Effect of Copper Sulphate Exposure on the Oxidative Stress, Gill Transcriptome and External Microbiota of Yellow Catfish, Pelteobagrus fulvidraco
Abstract
1. Introduction
2. Materials and Methods
2.1. Fish and Reagents
2.2. Experimental Design
2.3. Determination of Antioxidant Enzyme Activities
2.4. Transcriptomic Analysis
2.4.1. RNA Extraction and Sequencing
2.4.2. Bioinformatic Analyses
2.4.3. QRT-PCR Verification
2.5. Microbiota Analysis
2.5.1. DNA Extraction and Sequencing
2.5.2. Biodiversity Analysis
2.6. Statistical Analyses
3. Results
3.1. Behavior Observation and Antioxidant Enzyme Activity Analysis
3.2. Transcriptomic Analyses
3.2.1. Transcriptome Statistics
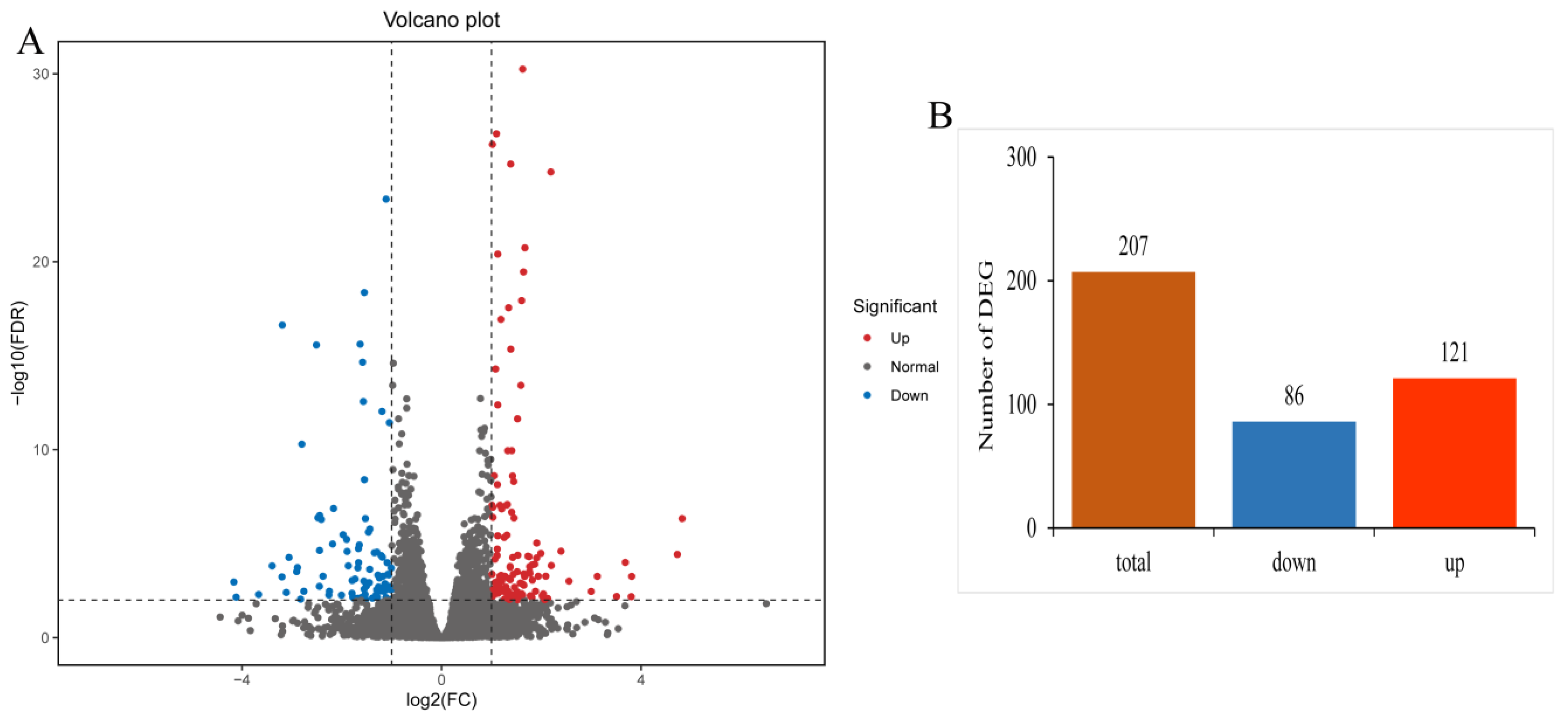
3.2.2. Identification of Differentially Expressed Genes
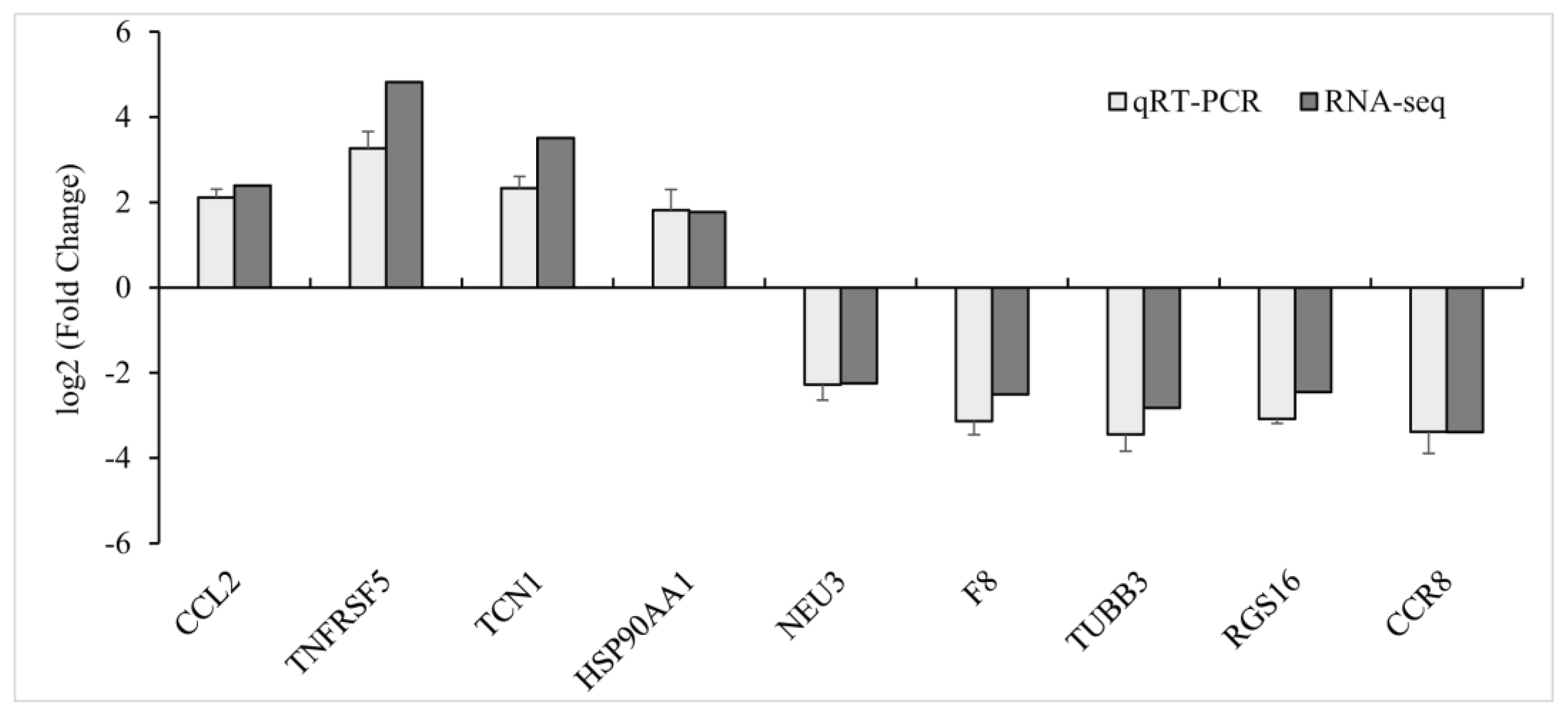
3.2.3. QRT-PCR Verification
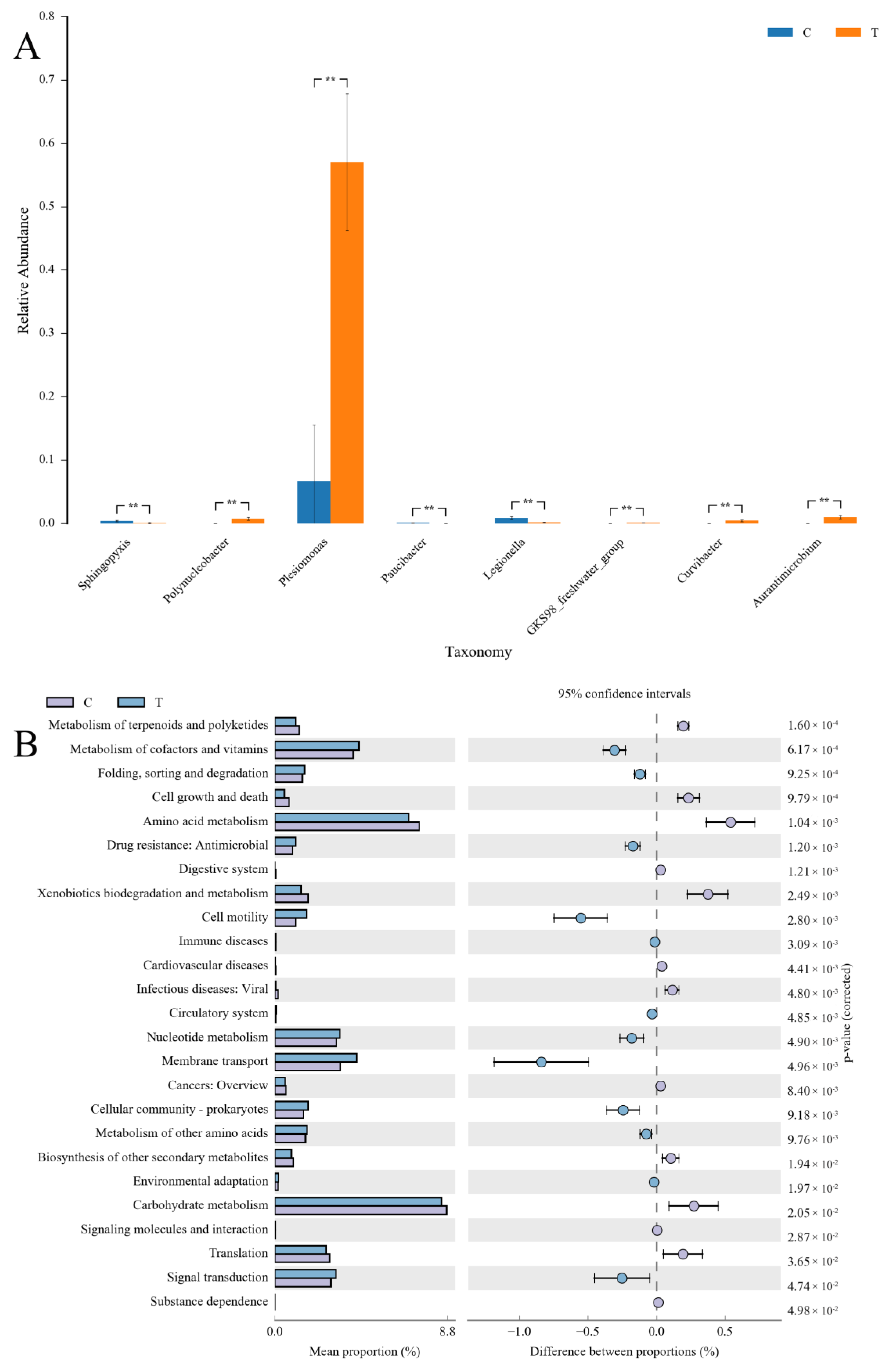
3.3. Microbiota Analysis
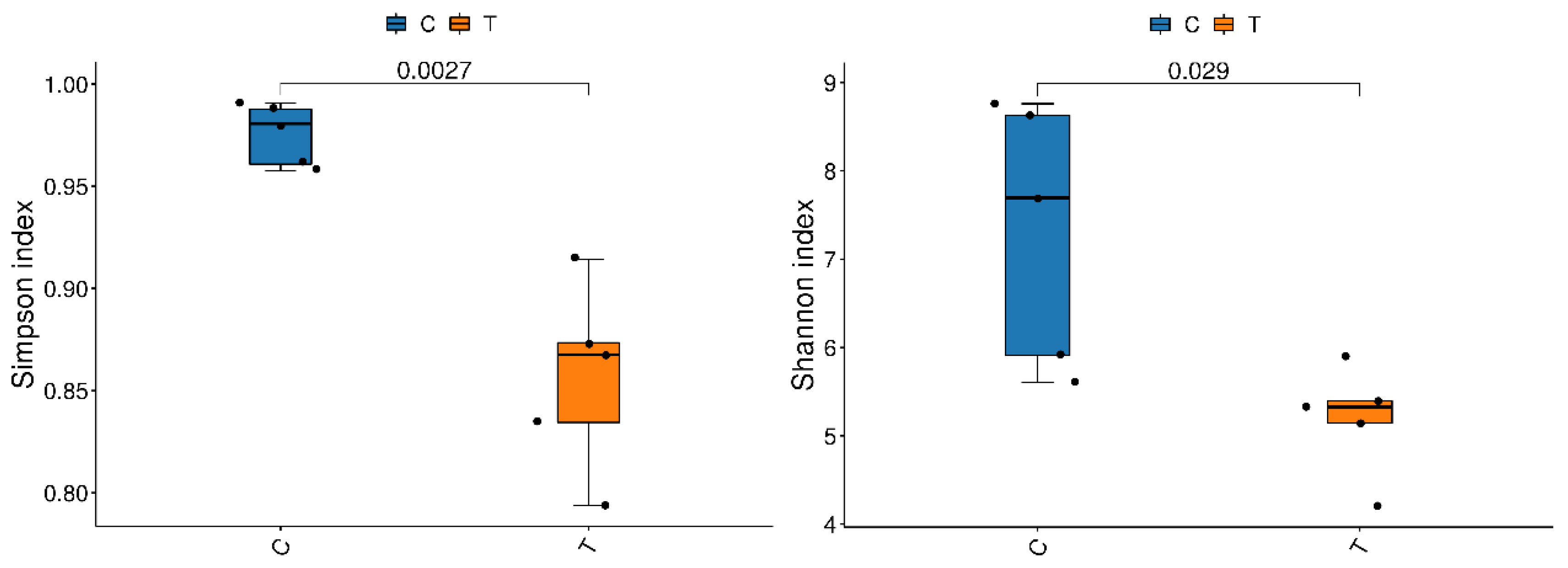
3.3.1. Sequencing Analysis and Taxonomic Annotation
