Comparison of Fungal Thermophilic and Mesophilic Catalase–Peroxidases for Their Antioxidative Properties
Abstract
1. Introduction
2. Materials and Methods
2.1. Cloning of Synthetic Genes for Thermostable and Mesophilic Catalase–Peroxidases
2.2. Site-Directed Mutagenesis
2.3. Heterologous Expression, Protein Purification, and Quantification
2.4. Nano-Differential Scanning Fluorimetry
2.5. Electronic Circular Dichroism Spectroscopy
2.6. Mass Spectrometry Analysis
2.7. Enzyme Kinetics
2.8. Protein Structure Modeling and Ligand Transport Analysis
3. Results
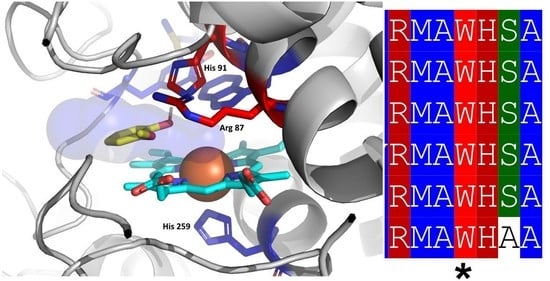
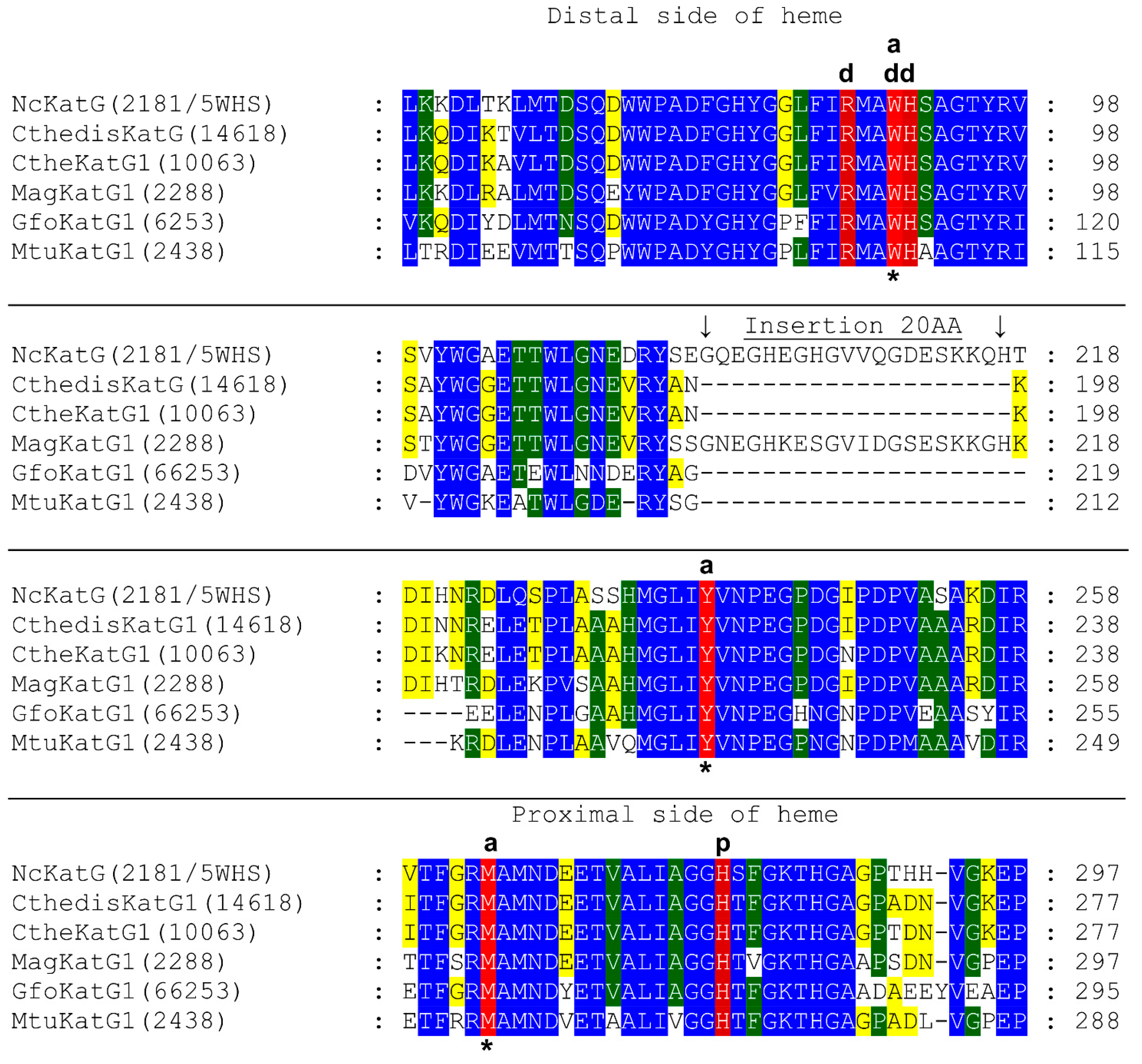
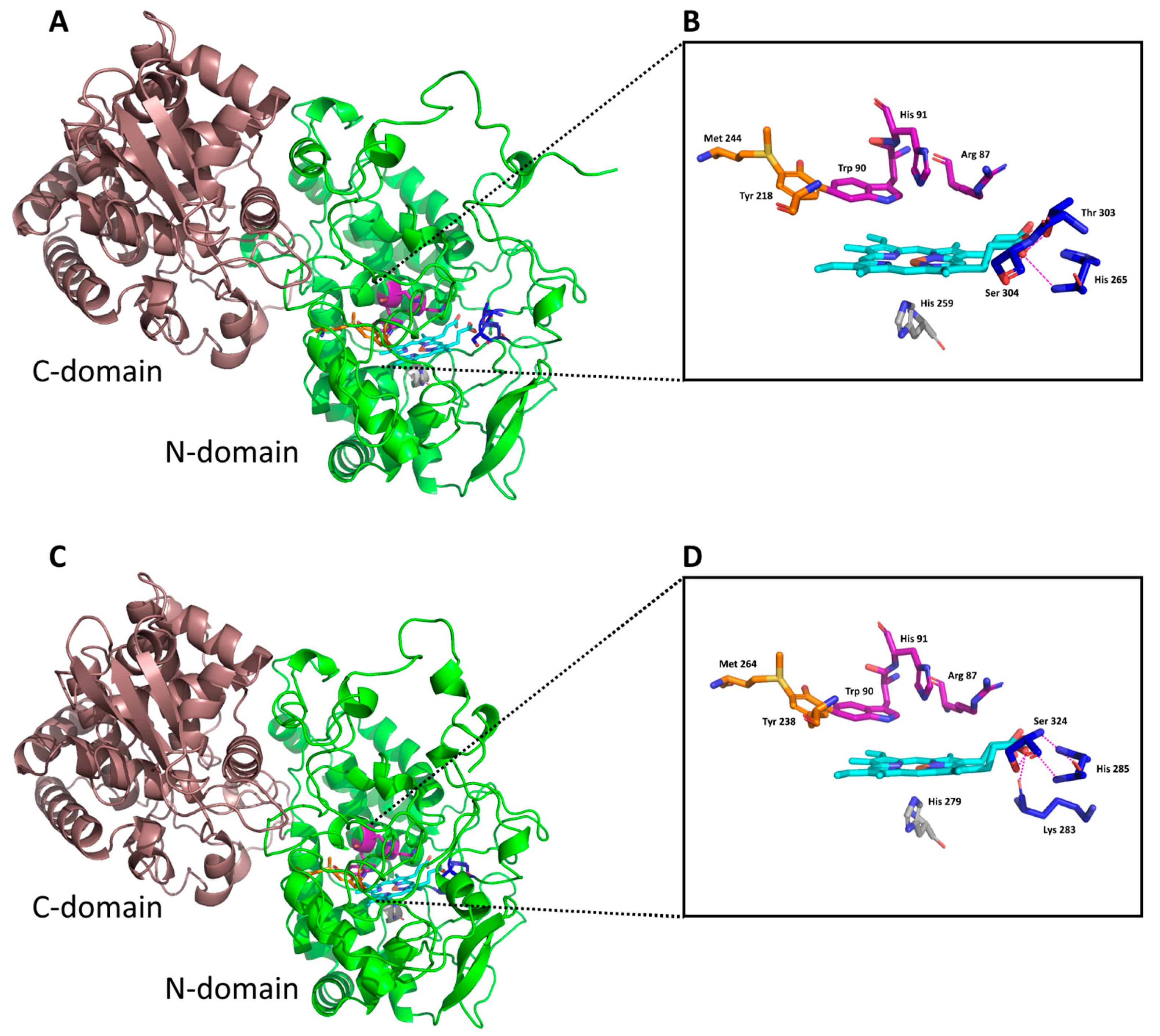
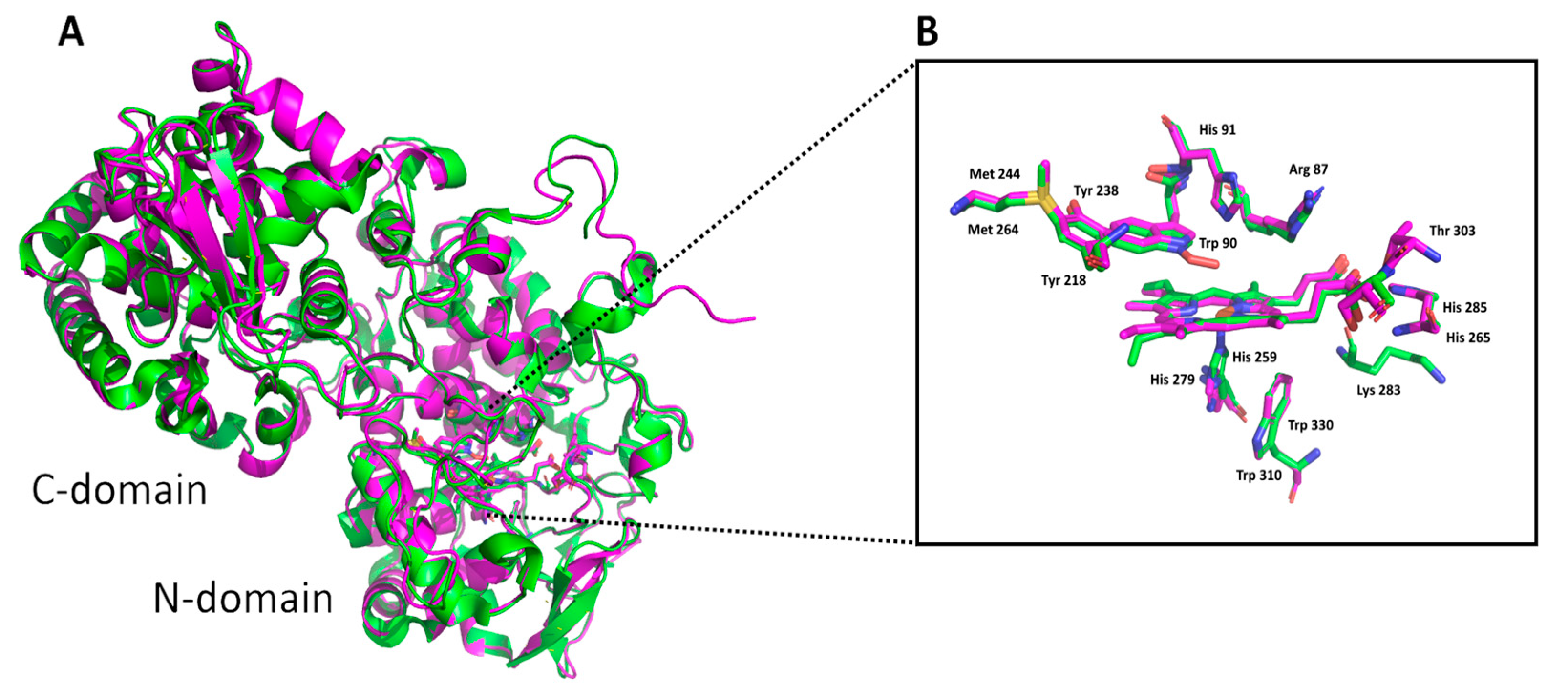
3.1. Insight in the Peculiar Structure of Bifunctional Catalase–Peroxidases
3.2. Investigation of Typical Covalent Adduct in Active Center of Fungal KatGs
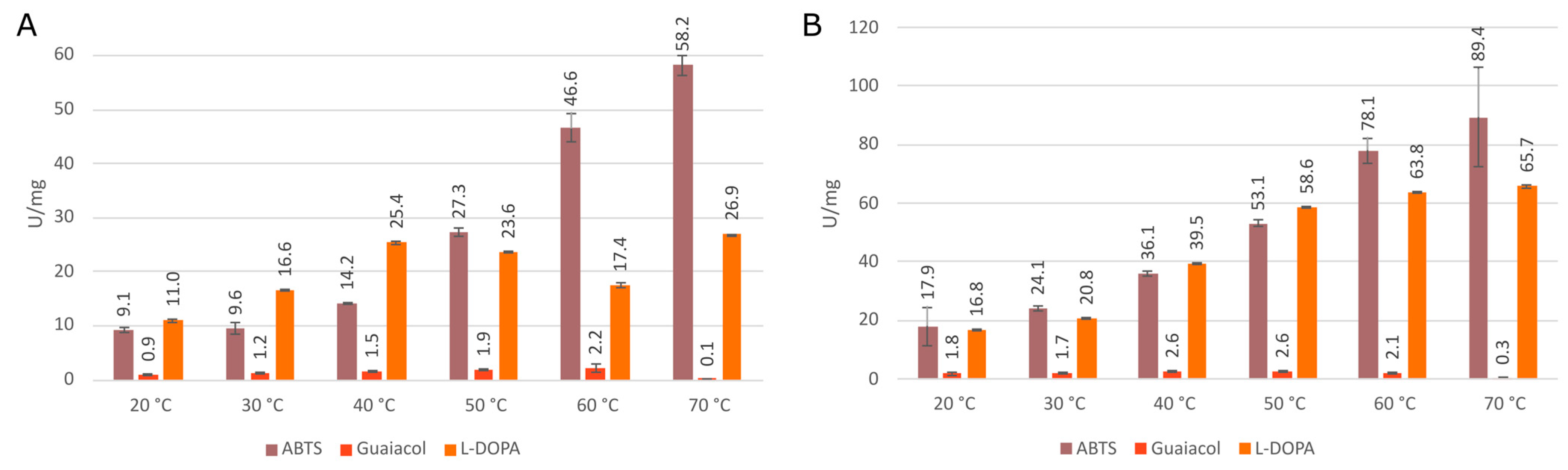
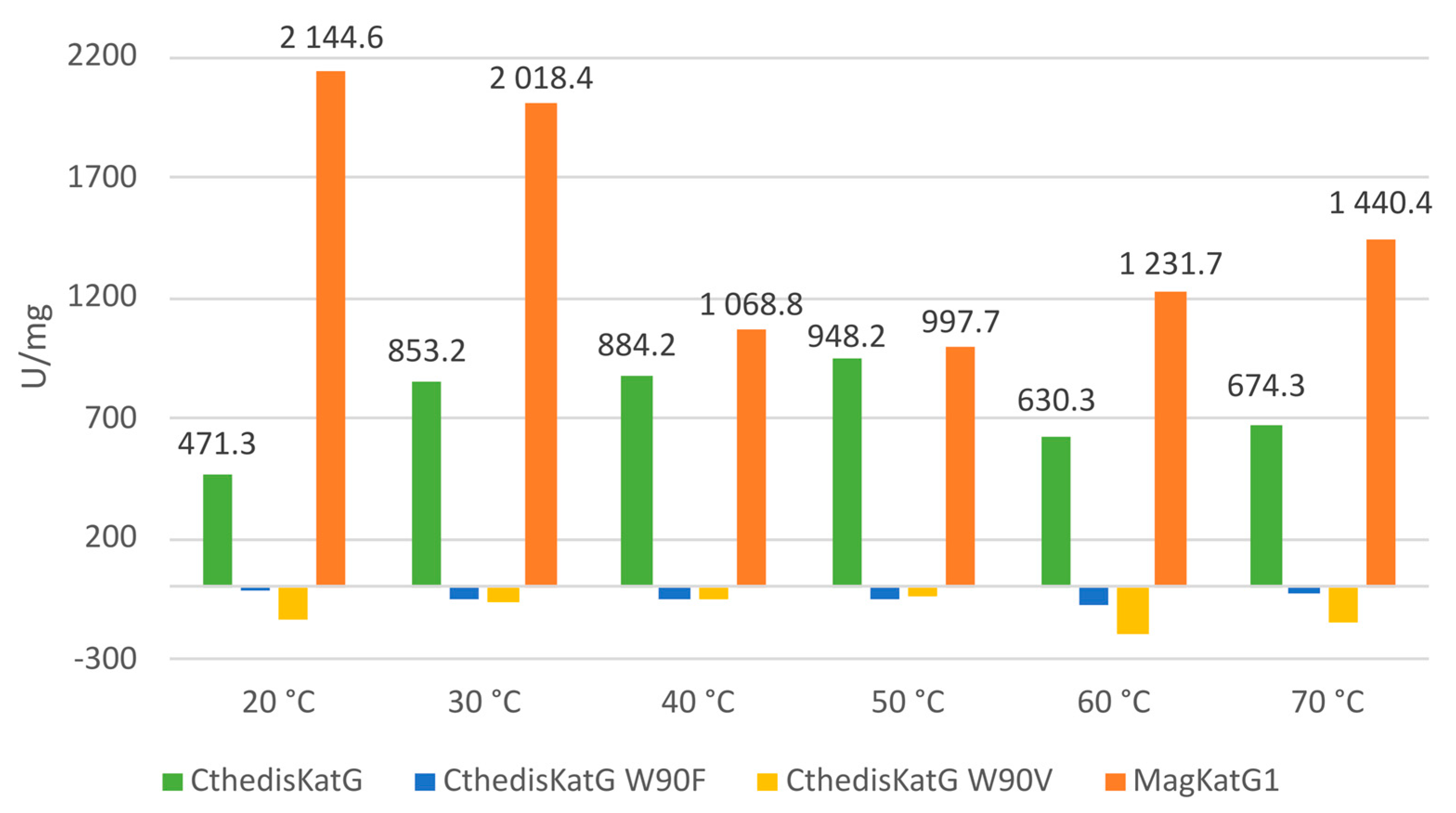
3.3. Comparison of Enzymatic Activities of Wild Type KatGs and Point Mutants
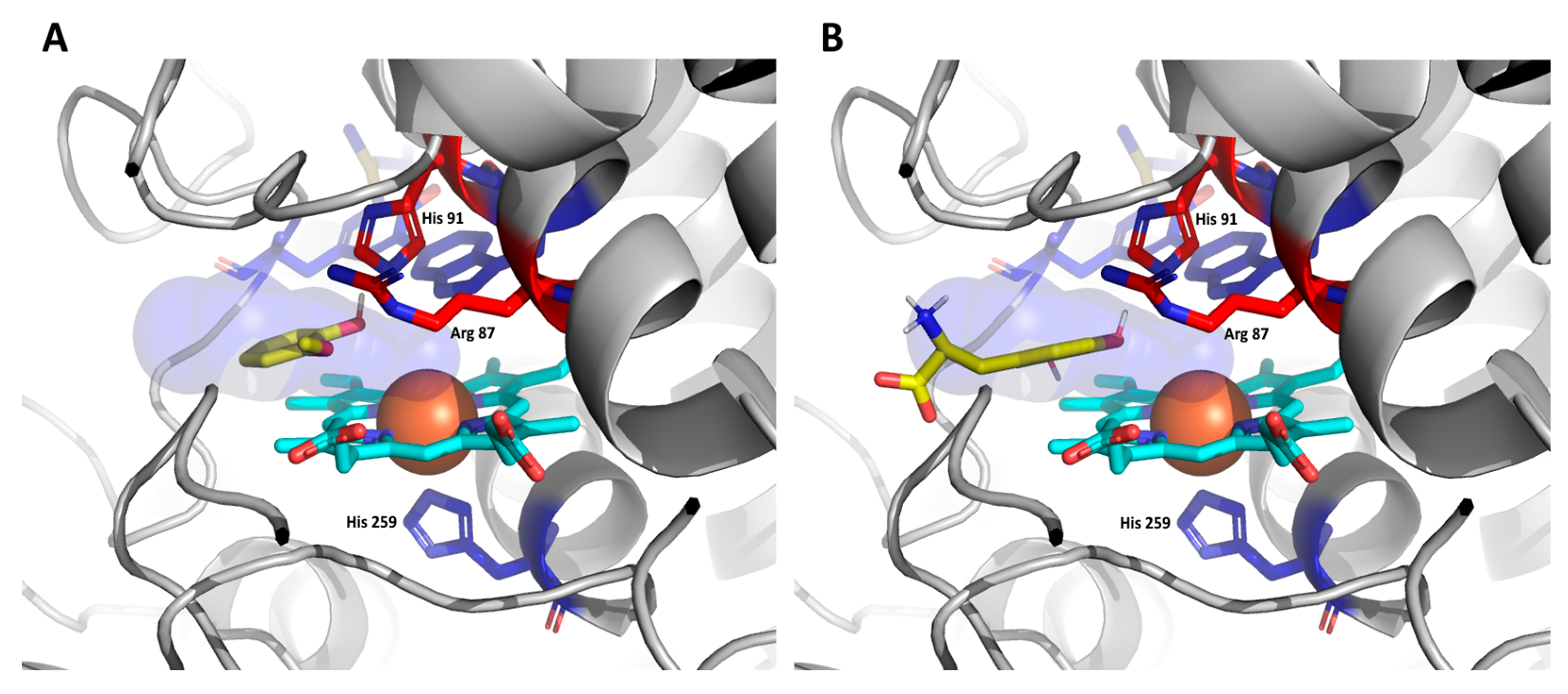
3.4. Docking of Substrates in the Active Center of Bifunctional Catalase–Peroxidase
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baker, A.; Lin, C.