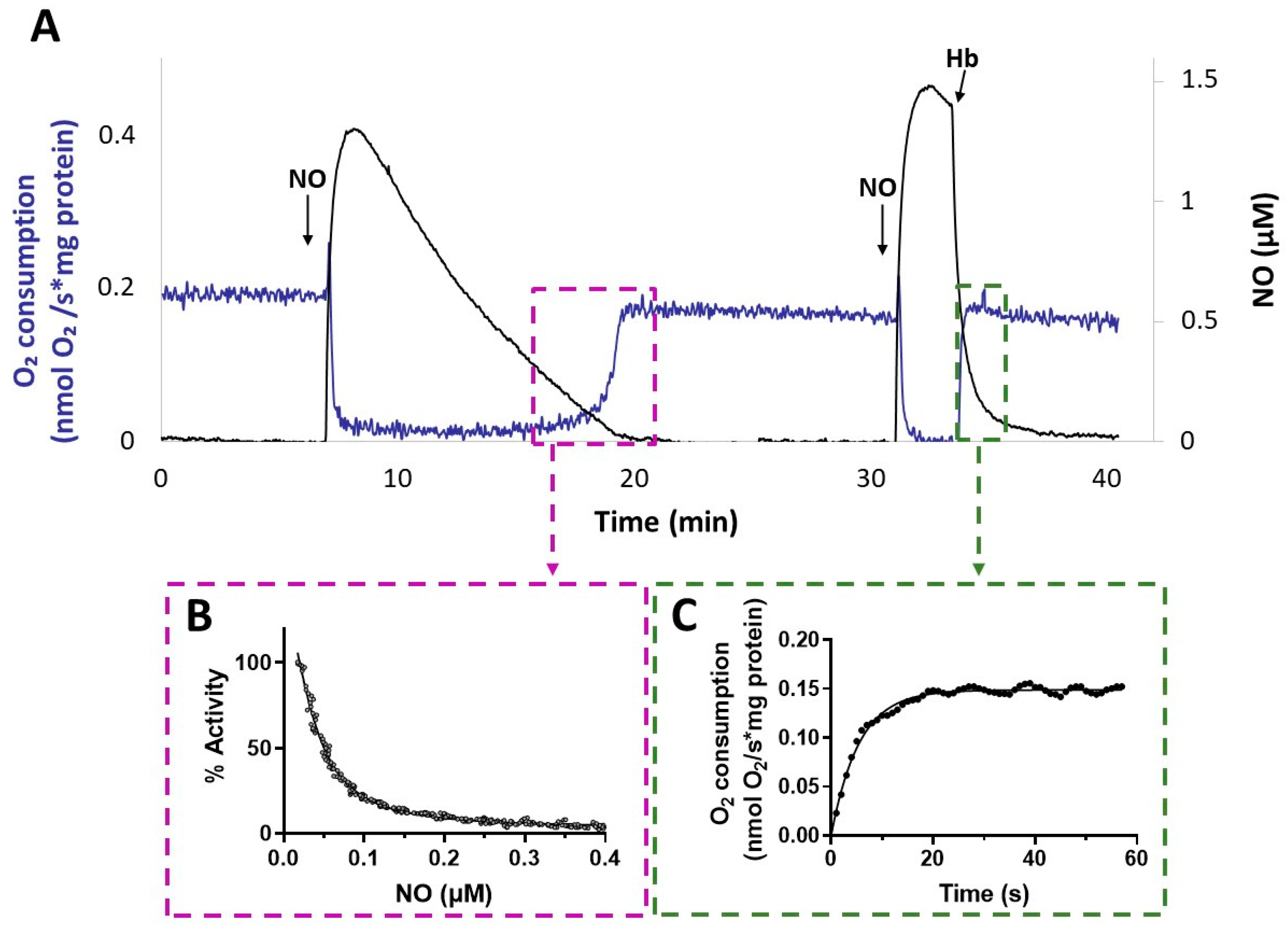
Interestingly, after NO vanished from the solution, the activity recovery was faster with membranes from the ΔcyoΔccoΔcox mutant compared to that with membranes from Δcio (Figure 5). This result suggests a relatively fast dissociation of NO from the reduced haem in the CIO active site during turnover. The kinetics of activity recovery from NO inhibition of ΔcyoΔccoΔcox membranes were investigated under two distinct experimental conditions: either allowing slow decay of NO levels upon reaction of NO with O2 in solution, or rapidly removing the NO in solution by reaction with excess oxyhemoglobin (HbO2). When all unbound NO was removed rapidly from the solution with fast NO scavengers, the recovery of respiration tended to proceed at the off-rate of NO from the haem in the oxidase active site [51]. These respirometric experiments were conducted in the presence of NADH and while adding NO at an [O2] level of about 100 µM. As shown in Figure 7, activity recovery in the presence of HbO2 was remarkably faster than in its absence (panel A), proceeding at 0.18 ± 0.01 s−1 (panel C), which represents a lower limit value for the off-rate of NO from CIO. This value is similar to those reported for isolated E. coli cytochrome bd in the reduced state (0.133 ± 0.05 s−1) [55] or as integrated in the native membrane preparations of an E. coli mutant strain lacking the haem-copper bo3 oxidase (0.163 s–1) [39]. The NO koff values reported for bd-type oxidases are about 30 times higher than the ones reported for the mitochondrial cytochrome c oxidase and the vast majority of haem proteins.