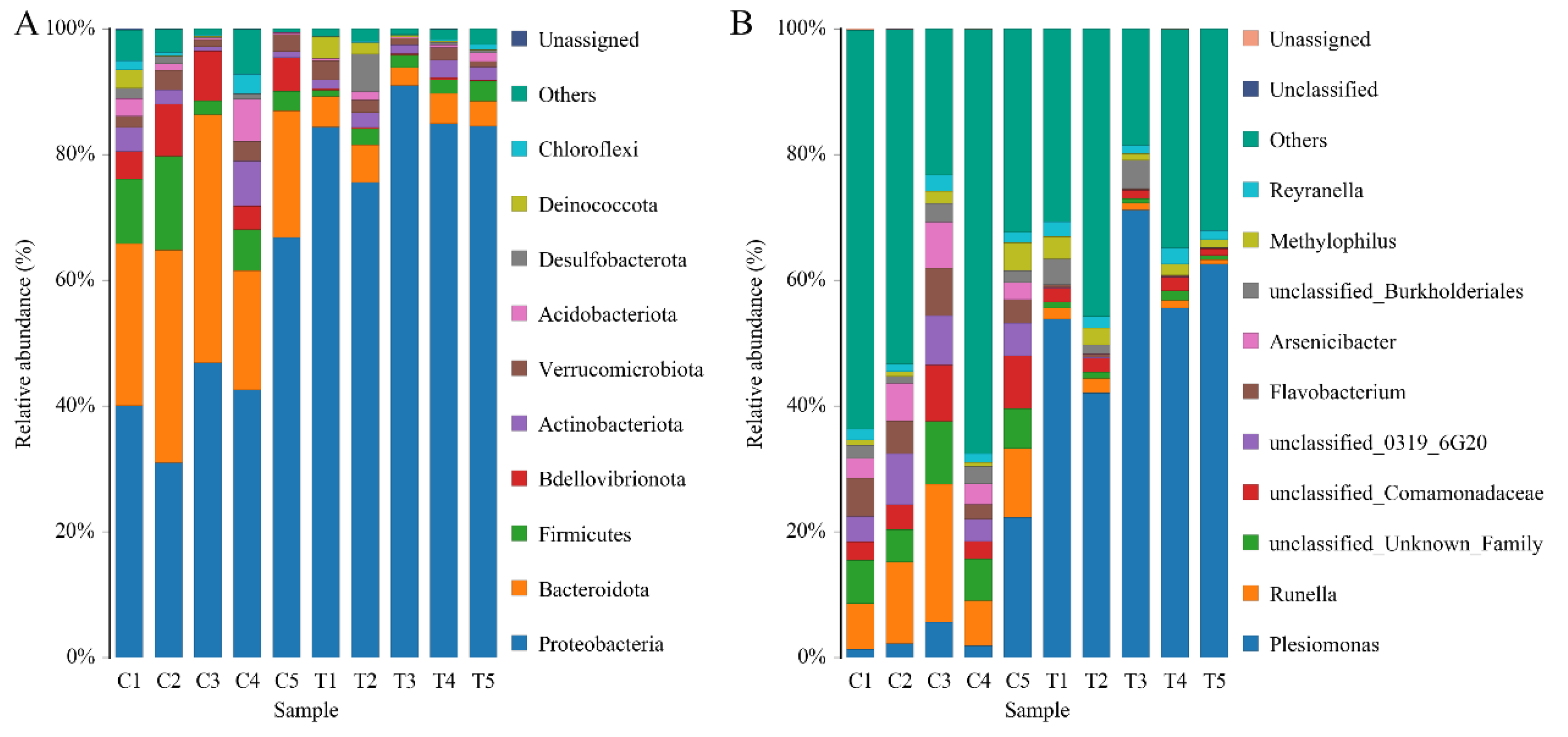
3.3.2. Microbial Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qiang, J.; Zhong, C.Y.; Bao, J.W.; Liang, M.; Liang, C.; Tao, Y.F.; Li, H.X.; He, J.; Xu, P. Synergistic effect of water temperature and dissolved oxygen concentration on rates of fertilization, hatching and deformity of hybrid yellow catfish (Tachysurus fulvidraco female x Pseudobagrus vachelliic male). J. Therm. Biol. 2019, 83, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations. The State of World Fisheries and Aquaculture 2020. Sustainability in Action; Food and Agriculture Organization of the United Nations: Rome, Italy, 2020. [Google Scholar]
- Wang, Z.; Zhou, T.; Gu, Z.M. New data of two trichodinid ectoparasites (Ciliophora: Trichodinidae) from farmed freshwater fishes in Hubei, China. Eur. J. Protistol. 2017, 60, 50–59. [Google Scholar] [CrossRef]