-C.; Lett, C.; Karpinska, B.; Wright, M.H.; Foyer, C.H. Catalase: A critical node in the regulation of cell fate. Free Radic. Biol. Med. 2023, 199, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Veitch, N.C. Horseradish peroxidase: A modern view of a classic enzyme. Phytochemistry 2004, 65, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Ivancich, A.; Donald, L.J.; Villanueva, J.; Wiseman, B.; Fita, I.; Loewen, P.C. Spectroscopic and Kinetic Investigation of the Reactions of Peroxyacetic Acid with Burkholderia pseudomallei Catalase-Peroxidase, KatG. Biochemistry 2013, 52, 7271–7282. [Google Scholar] [CrossRef]
- Savelli, B.; Li, Q.; Webber, M.; Jemmat, A.M.; Robitaille, A.; Zamocky, M.; Mathé, C.; Dunand, C. RedoxiBase: A database for ROS homeostasis regulated proteins. Redox Biol. 2019, 26, 101247. [Google Scholar] [CrossRef]
- Zamocky, M.; Furtmüller, P.G.; Bellei, M.; Battistuzzi, G.; Stadlmann, J.; Vlasits, J.; Obinger, C. Intracellular catalase/peroxidase from the phytopathogenic rice blast fungus Magnaporthe grisea: Expression analysis and biochemical characterization of the recombinant protein. Biochem. J. 2009, 418, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Chelikani, P.; Fita, I.; Loewen, P.C. Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 2004, 61, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Bakalovic, N.; Teixeira, F.K.; Margis-Pinheiro, M.; Penel, C.; Dunand, C. Prokaryotic origins of the non-animal peroxidase superfamily and organelle-mediated transmission to eukaryotes. Genomics 2007, 89, 567–579. [Google Scholar] [CrossRef]