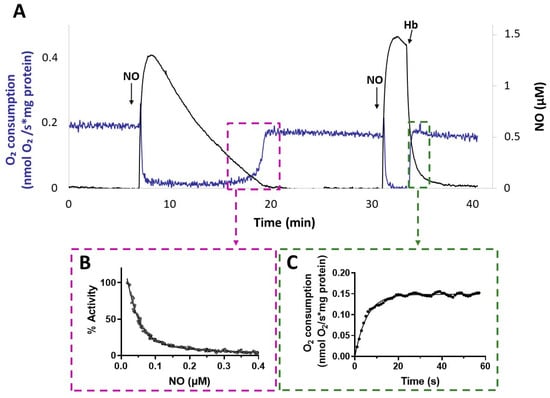
Figure 7.
NO inhibition of CIO. (A) O2 consumption rate (blue line) and NO concentration (black line) simultaneously recorded after addition of NO to ΔcyoΔccoΔcox membranes respiring O2 in the presence of NADH. Following the addition of the first aliquot of NO, administered at 100 μM O2, respiration is transiently and fully inhibited. As NO slowly decays by reaction with O2 in solution, respiration recovery takes place, eventually reaching the rate observed prior to inhibition. Afterwards, addition of a second aliquot of NO at 80 μM O2 inhibits respiration again. Following the addition of excess HbO2, which rapidly reacts with NO, fast activity recovery is observed upon NO disappearance from solution. Additions: NADH (1 mM), CIO-containing membranes (0.2 mg protein/mL), NO (1.3 µM), HbO2 (15 μM). (B) Percentage of control activity measured as NO vanishes from solution. Data from the relative boxed area depicted in panel A are plotted as a function of [NO], and data analysis yielded a half-maximal inhibitory concentration for NO value of 55 nM in this sample. The data do not intercept the y-axis, as invariably observed and tentatively explained by limitations in the NO electrode response at very low NO concentrations. (C) Respiration rates measured after addition of HbO2. Data from the relative boxed area depicted in panel (A) are plotted as a function of time and fitted to a single exponential (solid line), yielding an NO off-rate value of 0.18 s−1.
The slower activity recovery in the absence of HbO2 allowed us to investigate the dependence of the respiratory activity on [NO], from which we estimated a half-maximal inhibitory concentration value for NO (apparent IC50) of 49 ± 18 nM NO (panel B). An apparent IC50 value of about 100 nM NO at 70 μM O2 was previously reported for the E. coli purified bd-I [38].
4. Discussion
Respiratory flexibility is likely to be a major contributor to the success of P. aeruginosa as an opportunistic pathogen. The presence of several aerobic terminal oxidases with different affinities for O2 may be critical for microorganisms which need to thrive in environments characterized by steep O2 gradients, such as P. aeruginosa in biofilms. However, the mechanisms regulating the expression of terminal oxidases and their function appear to extend beyond O2 concentrations [29,56,57,58]. The view that oxidases with low or high O2 affinity are respectively expressed at high or low O2 concentration is not always pertinent. For instance, P. aeruginosa expresses the high-affinity cbb3-1 during aerobic growth even under high oxygen conditions [29]; in contrast, while the low-affinity aa3 oxidase is poorly expressed at high concentrations of O2, its expression is upregulated in the stationary phase and more significantly under starvation of carbon, nitrogen, or iron [29]. Arai et al. suggested that the aa3 oxidase is preferentially used under low nutrient conditions, as this oxidase has the highest proton translocation efficiency of all five P. aeruginosa terminal oxidases and consequently the highest efficiency in generating ATP [29]. Thus, each oxidase appears to be operative under specific conditions, suggesting that modulation of the P. aeruginosa respiratory chain is effectively influenced by environmental conditions and stresses.
Such adaptations are important for colonization of infection sites, particularly in the lungs of CF individuals, where bacteria are challenged not only by low O2 levels but by the paucity of nutrients and the presence of toxic species. The low-affinity CIO is preferentially expressed during the stationary phase, when O2 is relatively scarce, but secondary and toxic metabolites acting as signalling molecules or virulence factors are produced [59,60]. As the amount of H2S significantly increases in the stationary phase [61], and as bd-type oxidases, at least in E. coli, have been shown to be insensitive to H2S inhibition, we hypothesized that P. aeruginosa CIO could be insensitive to such long-known inhibitors of O2 respiration as well. Consistently, we proved that CIO activity is highly insensitive to sulfide and that, remarkably, its expression is enhanced in the presence of this agent, further confirming that the expression of terminal oxidases in this pathogen is dependent on environmental stressors [43,53].