- Wise, A.L.; LaFrentz, B.R.; Kelly, A.M.; Khoo, L.H.; Xu, T.; Liles, M.R.; Bruce, T.J. A review of bacterial co-infections in farmed catfish: Components, diagnostics, and treatment directions. Animals 2021, 11, 3240. [Google Scholar] [CrossRef]
- Tavares-Dias, M. Toxic, physiological, histomorphological, growth performance and antiparasitic effects of copper sulphate in fish aquaculture. Aquaculture 2021, 535, 736350. [Google Scholar] [CrossRef]
- Monteiro, S.M.; Rocha, E.; Fontainhas-Fernandes, A.; Sousa, M. Quantitative histopathology of Oreochromis niloticus gills after copper exposure. J. Fish Biol. 2008, 73, 1376–1392. [Google Scholar] [CrossRef]
- Malhotra, N.; Ger, T.R.; Uapipatanakul, B.; Huang, J.C.; Chen, K.H.C.; Hsiao, C.D. Review of copper and copper nanoparticle toxicity in fish. Nanomaterials 2020, 10, 1126. [Google Scholar] [CrossRef] [PubMed]
- Kirici, M.; Turk, C.; Caglayan, C.; Kirici, M. Toxic effects of copper sulphate pentahydrate on antioxidant enzyme activities and lipid peroxidation of freshwater fish Capoeta Umbla (Heckel, 1843) tissues. Appl. Ecol. Environ. Res. 2017, 15, 1685–1696. [Google Scholar] [CrossRef]