- Passardi, F.; Zamocky, M.; Favet, J.; Jakopitsch, C.; Penel, C.; Obinger, C.; Dunand, C. Phylogenetic distribution of catalase-peroxidases: Are there patches of order in chaos? Gene 2007, 397, 101–113. [Google Scholar] [CrossRef]
- Zámocký, M.; Furtmüller, P.G.; Obinger, C. Two distinct groups of fungal catalase/peroxidases. Biochem. Soc. Trans. 2009, 37, 772–777. [Google Scholar] [CrossRef]
- Jakopitsch, C.; Kolarich, D.; Petutschnig, G.; Furtmüller, P.G.; Obinger, C. Distal side tryptophan, tyrosine and methionine in catalase-peroxidases are covalently linked in solution. FEBS Lett. 2003, 552, 135–140. [Google Scholar] [CrossRef][Green Version]
- Zámocký, M.; García-Fernández, Q.; Gasselhuber, B.; Jakopitsch, C.; Furtmüller, P.G.; Loewen, P.C.; Fita, I.; Obinger, C.; Carpena, X. High Conformational Stability of Secreted Eukaryotic Catalase-peroxidases. J. Biol. Chem. 2012, 287, 32254–32262. [Google Scholar] [CrossRef] [PubMed]
- Wingfield, P.T. Protein Precipitation Using Ammonium Sulfate. Curr. Protoc. Protein Sci. 1998, 13, A.3F.1–A.3F.8. [Google Scholar] [CrossRef]
- Magnusson, A.O.; Szekrenyi, A.; Joosten, H.; Finnigan, J.; Charnock, S.; Fessner, W. nanoDSF as screening tool for enzyme libraries and biotechnology development. FEBS J. 2019, 286, 184–204. [Google Scholar] [CrossRef] [PubMed]