Importantly, these results strongly support the central role of CIO in P. aeruginosa growth and colonization potential in sulfide-rich environments, and agree with previous studies on other pathogens. Saini et al. reported that H2S promotes respiration and growth in Mycobacterium tuberculosis, Mycobacterium smegmatis, and Mycobacterium bovis BCG via cytochrome bd, with the other terminal oxidase cytochrome aa3 (forming a supercomplex with cytochromes bcc) being inhibited by host-derived H2S produced during infection [62]. Interestingly, endogenously produced H2S allows bioenergetic homeostasis by stimulating respiration, primarily via the bd-type terminal oxidase in multidrug-resistant and drug-susceptible clinical M. tuberculosis strains, but not in the non-pathogenic M. smegmatis [63]. While it has been known for more than 70 years that clinical strains of P. aeruginosa produce endogenous H2S [64], it was only in 2011 that a study linked H2S generation to P. aeruginosa pathogenicity [14]. In that work, the authors showed that endogenous H2S protects P. aeruginosa, as well as Staphylococcus aureus, E. coli, and Bacillus anthracis, from oxidative and antibiotics stress, proposing H2S as a general defence mechanism against antibiotic killing. However, while a number of studies have later confirmed this proposal [61,65,66], others have questioned the universality of this mechanism among bacteria [67,68], including P. aeruginosa [69]. H2S and its derived sulfane sulfur-containing species have been found to promote transcription and production of several virulence factors by activating LasR [70], which is a quorum sensing master regulator in P. aeruginosa PAO1. This finding points to a role of H2S in P. aeruginosa virulence and implies that CIO may be relevant during infection by allowing sulfide-resistant growth and energy production.
In accordance with the potential role of CIO in microbial pathogenicity, we report here that, under the tested conditions, CIO is more resistant to NO damage than the other P. aeruginosa respiratory oxidases. Experimental evidence supports that bd-type oxidases are key enzymes for host colonization under nitrosative stress. Deletion of cytochrome bd-I from a multi-drug resistant uropathogenic E. coli strain impaired the survival of bacteria in the mouse urinal tract [34]. The NO tolerance of this oxidase agrees with the enhanced expression of its coding genes observed under NO stress in E. coli. S. enterica ser. Typhimurium, S. aureus, M. tuberculosis, and Bacillus subtilis [25,71,72,73,74,75]. It has been proposed that the NO tolerance of bd oxidase is due to its ability to rapidly convert NO into nitrite during turnover and its unusually high NO dissociation rate from the active site [55]. As with most haem-copper oxidases [76,77], E. coli and Azotobacter vinelandii bd-type oxidases are rapidly, potently, and reversibly inhibited by NO through binding at reduced haem d [38]. Accordingly, we found that P. aeruginosa CIO inhibition by NO is complete and fully reversible, and that the recovery of activity is fast due to the high NO off rate [39,51,55,78].
We suggest that the fast recovery of CIO activity from NO inhibition could allow P. aeruginosa to cope with this potent inhibitor of aerobic respiration during infection, when the pathogen is exposed to the NO produced by eukaryotic host cells as part of the mammalian immune defence system [79,80]. To keep NO levels low and avoid its toxicity, P. aeruginosa is equipped with at least two efficient inducible NO detoxification mechanisms: a flavohemoglobin, which has NO dioxygenase activity under aerobic conditions and is induced by the NO donors sodium nitroprusside and S-nitrosoglutathione (GSNO) [81], and an NO-reductase, which removes NO under microaerobic conditions and allows intracellular survival in the NO-producing RAW 264.7 macrophage by counteracting the host’s defence systems [82,83]. Our data show that CIO may contribute to NO tolerance by allowing prompt recovery of energy production following NO stress, thereby conferring a physiological advantage to P. aeruginosa. Interestingly, transcription of the cyo gene coding for bo3 oxidase is induced by GSNO [81,84,85,86,87]; however, the role of the bo3 oxidase in NO resistance is unclear at present, and future studies are needed to clarify this issue. In light of the plasticity of the P. aeruginosa aerobic electron transfer chain and the fine-tuning of respiratory enzyme expression in response to environmental conditions, it is conceivable that other terminal oxidases are implicated in NO tolerance in addition to CIO. Indeed, as reported above, two oxidases appear to be involved in protection against cyanide toxicity under different environmental conditions [30,32].
The use of mutant strains may be a limitation of this study, as mutations can potentially influence other processes in the strain and introduce phenotypic changes. However, the growth and expression data on wild-type cells are consistent with those obtained with the mutant strains, giving us reason to believe that our conclusions are correct. Another limitation is the utilization of the PAO1 laboratory reference strain; future studies with clinical strains are needed in order to shed light on the effect of NO and H2S on terminal oxidase gene expression and the extent of tolerance to these gases in vivo.
5. Conclusions
Collectively, our data show that CIO provides P. aeruginosa with aerobic respiration tolerance to NO and H2S, noxious molecules to which this pathogen is exposed during infection, suggesting a role of CIO in P. aeruginosa virulence. Accordingly, we observed that exposure of P. aeruginosa PAO1 to H2S upregulates a gene coding for CIO. These new findings contribute to a better understanding of how different terminal complexes participate in the respiratory chain under various growth conditions. This study provides further evidence that high tolerance to stressors is one of the most distinctive and interesting features of bd-type oxidases, which have recently been recognised as attractive potential targets for the development of next-generation antimicrobials [25]. In the current scenario, where research and development on new strategies to treat P. aeruginosa infections has become essential, targeting bacterial energy metabolism by inhibition of respiratory chain components is a promising strategy [20].
Author Contributions
Conceptualization, E.F., A.G. and G.R.; methodology, M.R.N., L.C., F.G., M.M., G.R. and E.F.; investigation, M.R.N., L.C., F.G. and M.M.; data curation, M.R.N., L.C., F.G. and M.M.; writing—original draft preparation: E.F., M.R.N. and G.R.; writing—review and editing, M.R.N., E.F., A.G. and G.R.; funding acquisition, E.F., A.G. and G.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Italian Ministry of University and Research (PRIN 2022 grant 20224BYR59, Funded by the EU—NextGenerationEU to E.F., A.G. and G.R.), by EU funding within the NextGenerationEU-MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT to A.G.), and by Sapienza University of Rome (grants RP12117A8AA5B0E7 and RM122181698FC99 to E.F.).