- Delahaut, V.; Raskovic, B.; Salvado, M.S.; Bervoets, L.; Blust, R.; De Boeck, G. Toxicity and bioaccumulation of Cadmium, Copper and Zinc in a direct comparison at equitoxic concentrations in common carp (Cyprinus carpio) juveniles. PLoS ONE 2020, 15, e0220485. [Google Scholar] [CrossRef]
- Shaw, B.J.; Al-Bairuty, G.; Handy, R.D. Effects of waterborne copper nanoparticles and copper sulphate on rainbow trout, (Oncorhynchus mykiss): Physiology and accumulation. Aquat. Toxicol. 2012, 116, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Embar-Gopinath, S.; Butler, R.; Nowak, B. Influence of salmonid gill bacteria on development and severity of amoebic gill disease. Dis. Aquat. Org. 2005, 67, 55–60. [Google Scholar] [CrossRef]
- Reverter, M.; Tapissier-Bontemps, N.; Lecchini, D.; Banaigs, B.; Sasal, P. Biological and ecological roles of external fish mucus: A review. Fishes 2018, 3, 41. [Google Scholar] [CrossRef]
- Zhang, R.; Wu, G.T.; Zhu, J.Y.; Wang, X.W.; Liu, L.L.; Li, H.J.; Zhu, H. Povidone iodine exposure alters the immune response and microbiota of the gill and skin in koi carp, Cyprinus carpio. Aquaculture 2023, 563, 738926. [Google Scholar] [CrossRef]
- Quezada-Rodriguez, P.R.; Taylor, R.S.; Samsing, F.; Rigby, M.; Wood, A.T.; Nowak, B.F.; Wynne, J.W. Effect of a prophylactic treatment with chloramine-T on gill histology and microbiome of Atlantic salmon (Salmo salar) under commercial conditions. Aquaculture 2022, 546, 737319. [Google Scholar] [CrossRef]
- Mohammed, H.H.; Arias, C.R. Potassium permanganate elicits a shift of the external fish microbiome and increases host susceptibility to columnaris disease. Vet. Res. 2015, 46, 82. [Google Scholar] [CrossRef]
- SC/T 1132-2016; Fishery Drugs Use Standard. Ministry of Agriculture and Rural Affairs of the People’s Republic of China, China Agriculture Press: Beijing, China, 2016.