- Michalski, A.; Damoc, E.; Lange, O.; Denisov, E.; Nolting, D.; Müller, M.; Viner, R.; Schwartz, J.; Remes, P.; Belford, M.; et al. Ultra High Resolution Linear Ion Trap Orbitrap Mass Spectrometer (Orbitrap Elite) Facilitates Top Down LC MS/MS and Versatile Peptide Fragmentation Modes. Mol. Cell. Proteom. 2012, 11, O111.013698. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Barr, D.; Aust, S. On the Mechanism of Peroxidase-Catalyzed Oxygen Production. Arch. Biochem. Biophys. 1993, 303, 377–382. [Google Scholar] [CrossRef]
- Mutsuda, M.; Ishikawa, T.; Takeda, T.; Shigeoka, S. The catalase-peroxidase of Synechococcus PCC 7942: Purification, nucleotide sequence analysis and expression in Escherichia coli. Biochem. J. 1996, 316, 251–257. [Google Scholar] [CrossRef]
- Rodríguez-López, J.; Bañón-Arnao, M.; Martinez-Ortiz, F.; Tudela, J.; Acosta, M.; Varón, R.; García-Cánovas, F. Catalytic oxidation of 2,4,5-trihydroxyphenylalanine by tyrosinase: Identification and evolution of intermediates. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzym. 1992, 1160, 221–228. [Google Scholar] [CrossRef]
- Martinez, M.J.; Ruiz-Duenas, F.J.; Guillen, F.; Martinez, A.T. Purification and Catalytic Properties of Two Manganese Peroxidase Isoenzymes from Pleurotus eryngii. JBIC J. Biol. Inorg. Chem. 1996, 237, 424–432. [Google Scholar] [CrossRef]