Data Availability Statement
Data is contained within the article.
Acknowledgments
We thank Paolo Visca (University Roma Tre, Rome, Italy) and Emanuela Frangipani (University of Urbino Carlo Bo, Urbino, Italy) for providing the PAO1 mutant strains ∆cio and ∆cyo∆cco∆cox.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Szabo, C. A Timeline of Hydrogen Sulfide (H2S) Research: From Environmental Toxin to Biological Mediator. Biochem. Pharmacol. 2018, 149, 5–19. [Google Scholar] [CrossRef]
- Deshmukh, R.; Harwansh, R.K.; Bandyopadhyay, N.; Bandopadhyay, S.; Kumar, P. Pharmacology of Gasotransmitters (Nitric Oxide and Carbon Monoxide) and Their Action. In Frontiers in Pharmacology of Neurotransmitters; Springer: Singapore, 2020; pp. 579–617. [Google Scholar]
- Lo Faro, M.L.; Fox, B.; Whatmore, J.L.; Winyard, P.G.; Whiteman, M. Hydrogen Sulfide and Nitric Oxide Interactions in Inflammation. Nitric Oxide 2014, 41, 38–47. [Google Scholar] [CrossRef]
- Mancuso, C.; Navarra, P.; Preziosi, P. Roles of Nitric Oxide, Carbon Monoxide, and Hydrogen Sulfide in the Regulation of the Hypothalamic–Pituitary–Adrenal Axis. J. Neurochem. 2010, 113, 563–575. [Google Scholar] [CrossRef]
- Farrugia, G.; Szurszewski, J.H. Carbon Monoxide, Hydrogen Sulfide, and Nitric Oxide as Signaling Molecules in the Gastrointestinal Tract. Gastroenterology 2014, 147, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Olson, K.R.; Donald, J.A. Nervous Control of Circulation--the Role of Gasotransmitters, NO, CO, and H2S. Acta Histochem. 2009, 111, 244–256. [Google Scholar] [CrossRef] [PubMed]
- Wareham, L.K.; Southam, H.M.; Poole, R.K. Do Nitric Oxide, Carbon Monoxide and Hydrogen Sulfide Really Qualify as “gasotransmitters” in Bacteria? Biochem. Soc. Trans. 2018, 46, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Jesse, H.E.; Poole, R.K.; Tinajero-Trejo, M. Gasotransmitters, Poisons, and Antimicrobials: It’s a Gas, Gas, Gas! F1000Prime Rep. 2013, 5, 28. [Google Scholar] [CrossRef]
- Toliver-Kinsky, T.; Cui, W.; Törö, G.; Lee, S.J.; Shatalin, K.; Nudler, E.; Szaboa, C. H2S, a Bacterial Defense Mechanism against the Host Immune Response. Infect. Immun. 2018, 87, e00272-18. [Google Scholar] [CrossRef] [PubMed]
- Luhachack, L.; Nudler, E. Bacterial Gasotransmitters: An Innate Defense against Antibiotics. Curr. Opin. Microbiol. 2014, 21, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Mironov, A.; Seregina, T.; Nagornykh, M.; Luhachack, L.G.; Korolkova, N.; Lopes, L.E.; Kotova, V.; Zavilgelsky, G.; Shakulov, R.; Shatalin, K.; et al. Mechanism of H2S-Mediated Protection against Oxidative Stress in Escherichia coli. Proc. Natl. Acad. Sci. USA 2017, 114, 6022–6027. [Google Scholar] [CrossRef]
- Bryan, N.S.; Lefer, D.J. Update on Gaseous Signaling Molecules Nitric Oxide and Hydrogen Sulfide: Strategies to Capture Their Functional Activity for Human Therapeutics. Mol. Pharmacol. 2019, 96, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Mendes, S.S.; Miranda, V.; Saraiva, L.M. Hydrogen Sulfide and Carbon Monoxide Tolerance in Bacteria. Antioxidants 2021, 10, 729. [Google Scholar] [CrossRef] [PubMed]
- Shatalin, K.; Shatalina, E.; Mironov, A.; Nudler, E. H2S: A Universal Defense against Antibiotics in Bacteria. Science 2011, 334, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Gusarov, I.; Shatalin, K.; Starodubtseva, M.; Nudler, E. Endogenous Nitric Oxide Protects Bacteria against a Wide Spectrum of Antibiotics. Science 2009, 325, 1380–1384. [Google Scholar] [CrossRef]
- Walsh, B.J.C.; Giedroc, D.P. H2S and Reactive Sulfur Signaling at the Host-Bacterial Pathogen Interface. J. Biol. Chem. 2020, 295, 13150–13168. [Google Scholar] [CrossRef] [PubMed]
- Pal, V.K.; Bandyopadhyay, P.; Singh, A. Hydrogen Sulfide in Physiology and Pathogenesis of Bacteria and Viruses. IUBMB Life 2018, 70, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Shen, J.; Edmonds, K.A.; Luebke, J.L.; Hickey, A.K.; Palmer, L.D.; Chang, F.-M.J.; Bruce, K.A.; Kehl-Fie, T.E.; Skaar, E.P.; et al. Sulfide Homeostasis and Nitroxyl Intersect via Formation of Reactive Sulfur Species in Staphylococcus aureus. mSphere 2017, 2, e00082-17. [Google Scholar] [CrossRef] [PubMed]
- Kaila, V.R.I.; Wikström, M. Architecture of Bacterial Respiratory Chains. Nat. Rev. Microbiol. 2021, 19, 319–330. [Google Scholar] [CrossRef]
- Hards, K.; Cook, G.M. Targeting Bacterial Energetics to Produce New Antimicrobials. Drug Resist. Updat. 2018, 36, 1–12. [Google Scholar] [CrossRef]
- Folkesson, A.; Jelsbak, L.; Yang, L.; Johansen, H.K.; Ciofu, O.; Hoiby, N.; Molin, S. Adaptation of Pseudomonas aeruginosa to the Cystic Fibrosis Airway: An Evolutionary Perspective. Nat. Rev. Microbiol. 2012, 10, 841–851. [Google Scholar] [CrossRef]
- Winstanley, C.; O’Brien, S.; Brockhurst, M.A. Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends Microbiol. 2016, 24, 327–337. [Google Scholar] [CrossRef]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 27 February 2017).