- Peskin, A.V.; Winterbourn, C.C. A microtiter plate assay for superoxide dismutase using a water-soluble tetrazolium salt (WST-1). Clin. Chim. Acta 2000, 293, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Góth, L. A simple method for determination of serum catalase activity and revision of reference range. Clin. Chim. Acta 1991, 196, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Mannervik, B.; Guthenberg, C. Glutathione transferase (human placenta). Methods Enzymol. 1981, 77, 231–235. [Google Scholar]
- Ohkawa, H.; Ohishi, N.; Yagi, K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 1979, 95, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, Y.; Dong, J.; Yang, Q.; Xu, N.; Yang, Y.; Gu, Z.; Ai, X. Transcriptome analysis of goldfish (Carassius auratus) in response to Gyrodactylus kobayashii infection. Parasitol. Res. 2021, 120, 161–171. [Google Scholar] [CrossRef]
- Shekh, K.; Tang, S.; Niyogi, S.; Hecker, M. Expression stability and selection of optimal reference genes for gene expression normalization in early life stage rainbow trout exposed to cadmium and copper. Aquat. Toxicol. 2017, 190, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Luo, Z.; Chen, G.H.; Wei, C.C.; Zhuo, M.Q. Identification of eight copper (Cu) uptake related genes from yellow catfish Pelteobagrus fulvidraco, and their tissue expression and transcriptional responses to dietborne Cu exposure. J. Trace Elem. Med. Biol. 2017, 44, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Mu, D.Y.; Meng, J.H.; Bo, X.X.; Wu, M.Y.; Xiao, H.; Wang, H.Y. The effect of cadmium exposure on diversity of intestinal microbial community of Rana chensinensis tadpoles. Ecotoxicol. Environ. Saf. 2018, 154, 6–12. [Google Scholar] [CrossRef]
- Farmer, B.D.; Straus, D.L.; Beck, B.H.; Mitchell, A.J.; Freeman, D.; Meinelt, T. Effectiveness of copper sulphate, potassium permanganate and peracetic acid to reduce mortality and infestation of Ichthyobodo necator in channel catfish Ictalurus punctatus (Rafinesque 1818). Aquac. Res. 2013, 44, 1103–1109. [Google Scholar] [CrossRef]
- Farmer, B.D.; Fuller, S.A.; Mitchell, A.J.; Straus, D.L.; Bullard, S.A. Efficacy of bath treatments of formalin and copper sulfate on cultured white bass, Morone chrysops, concurrently infected by Onchocleidus mimus and Ichthyophthirius multifiliis. J. World Aquac. Soc. 2013, 44, 305–310. [Google Scholar] [CrossRef]
- Hoseinifar, S.H.; Yousefi, S.; Van Doan, H.; Ashouri, G.; Gioacchini, G.; Maradonna, F.; Carnevali, O. Oxidative stress and antioxidant defense in fish: The implications of probiotic, prebiotic, and synbiotics. Rev. Fish. Sci. Aquac. 2020, 29, 198–217. [Google Scholar] [CrossRef]
- Outama, P.; Linh, N.V.; Xuan, C.L.; Wannavijit, S.; Tongsiri, S.; Chitmanat, C.; Montha, N.; Van Doan, H. Passionfruit (Passiflora edulis) peel powder stimulates the immune and antioxidant defense system in nile tilapia, Oreochromis niloticus, cultivated in a biofloc system. Fishes 2022, 7, 233. [Google Scholar] [CrossRef]
- Trivedi, M.H.; Sangai, N.P.; Renuka, A. Assessment of toxicity of copper sulphate pentahydrate on oxidative stress indicators on liver of goldfish (Carassius auratus). Bull. Environ. Pharmacol. Life Sci. 2012, 1, 52–57. [Google Scholar]
- Yonar, M.E.; Ispir, U.; Yonar, S.M.; Kirici, M. Effect of copper sulphate on the antioxidant parameters in the rainbow trout fry, Oncorhynchus mykiss. Cell. Mol. Biol. 2016, 62, 55–58. [Google Scholar]
- Sanchez, W.; Palluel, O.; Meunier, L.; Coquery, M.; Porcher, J.M.; Ait-Aissa, S. Copper-induced oxidative stress in three-spined stickleback: Relationship with hepatic metal levels. Environ. Toxicol. Pharmacol. 2005, 19, 177–183. [Google Scholar] [CrossRef]