- Hildebrandt, A.G.; Roots, I. Reduced nicotinamide adenine dinucleotide phosphate (NADPH)-dependent formation and breakdown of hydrogen peroxide during mixed function oxidation reactions in liver microsomes. Arch. Biochem. Biophys. 1975, 171, 385–397. [Google Scholar] [CrossRef]
- Hekkelman, M.L.; de Vries, I.; Joosten, R.P.; Perrakis, A. AlphaFill: Enriching AlphaFold models with ligands and cofactors. Nat. Methods 2023, 20, 205–213. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, D.; Nowotny, J.; Cao, R.; Cheng, J. 3Drefine: An interactive web server for efficient protein structure refinement. Nucleic Acids Res. 2016, 44, W406–W409. [Google Scholar] [CrossRef] [PubMed]
- Stourac, J.; Vavra, O.; Kokkonen, P.; Filipovic, J.; Pinto, G.; Brezovsky, J.; Damborsky, J.; Bednar, D. Caver Web 1.0: Identification of tunnels and channels in proteins and analysis of ligand transport. Nucleic Acids Res. 2019, 47, W414–W422. [Google Scholar] [CrossRef]
- Chovancova, E.; Pavelka, A.; Benes, P.; Strnad, O.; Brezovsky, J.; Kozlikova, B.; Gora, A.; Sustr, V.; Klvana, M.; Medek, P.; et al. CAVER 3.0: A Tool for the Analysis of Transport Pathways in Dynamic Protein Structures. PLOS Comput. Biol. 2012, 8, e1002708. [Google Scholar] [CrossRef]
- Vavra, O.; Filipovic, J.; Plhak, J.; Bednar, D.; Marques, S.M.; Brezovsky, J.; Stourac, J.; Matyska, L.; Damborsky, J. CaverDock: A molecular docking-based tool to analyse ligand transport through protein tunnels and channels. Bioinformatics 2019, 35, 4986–4993. [Google Scholar] [CrossRef]