- Borisov, V.B.; Siletsky, S.A.; Paiardini, A.; Hoogewijs, D.; Forte, E.; Giuffrè, A.; Poole, R.K. Bacterial Oxidases of the Cytochrome bd Family: Redox Enzymes of Unique Structure, Function, and Utility as Drug Targets. Antioxid. Redox Signal 2021, 34, 1280–1318. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, L.; Pitt, M.; Williams, H.D. The CioAB Genes from Pseudomonas aeruginosa Code for a Novel Cyanide-Insensitive Terminal Oxidase Related to the Cytochrome bd Quinol Oxidases. Mol. Microbiol. 1997, 24, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Arai, H. Regulation and Function of Versatile Aerobic and Anaerobic Respiratory Metabolism in Pseudomonas aeruginosa. Front. Microbiol. 2011, 2, 103. [Google Scholar] [CrossRef]
- Alvarez-Ortega, C.; Harwood, C.S. Responses of Pseudomonas aeruginosa to Low Oxygen Indicate That Growth in the Cystic Fibrosis Lung Is by Aerobic Respiration. Mol. Microbiol. 2007, 65, 153–165. [Google Scholar] [CrossRef]
- Arai, H.; Kawakami, T.; Osamura, T.; Hirai, T.; Sakai, Y.; Ishii, M. Enzymatic Characterization and in vivo Function of Five Terminal Oxidases in Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 4206–4215. [Google Scholar] [CrossRef]
- Zlosnik, J.E.A.; Tavankar, G.R.; Bundy, J.G.; Mossialos, D.; O’Toole, R.; Williams, H.D. Investigation of the Physiological Relationship between the Cyanide-Insensitive Oxidase and Cyanide Production in Pseudomonas aeruginosa. Microbiology 2006, 152, 1407–1415. [Google Scholar] [CrossRef]
- Cunningham, L.; Williams, H.D. Isolation and Characterization of Mutants Defective in the Cyanide-Insensitive Respiratory Pathway of Pseudomonas aeruginosa. J. Bacteriol. 1995, 177, 432–438. [Google Scholar] [CrossRef][Green Version]
- Cooper, M.; Tavankar, G.R.; Williams, H.D. Regulation of Expression of the Cyanide-Insensitive Terminal Oxidase in Pseudomonas aeruginosa. Microbiology 2003, 149, 1275–1284. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hirai, T.; Osamura, T.; Ishii, M.; Arai, H. Expression of Multiple cbb3 Cytochrome c Oxidase Isoforms by Combinations of Multiple Isosubunits in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2016, 113, 12815–12819. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, M.; Achard, M.E.S.; Idris, A.; Totsika, M.; Phan, M.D.; Peters, K.M.; Sarkar, S.; Ribeiro, C.A.; Holyoake, L.V.; Ladakis, D.; et al. The Cytochrome bd-I Respiratory Oxidase Augments Survival of Multidrug-Resistant Escherichia coli during Infection. Sci. Rep. 2016, 6, 35285. [Google Scholar] [CrossRef]
- Seregina, T.A.; Lobanov, K.V.; Shakulov, R.S.; Mironov, A.S. Inactivation of terminal oxidase bd-I leads to supersensitivity of E. coli to quinolone and beta-lactam antibiotics. Mol. Biol. (Mosk) 2022, 56, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Forte, E.; Siletsky, S.A.; Sarti, P.; Giuffrè, A. Cytochrome bd from Escherichia coli Catalyzes Peroxynitrite Decomposition. Biochim. Biophys. Acta Bioenerg. 2015, 1847, 182–188. [Google Scholar] [CrossRef]
- Borisov, V.B.; Forte, E. Bioenergetics and Reactive Nitrogen Species in Bacteria. Int. J. Mol. Sci. 2022, 23, 7321. [Google Scholar] [CrossRef]
- Borisov, V.B.; Forte, E.; Konstantinov, A.A.; Poole, R.K.; Sarti, P.; Giuffrè, A. Interaction of the Bacterial Terminal Oxidase Cytochrome bd with Nitric Oxide. FEBS Lett. 2004, 576, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Mason, M.G.; Shepherd, M.; Nicholls, P.; Dobbin, P.S.; Dodsworth, K.S.; Poole, R.K.; Cooper, C.E. Cytochrome bd Confers Nitric Oxide Resistance to Escherichia coli. Nat. Chem. Biol. 2009, 5, 94–96. [Google Scholar] [CrossRef]
- Borisov, V.B.; Forte, E.; Davletshin, A.; Mastronicola, D.; Sarti, P.; Giuffrè, A. Cytochrome bd Oxidase from Escherichia coli Displays High Catalase Activity: An Additional Defense against Oxidative Stress. FEBS Lett. 2013, 587, 2214–2218. [Google Scholar] [CrossRef]