- Kumar, M.; Singh, S.; Dwivedi, S.; Trivedi, A.; Dubey, I.; Trivedi, S.P. Copper-induced genotoxicity, oxidative stress, and alteration in transcriptional level of autophagy-associated genes in snakehead fish Channa punctatus. Biol. Trace Elem. Res. 2023, 201, 2022–2035. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, M.M.; EL-Fiky, S.A.; Soheir, Y.M.; Abeer, A.I. Cytogenetic studies on the effect of copper sulfate and lead acetate pollution on Oreochromis niloticus fish. Asian J. Cell Biol. 2008, 3, 51–60. [Google Scholar] [CrossRef]
- Nekoubin, H.; Gharedaashi, E.; Hatefi, S.; Sudagar, M.; Shahriari, R.; Asgharimoghadam, A. Determination of LC50 of copper sulfate and lead(II) nitrate and behavioral responses of grass carp (Ctenopharyngodon Idella). Walailak J. Sci. Technol. 2012, 9, 333–340. [Google Scholar]
- Mokhtar, D.M.; Zaccone, G.; Alesci, A.; Kuciel, M.; Hussein, M.T.; Sayed, R.K.A. Main components of fish immunity: An overview of the fish immune system. Fishes 2023, 8, 93. [Google Scholar] [CrossRef]
- Bowden, T.J. Modulation of the immune system of fish by their environment. Fish Shellfish Immunol. 2008, 25, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Reis, M.I.R.; do Vale, A.; Pereira, P.J.B.; Azevedo, J.E.; dos Santos, N.M.S. Caspase-1 and IL-1 beta processing in a teleost fish. PLoS ONE 2012, 7, e50450. [Google Scholar] [CrossRef]
- Sukegawa, S.; Miyagi, E.; Bouamr, F.; Farkasova, H.; Strebel, K. Mannose receptor 1 restricts HIV particle release from infected macrophages. Cell Rep. 2018, 22, 786–795. [Google Scholar] [CrossRef]
- Wu, Y.; Huang, M.; Lu, Y.; Huang, Y.; Jian, J. Molecular characterization and functional analysis of CD209E from Nile Tilapia (Oreochromis Niloticus) involved in immune response to bacterial infection. Fish Shellfish Immunol. 2023, 136, 108718. [Google Scholar] [CrossRef]
- Razmara, P.; Imbery, J.J.; Koide, E.; Helbing, C.C.; Wiseman, S.B.; Gauthier, P.T.; Bray, D.F.; Needham, M.; Haight, T.; Zovoilis, A.; et al. Mechanism of copper nanoparticle toxicity in rainbow trout olfactory mucosa. Environ. Pollut. 2021, 284, 117141. [Google Scholar] [CrossRef]
- Wang, T.; Long, X.H.; Liu, Z.P.; Cheng, Y.Z.; Yan, S.H. Effect of copper nanoparticles and copper sulphate on oxidation stress, cell apoptosis and immune responses in the intestines of juvenile Epinephelus coioides. Fish Shellfish Immunol. 2015, 44, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Hoseini, S.M.; Khalili, M.; Rajabiesterabadi, H.; Hoseinifar, S.H.; Doan, H.V. Effects of dietary monoterpene, myrcene, administration on immune- and health-related genes expression in common carp gill following exposure to copper sulfate. Fish Shellfish Immunol. 2020, 98, 438–445. [Google Scholar] [CrossRef] [PubMed]
- Legrand, T.P.R.A.; Wynne, J.W.; Weyrich, L.S.; Oxley, A.P.A. A microbial sea of possibilities: Current knowledge and prospects for an improved understanding of the fish microbiome. Rev. Aquac. 2020, 12, 1101–1134. [Google Scholar] [CrossRef]
- Pratte, Z.A.; Besson, M.; Hollman, R.D.; Stewart, F.J. The gills of reef fish support a distinct microbiome influenced by host-specific factors. Appl. Environ. Microbiol. 2018, 84, e00063-18. [Google Scholar] [CrossRef]
- Brown, R.; Moore, L.; Mani, A.; Patel, S.; Salinas, I. Effects of ploidy and salmonid alphavirus infection on the skin and gill microbiome of Atlantic salmon (Salmo salar). PLoS ONE 2021, 16, e0243684. [Google Scholar] [CrossRef] [PubMed]
- Rosado, D.; Perez-Losada, M.; Severino, R.; Cable, J.; Xavier, R. Characterization of the skin and gill microbiomes of the farmed seabass (Dicentrarchus labrax) and seabream (Sparus aurata). Aquaculture 2019, 500, 57–64. [Google Scholar] [CrossRef]
- Zhou, D.; Zhang, T.; Ren, L.; Fang, D.A.; Xu, D.P. Differential study of microbiota in the gill and intestine of silver carp (Hypophthalmichthys molitrix) from the algae-dominated and hydrophyte-dominated areas of Taihu Lake, China. Fishes 2022, 7, 304. [Google Scholar] [CrossRef]