- Yuan, F.; Yin, S.; Xu, Y.; Xiang, L.; Wang, H.; Li, Z.; Fan, K.; Pan, G. The Richness and Diversity of Catalases in Bacteria. Front. Microbiol. 2021, 12, 645477. [Google Scholar] [CrossRef]
- Ślesak, I.; Kula, M.; Ślesak, H.; Miszalski, Z.; Strzałka, K. How to define obligatory anaerobiosis? An evolutionary view on the antioxidant response system and the early stages of the evolution of life on Earth. Free. Radic. Biol. Med. 2019, 140, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Salar, R. Thermophilic Fungi: Basic Concepts and Biotechnological Applications; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Gold, M.H.; Youngs, H.L.; Gelpke, M.D. Manganese peroxidase. Met. Ions Biol. Syst. 2000, 37, 559–586. [Google Scholar]
- Colin, J.; Wiseman, B.; Switala, J.; Loewen, P.C.; Ivancich, A. Distinct Role of Specific Tryptophans in Facilitating Electron Transfer or as [Fe(IV)=O Trp•] Intermediates in the Peroxidase Reaction of Bulkholderia pseudomallei Catalase-Peroxidase: A Multifrequency EPR Spectroscopy Investigation. J. Am. Chem. Soc. 2009, 131, 8557–8563. [Google Scholar] [CrossRef] [PubMed]
- Casella, L.; Monzani, E.; Nicolis, S. Potential Applications of Peroxidases in the Fine Chemical Industries BT–Biocatalysis Based on Heme Peroxidases: Peroxidases as Potential Industrial Biocatalysts; Torres, E., Ayala, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 111–153. [Google Scholar]