- Borisov, V.B.; Nastasi, M.R.; Forte, E. Cytochrome bd as Antioxidant Redox Enzyme. Mol. Biol. 2023, 57, 1077–1084. [Google Scholar] [CrossRef]
- Al-Attar, S.; Yu, Y.; Pinkse, M.; Hoeser, J.; Friedrich, T.; Bald, D.; De Vries, S. Cytochrome bd Displays Significant Quinol Peroxidase Activity. Sci. Rep. 2016, 6, 27631. [Google Scholar] [CrossRef]
- Forte, E.; Borisov, V.B.; Falabella, M.; Colaço, H.G.; Tinajero-Trejo, M.; Poole, R.K.; Vicente, J.B.; Sarti, P.; Giuffrè, A. The Terminal Oxidase Cytochrome bd Promotes Sulfide-Resistant Bacterial Respiration and Growth. Sci. Rep. 2016, 6, 23788. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Falcone, M.; Molin, S.; Johansen, H.K. High-Resolution in Situ Transcriptomics of Pseudomonas aeruginosa Unveils Genotype Independent Patho-Phenotypes in Cystic Fibrosis Lungs. Nat. Commun. 2018, 9, 3459. [Google Scholar] [CrossRef]
- Cowley, E.S.; Kopf, S.H.; Lariviere, A.; Ziebis, W.; Newman, D.K. Pediatric Cystic Fibrosis Sputum Can Be Chemically Dynamic, Anoxic, and Extremely Reduced Due to Hydrogen Sulfide Formation. mBio 2015, 6, e00767-15. [Google Scholar] [CrossRef]
- Nagy, P.; Pálinkás, Z.; Nagy, A.; Budai, B.; Tóth, I.; Vasas, A. Chemical Aspects of Hydrogen Sulfide Measurements in Physiological Samples. Biochim. Biophys. Acta 2014, 1840, 876–891. [Google Scholar] [CrossRef] [PubMed]
- Nashef, A.S.; Osuga, D.T.; Feeney, R.E. Determination of Hydrogen Sulfide with 5,5′-Dithiobis-(2-Nitrobenzoic Acid), N-Ethylmaleimide, and Parachloromercuribenzoate. Anal. Biochem. 1977, 79, 394–405. [Google Scholar] [CrossRef]
- Stubauer, G.; Giuffrè, A.; Brunori, M.; Sarti, P. Cytochrome c Oxidase Does Not Catalyze the Anaerobic Reduction of NO. Biochem. Biophys. Res. Commun. 1998, 245, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Hijazi, S.; Visca, P.; Frangipani, E. Gallium-Protoporphyrin IX Inhibits Pseudomonas aeruginosa Growth by Targeting Cytochromes. Front. Cell Infect. Microbiol. 2017, 7, 12. [Google Scholar] [CrossRef]
- Frangipani, E.; Slaveykova, V.I.; Reimmann, C.; Haas, D. Adaptation of Aerobically Growing Pseudomonas aeruginosa to Copper Starvation. J. Bacteriol. 2008, 190, 6706–6717. [Google Scholar] [CrossRef]
- Sarti, P.; Giuffré, A.; Forte, E.; Mastronicola, D.; Barone, M.C.; Brunori, M. Nitric Oxide and Cytochrome c Oxidase: Mechanisms of Inhibition and NO Degradation. Biochem. Biophys. Res. Commun. 2000, 274, 183–187. [Google Scholar] [CrossRef]
- Letizia, M.; Mellini, M.; Fortuna, A.; Visca, P.; Imperi, F.; Leoni, L.; Rampioni, G. PqsE Expands and Differentially Modulates the RhlR Quorum Sensing Regulon in Pseudomonas aeruginosa. Microbiol. Spectr. 2022, 10, e0096122. [Google Scholar] [CrossRef]
- Nicholls, P.; Marshall, D.C.; Cooper, C.E.; Wilson, M.T. Sulfide Inhibition of and Metabolism by Cytochrome c Oxidase. Biochem. Soc. Trans. 2013, 41, 1312–1316. [Google Scholar] [CrossRef]
- Kawakami, T.; Kuroki, M.; Ishii, M.; Igarashi, Y.; Arai, H. Differential Expression of Multiple Terminal Oxidases for Aerobic Respiration in Pseudomonas aeruginosa. Env. Environ. Microbiol. 2010, 12, 1399–1412. [Google Scholar] [CrossRef]
- Borisov, V.B.; Forte, E.; Sarti, P.; Brunori, M.; Konstantinov, A.A.; Giuffrè, A. Redox Control of Fast Ligand Dissociation from Escherichia coli Cytochrome bd. Biochem. Biophys. Res. Commun. 2007, 355, 97–102. [Google Scholar] [CrossRef]
- Le Laz, S.; Kpebe, A.; Bauzan, M.; Lignon, S.; Rousset, M.; Brugna, M. Expression of Terminal Oxidases under Nutrient-Starved Conditions in Shewanella oneidensis: Detection of the A-Type Cytochrome c Oxidase. Sci. Rep. 2016, 6, 19726. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Garcia-Robledo, E.; Lund, M.B.; Lehner, P.; Borisov, S.M.; Klimant, I.; Revsbech, N.P.; Schramm, A. Gene Expression of Terminal Oxidases in Two Marine Bacterial Strains Exposed to Nanomolar Oxygen Concentrations. FEMS Microbiol. Ecol. 2018, 94, fiy072. [Google Scholar] [CrossRef]