- Rosado, D.; Xavier, R.; Severino, R.; Tavares, F.; Cable, J.; Perez-Losada, M. Effects of disease, antibiotic treatment and recovery trajectory on the microbiome of farmed sea bass (Dicentrarchus labrax). Sci. Rep. 2019, 9, 18946. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef]
- Litvak, Y.; Byndloss, M.X.; Tsolis, R.M.; Baumler, A.J. Dysbiotic Proteobacteria expansion: A microbial signature of epithelial dysfunction. Curr. Opin. Microbiol. 2017, 39, 1–6. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L.; McIver, C.J. Plesiomonas shigelloides revisited. Clin. Microbiol. Rev. 2016, 29, 349–374. [Google Scholar] [CrossRef]
- Behera, B.K.; Bera, A.K.; Paria, P.; Das, A.; Parida, P.K.; Kumari, S.; Bhowmick, S.; Das, B.K. Identification and pathogenicity of Plesiomonas shigelloides in Silver Carp. Aquaculture 2018, 493, 314–318. [Google Scholar] [CrossRef]
- Nisha, R.G.; Rajathi, V.; Manikandan, R.; Prabhu, N.M. Isolation of Plesiomonas shigelloides from infected cichlid fishes using 16S rRNA characterization and its control with probiotic Pseudomonas sp. Acta Sci. Vet. 2014, 42, 1–7. [Google Scholar]
- Perez, J.; Contreras-Moreno, F.J.; Marcos-Torres, F.J.; Moraleda-Munoz, A.; Munoz-Dorado, J. The antibiotic crisis: How bacterial predators can help. Comput. Struct. Biotechnol. J. 2020, 18, 2547–2555. [Google Scholar] [CrossRef]
- Willis, A.R.; Moore, C.; Mazon-Moya, M.; Krokowski, S.; Lambert, C.; Till, R.; Mostowy, S.; Sockett, R.E. Injections of predatory bacteria work alongside host immune cells to treat Shigella infection in zebrafish larvae. Curr. Biol. 2016, 26, 3343–3351. [Google Scholar] [CrossRef]
- Tarnecki, A.M.; Levi, N.J.; Resley, M.; Main, K. Effect of copper sulfate on the external microbiota of adult common snook (Centropomus undecimalis). Anim. Microbiome 2021, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Stojanov, S.; Berlec, A.; Strukelj, B. The influence of probiotics on the Firmicutes/Bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef] [PubMed]
- Magne, F.; Gotteland, M.; Gauthier, L.; Zazueta, A.; Pesoa, S.; Navarrete, P.; Balamurugan, R. The Firmicutes/Bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients 2020, 12, 1474. [Google Scholar] [CrossRef] [PubMed]

| Treatment | SOD (U/mg Protein) | CAT (U/mg Protein) | GSH-Px (U/mg Protein) | MDA (nmol/mg Protein) |
|---|---|---|---|---|
| Control group | 55.45 ± 3.19 | 6.84 ± 0.53 | 6.63 ± 0.7 | 5.91 ± 1.51 |
| Exposed group | 39.19 ± 3.15 * | 4.8 ± 0.6 * | 4.69 ± 0.68 * | 10.94 ± 1.36 * |
| Phylum | Relative Abundance (%) | |
|---|---|---|
| 0 mg/L Copper Sulphate | 0.7 mg/L Copper Sulphate | |
| Bacteroidotas | 27.64 ± 3.96 | 4.51 ± 0.51 * |
| Bdellovibrionota | 5.93 ± 0.92 | 0.26 ± 0.04 * |
| Proteobacteria | 45.46 ± 5.93 | 84.07 ± 2.46 * |
| Firmicutes | 7.4 ± 2.35 | 2.16 ± 5.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, S.; Yang, Q.; Song, Y.; Cheng, B.; Ai, X. Effect of Copper Sulphate Exposure on the Oxidative Stress, Gill Transcriptome and External Microbiota of Yellow Catfish, Pelteobagrus fulvidraco. Antioxidants 2023, 12, 1288. https://doi.org/10.3390/antiox12061288
Zhou S, Yang Q, Song Y, Cheng B, Ai X. Effect of Copper Sulphate Exposure on the Oxidative Stress, Gill Transcriptome and External Microbiota of Yellow Catfish, Pelteobagrus fulvidraco. Antioxidants. 2023; 12(6):1288. https://doi.org/10.3390/antiox12061288
Chicago/Turabian StyleZhou, Shun, Qiuhong Yang, Yi Song, Bo Cheng, and Xiaohui Ai. 2023. "Effect of Copper Sulphate Exposure on the Oxidative Stress, Gill Transcriptome and External Microbiota of Yellow Catfish, Pelteobagrus fulvidraco" Antioxidants 12, no. 6: 1288. https://doi.org/10.3390/antiox12061288
APA StyleZhou, S., Yang, Q., Song, Y., Cheng, B., & Ai, X. (2023). Effect of Copper Sulphate Exposure on the Oxidative Stress, Gill Transcriptome and External Microbiota of Yellow Catfish, Pelteobagrus fulvidraco. Antioxidants, 12(6), 1288. https://doi.org/10.3390/antiox12061288