- Farkas, Z.; Puškárová, A.; Šišková, A.O.; Poljovka, A.; Zámocký, M.; Vadkerti, E.; Urík, M.; Farkas, B.; Bučková, M.; Kraková, L.; et al. Evaluation of enzymatic stamp removal strategies on handmade (cellulose-based) and machine-made (lignin-containing) papers. Int. J. Biol. Macromol. 2023, 242, 124599. [Google Scholar] [CrossRef] [PubMed]
- Zámocký, M.; Kamlárová, A.; Maresch, D.; Chovanová, K.; Harichová, J.; Furtmüller, P.G. Hybrid Heme Peroxidases from Rice Blast Fungus Magnaporthe oryzae Involved in Defence against Oxidative Stress. Antioxidants 2020, 9, 655. [Google Scholar] [CrossRef] [PubMed]
- Zámocký, M.; Musil, M.; Danchenko, M.; Ferianc, P.; Chovanová, K.; Baráth, P.; Poljovka, A.; Bednář, D. Deep Insights into the Specific Evolution of Fungal Hybrid B Heme Peroxidases. Biology 2022, 11, 459. [Google Scholar] [CrossRef] [PubMed]
- Zámocký, M.; Harichová, J. Evolution of Heme Peroxygenases: Ancient Roots and Later Evolved Branches. Antioxidants 2022, 11, 1011. [Google Scholar] [CrossRef]

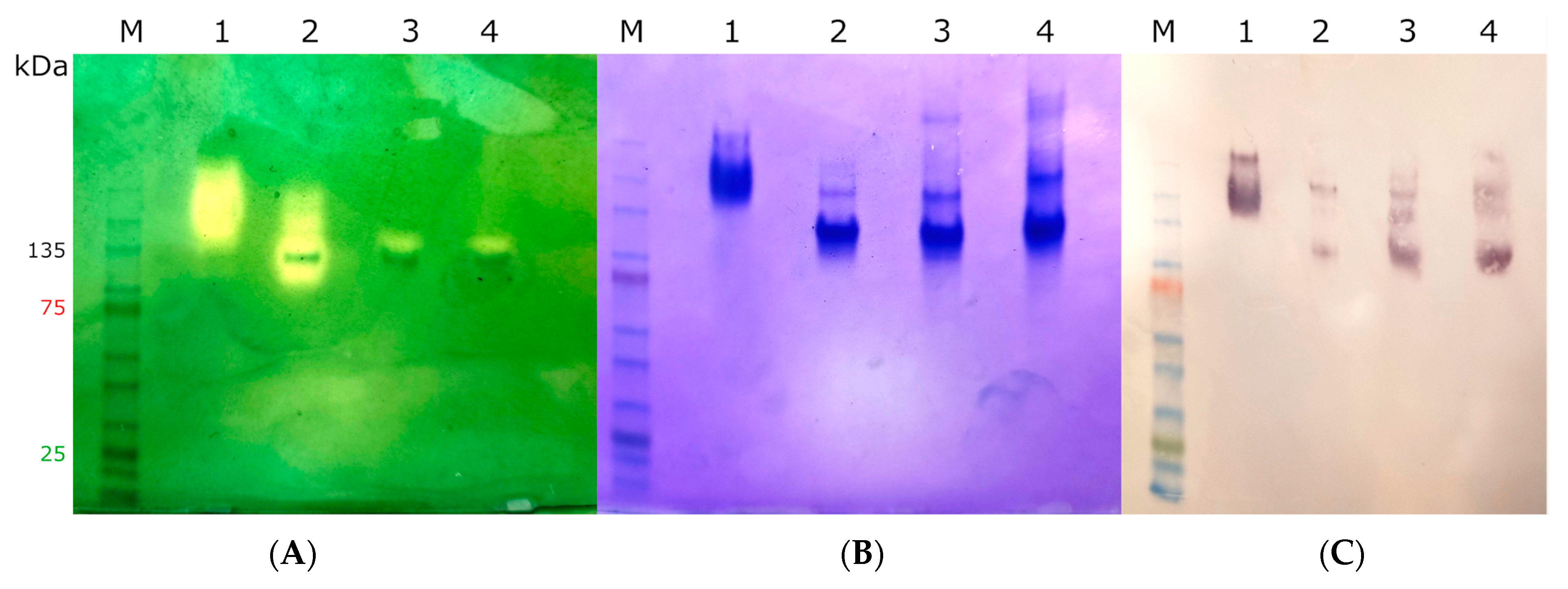
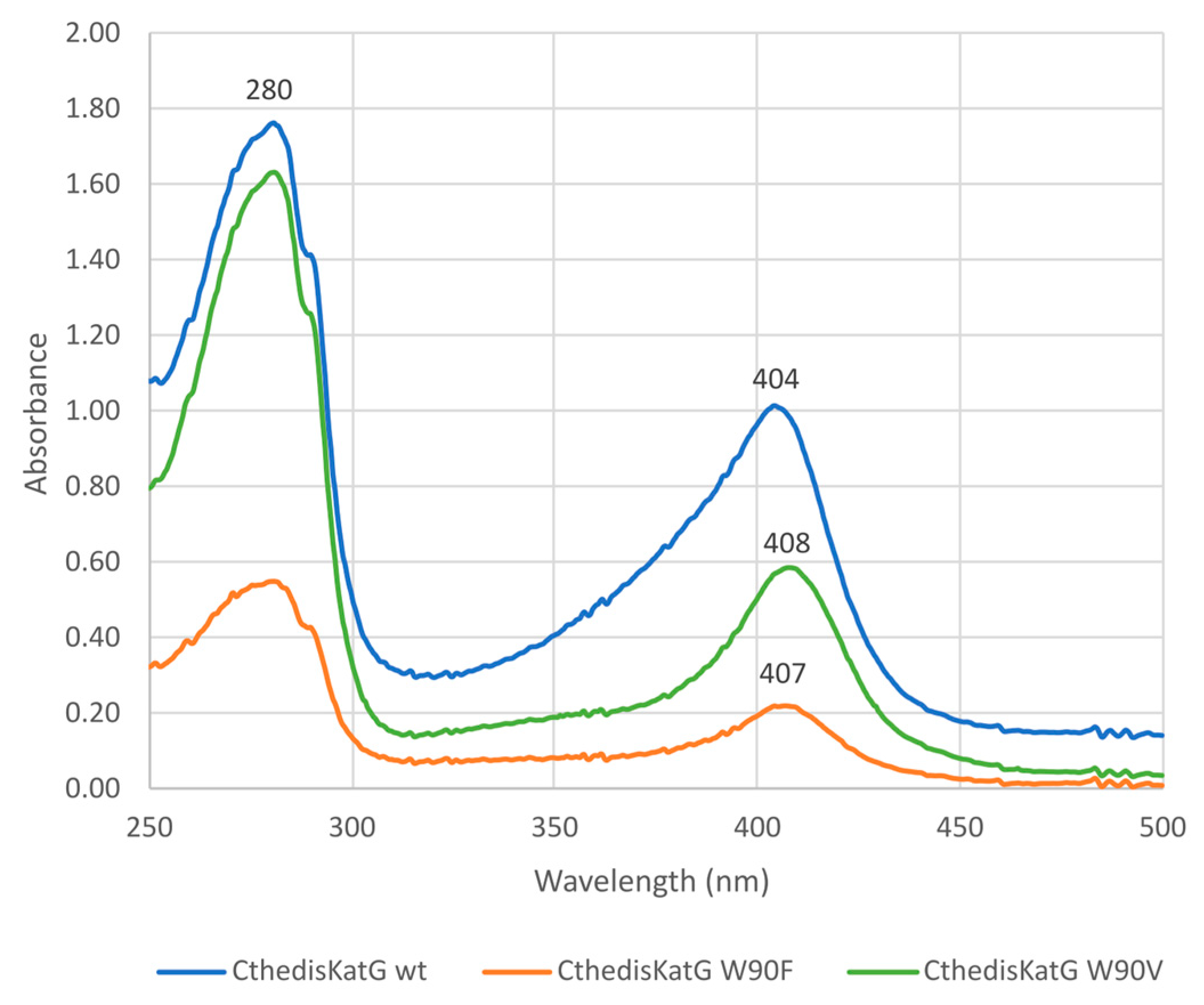
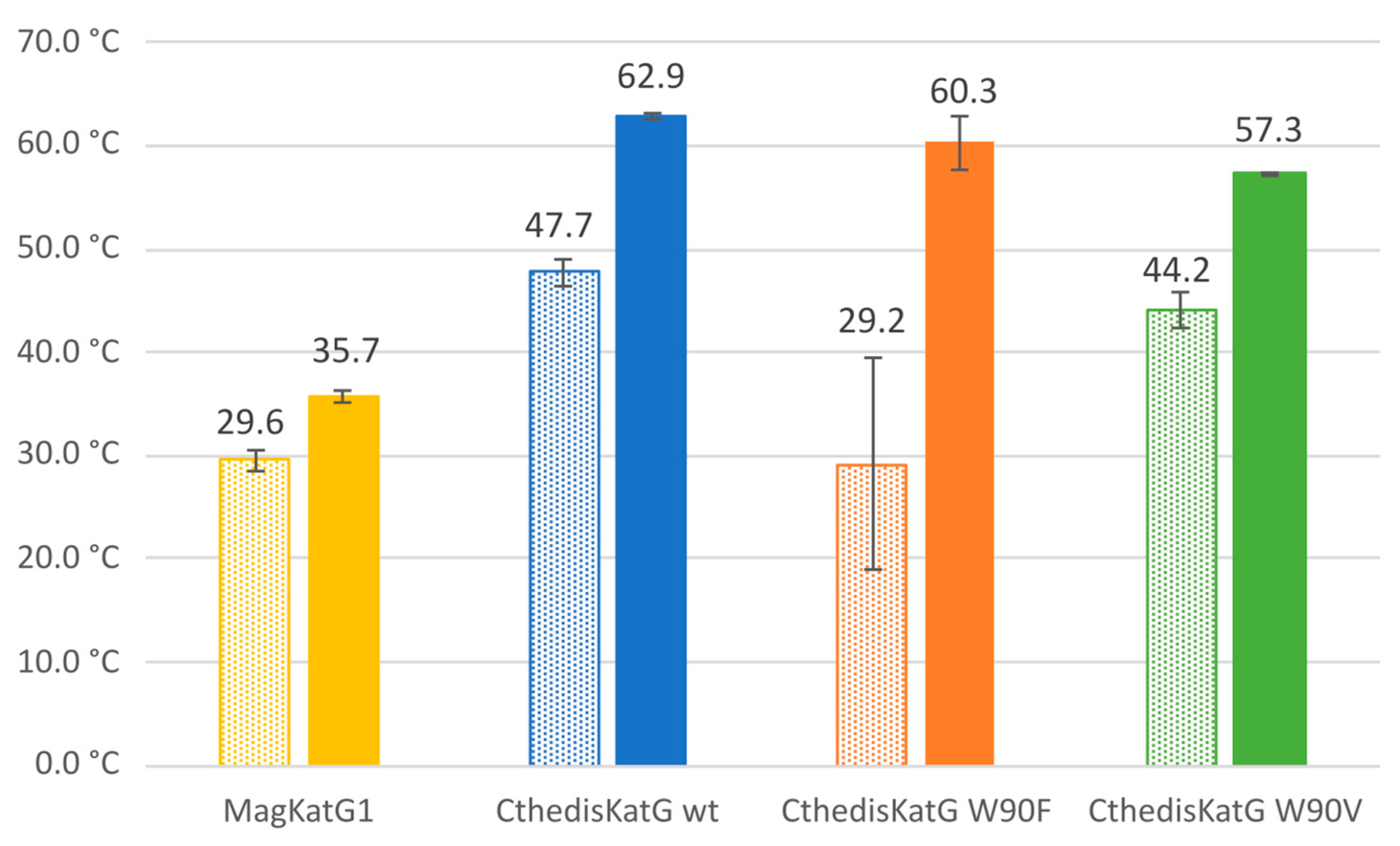

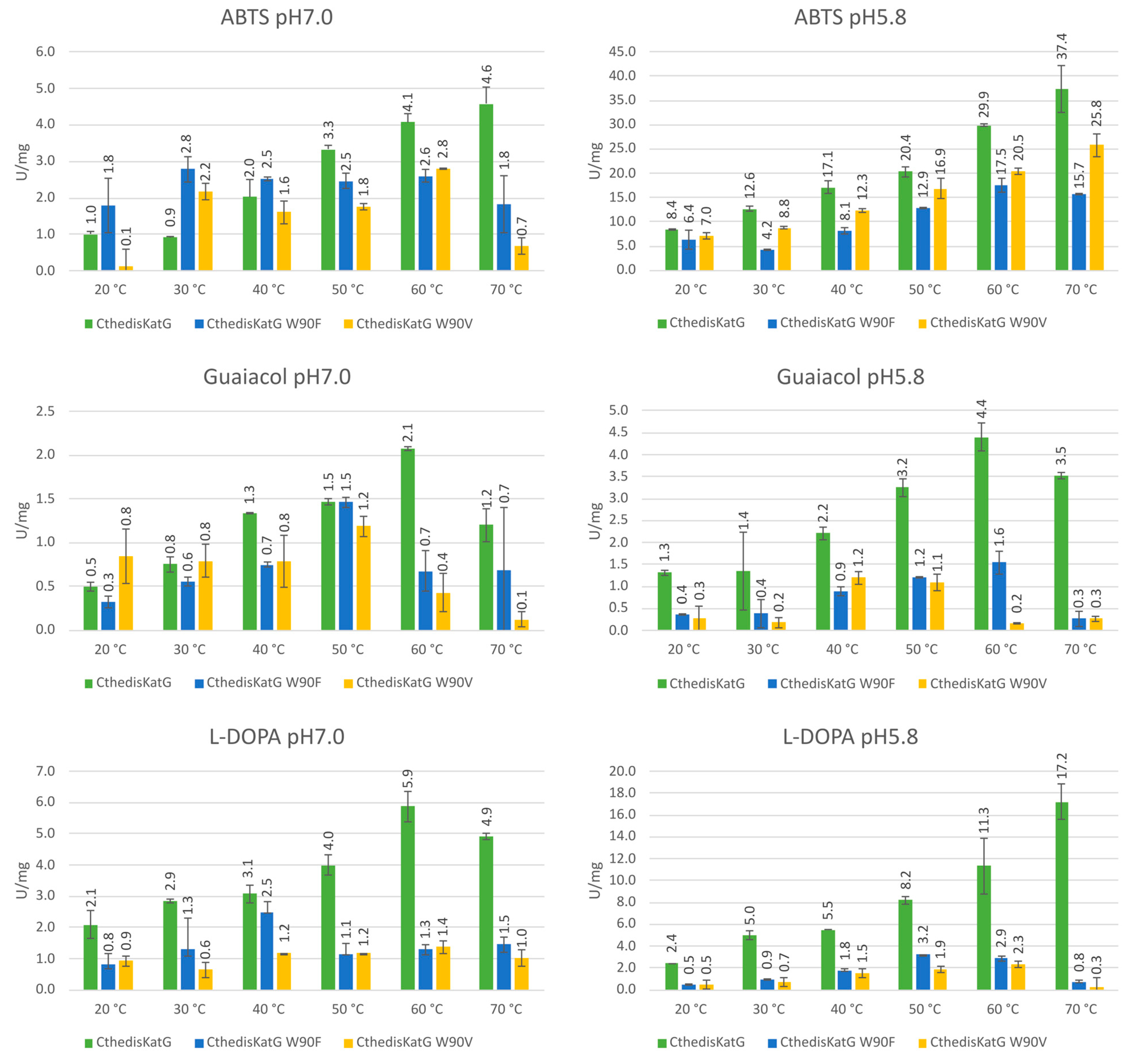


| Protein | MagKatG1 | CthedisKatG |
|---|---|---|
| α-helix | 29.6% | 32.5% |
| β-strand | 5.1% | 4.6% |