- Trojan, D.; Garcia-Robledo, E.; Meier, D.V.; Hausmann, B.; Revsbech, N.P.; Eichorst, S.A.; Woebken, D. Microaerobic Lifestyle at Nanomolar O2 Concentrations Mediated by Low-Affinity Terminal Oxidases in Abundant Soil Bacteria. mSystems 2021, 6, e00250-21. [Google Scholar] [CrossRef]
- Lau, G.W.; Hassett, D.J.; Ran, H.; Kong, F. The Role of Pyocyanin in Pseudomonas aeruginosa Infection. Trends Mol. Med. 2004, 10, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, L.E.P.; Price-Whelan, A.; Petersen, A.; Whiteley, M.; Newman, D.K. The Phenazine Pyocyanin Is a Terminal Signalling Factor in the Quorum Sensing Network of Pseudomonas aeruginosa. Mol. Microbiol. 2006, 61, 1308–1321. [Google Scholar] [CrossRef]
- Shatalin, K.; Nuthanakanti, A.; Kaushik, A.; Shishov, D.; Peselis, A.; Shamovsky, I.; Pani, B.; Lechpammer, M.; Vasilyev, N.; Shatalina, E.; et al. Inhibitors of Bacterial H2S Biogenesis Targeting Antibiotic Resistance and Tolerance. Science 2021, 372, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Saini, V.; Chinta, K.C.; Reddy, V.P.; Glasgow, J.N.; Stein, A.; Lamprecht, D.A.; Rahman, M.A.; Mackenzie, J.S.; Truebody, B.E.; Adamson, J.H.; et al. Hydrogen Sulfide Stimulates Mycobacterium tuberculosis Respiration, Growth and Pathogenesis. Nat. Commun. 2020, 11, 557. [Google Scholar] [CrossRef]
- Kunota, T.T.R.; Rahman, M.A.; Truebody, B.E.; Mackenzie, J.S.; Saini, V.; Lamprecht, D.A.; Adamson, J.H.; Sevalkar, R.R.; Lancaster, J.R.; Berney, M.; et al. Mycobacterium tuberculosis H2S Functions as a Sink to Modulate Central Metabolism, Bioenergetics, and Drug Susceptibility. Antioxidants 2021, 10, 1285. [Google Scholar] [CrossRef]
- Rozansky, R.; Weber, D.; Gurevitch, J. Production of Hydrogen Sulfide by Pseudomonas aeruginosa. Am. J. Clin. Pathol. 1950, 20, 1090–1091. [Google Scholar] [CrossRef]
- Chen, Y.-W.; Camacho, M.I.; Chen, Y.; Bhat, A.H.; Chang, C.; Peluso, E.A.; Wu, C.; Das, A.; Ton-That, H. Genetic Determinants of Hydrogen Sulfide Biosynthesis in Fusobacterium nucleatum Are Required for Bacterial Fitness, Antibiotic Sensitivity, and Virulence. mBio 2022, 13, e01936-22. [Google Scholar] [CrossRef]
- Seregina, T.A.; Lobanov, K.V.; Shakulov, R.S.; Mironov, A.S. Enhancement of the Bactericidal Effect of Antibiotics by Inhibition of Enzymes Involved in Production of Hydrogen Sulfide in Bacteria. Mol. Biol. 2022, 56, 638–648. [Google Scholar] [CrossRef]
- Weikum, J.; Ritzmann, N.; Jelden, N.; Klöckner, A.; Herkersdorf, S.; Josten, M.; Sahl, H.-G.; Grein, F. Sulfide Protects Staphylococcus aureus from Aminoglycoside Antibiotics but Cannot Be Regarded as a General Defense Mechanism against Antibiotics. Antimicrob. Agents Chemother. 2018, 62, e00602-18. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.Y.; Ong, K.X.; Surendran, S.T.; Sinha, A.; Lai, J.J.H.; Chen, J.; Liang, J.; Tay, L.K.S.; Cui, L.; Loo, H.L.; et al. Hydrogen Sulfide Sensitizes Acinetobacter baumannii to Killing by Antibiotics. Front. Microbiol. 2020, 11, 563226. [Google Scholar] [CrossRef] [PubMed]
- Caruso, L.; Mellini, M.; Catalano Gonzaga, O.; Astegno, A.; Forte, E.; Di Matteo, A.; Giuffrè, A.; Visca, P.; Imperi, F.; Leoni, L.; et al. Hydrogen Sulfide Production Does Not Affect Antibiotic Resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2024, 6, e0007524. [Google Scholar] [CrossRef] [PubMed]
- Xuan, G.; Lü, C.; Xu, H.; Li, K.; Liu, H.; Xia, Y.; Xun, L. Sulfane Sulfur Regulates LasR-Mediated Quorum Sensing and Virulence in Pseudomonas aeruginosa PAO1. Antioxidants 2021, 10, 1498. [Google Scholar] [CrossRef] [PubMed]
- Beebout, C.J.; Eberly, A.R.; Werby, S.H.; Reasoner, S.A.; Brannon, J.R.; De, S.; Fitzgerald, M.J.; Huggins, M.M.; Clayton, D.B.; Cegelski, L.; et al. Respiratory Heterogeneity Shapes Biofilm Formation and Host Colonization in Uropathogenic Escherichia coli. mBio 2019, 10, e02400-18. [Google Scholar] [CrossRef] [PubMed]