| Mw subunit | 83.476 kDa | 80.819 kDa |
| ε280nm | 165.81 | 149.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Poljovka, A.; Musil, M.; Bednář, D.; Chovanová, K.; Bauerová-Hlinková, V.; Bellová, J.; Kohútová, L.; Baráth, P.; Zámocký, M. Comparison of Fungal Thermophilic and Mesophilic Catalase–Peroxidases for Their Antioxidative Properties. Antioxidants 2023, 12, 1382. https://doi.org/10.3390/antiox12071382
Poljovka A, Musil M, Bednář D, Chovanová K, Bauerová-Hlinková V, Bellová J, Kohútová L, Baráth P, Zámocký M. Comparison of Fungal Thermophilic and Mesophilic Catalase–Peroxidases for Their Antioxidative Properties. Antioxidants. 2023; 12(7):1382. https://doi.org/10.3390/antiox12071382
Chicago/Turabian StylePoljovka, Andrej, Miloš Musil, David Bednář, Katarína Chovanová, Vladena Bauerová-Hlinková, Jana Bellová, Lenka Kohútová, Peter Baráth, and Marcel Zámocký. 2023. "Comparison of Fungal Thermophilic and Mesophilic Catalase–Peroxidases for Their Antioxidative Properties" Antioxidants 12, no. 7: 1382. https://doi.org/10.3390/antiox12071382
APA StylePoljovka, A., Musil, M., Bednář, D., Chovanová, K., Bauerová-Hlinková, V., Bellová, J., Kohútová, L., Baráth, P., & Zámocký, M. (2023). Comparison of Fungal Thermophilic and Mesophilic Catalase–Peroxidases for Their Antioxidative Properties. Antioxidants, 12(7), 1382. https://doi.org/10.3390/antiox12071382