- Jones-Carson, J.; Husain, M.; Liu, L.; Orlicky, D.J.; Vázquez-Torres, A. Cytochrome bd -Dependent Bioenergetics and Antinitrosative Defenses in Salmonella Pathogenesis. mBio 2016, 7, e02052-16. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.R.; Dunman, P.M.; Fang, F.C. The Nitrosative Stress Response of Staphylococcus aureus Is Required for Resistance to Innate Immunity. Mol. Microbiol. 2006, 61, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Sohaskey, C.D.; Kana, B.D.; Dawes, S.; North, R.J.; Mizrahi, V.; Gennaro, M.L. Changes in Energy Metabolism of Mycobacterium tuberculosis in Mouse Lung and under in vitro Conditions Affecting Aerobic Respiration. Proc. Natl. Acad. Sci. USA 2005, 102, 15629–15634. [Google Scholar] [CrossRef]
- Moore, C.M.; Nakano, M.M.; Wang, T.; Ye, R.W.; Helmann, J.D. Response of Bacillus subtilis to Nitric Oxide and the Nitrosating Agent Sodium Nitroprusside. J. Bacteriol. 2004, 186, 4655–4664. [Google Scholar] [CrossRef] [PubMed]
- Arjona, D.; Wikström, M.; Ädelroth, P. Nitric Oxide Is a Potent Inhibitor of the cbb3-Type Heme-Copper Oxidases. FEBS Lett. 2015, 589, 1214–1218. [Google Scholar] [CrossRef]
- Brown, G.C.; Cooper, C.E. Nanomolar Concentrations of Nitric Oxide Reversibly Inhibit Synaptosomal Respiration by Competing with Oxygen at Cytochrome Oxidase. FEBS Lett. 1994, 356, 295–298. [Google Scholar] [CrossRef] [PubMed]
- Giuffrè, A.; Borisov, V.B.; Mastronicola, D.; Sarti, P.; Forte, E. Cytochrome bd Oxidase and Nitric Oxide: From Reaction Mechanisms to Bacterial Physiology. FEBS Lett. 2012, 586, 622–629. [Google Scholar] [CrossRef]
- Fang, F.C. Perspectives Series: Host/Pathogen Interactions. Mechanisms of Nitric Oxide-Related Antimicrobial Activity. J. Clin. Investig. 1997, 99, 2818–2825. [Google Scholar] [CrossRef] [PubMed]
- Cutruzzolà, F.; Frankenberg-Dinkel, N. Origin and Impact of Nitric Oxide in Pseudomonas aeruginosa Biofilms. J. Bacteriol. 2016, 198, 55–65. [Google Scholar] [CrossRef]
- Arai, H.; Hayashi, M.; Kuroi, A.; Ishii, M.; Igarashi, Y. Transcriptional Regulation of the Flavohemoglobin Gene for Aerobic Nitric Oxide Detoxification by the Second Nitric Oxide-Responsive Regulator of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 3960–3968. [Google Scholar] [CrossRef]
- Kakishima, K.; Shiratsuchi, A.; Taoka, A.; Nakanishi, Y.; Fukumori, Y. Participation of Nitric Oxide Reductase in Survival of Pseudomonas aeruginosa in LPS-Activated Macrophages. Biochem. Biophys. Res. Commun. 2007, 355, 587–591. [Google Scholar] [CrossRef]
- Arai, H.; Kodama, T.; Igarashi, Y. Effect of Nitrogen Oxides on Expression of the nir and nor Genes for Denitrification in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 1999, 170, 19–24. [Google Scholar] [CrossRef]
- Friedrich, T.; Wohlwend, D.; Borisov, V.B. Recent Advances in Structural Studies of Cytochrome bd and Its Potential Application as a Drug Target. Int. J. Mol. Sci. 2022, 23, 3166. [Google Scholar] [CrossRef]
- Harikishore, A.; Mathiyazakan, V.; Pethe, K.; Gruber, G. Novel targets and inhibitors of the Mycobacterium tuberculosis cytochrome bd oxidase to foster anti-tuberculosis drug discovery. Expert Opin. Drug Discov. 2023, 18, 917–927. [Google Scholar] [CrossRef]
- Henry, S.A.; Webster, C.M.; Shaw, L.N.; Torres, N.J.; Jobson, M.E.; Totzke, B.C.; Jackson, J.K.; McGreig, J.E.; Wass, M.N.; Robinson, G.K.; et al. Steroid drugs inhibit bacterial respiratory oxidases and are lethal toward methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 2024, jiad540. [Google Scholar] [CrossRef] [PubMed]
- Saha, P.; Das, S.; Indurthi, H.K.; Kumar, R.; Roy, A.; Kalia, N.P.; Sharma, D.K. Cytochrome bd oxidase: An emerging anti-tubercular drug target. RSC Med. Chem. 2024, 15, 769–787. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).