Prenylcysteine Oxidase 1 Is a Key Regulator of Adipogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Induction of Differentiation
2.2. PCYOX1 Overexpression in CHO Cells
2.3. Stable Transfection with Short Hairpin RNA (shRNA)
2.4. Oil Red O Staining
2.5. Real-Time Quantitative Reverse Transcriptase PCR (qRT-PCR)
2.6. Oxidized Low Density Lipoprotein (oxLDL) Assay
2.7. PCOYX1 Activity Assay
2.8. Label-Free Mass Spectrometry (LC-MSE) Analysis
2.9. GO Analysis
2.10. Mass Spectrometry-Based Quantification of PCYOX1
2.11. Mice and Diets
2.12. Statistical Analysis
3. Results
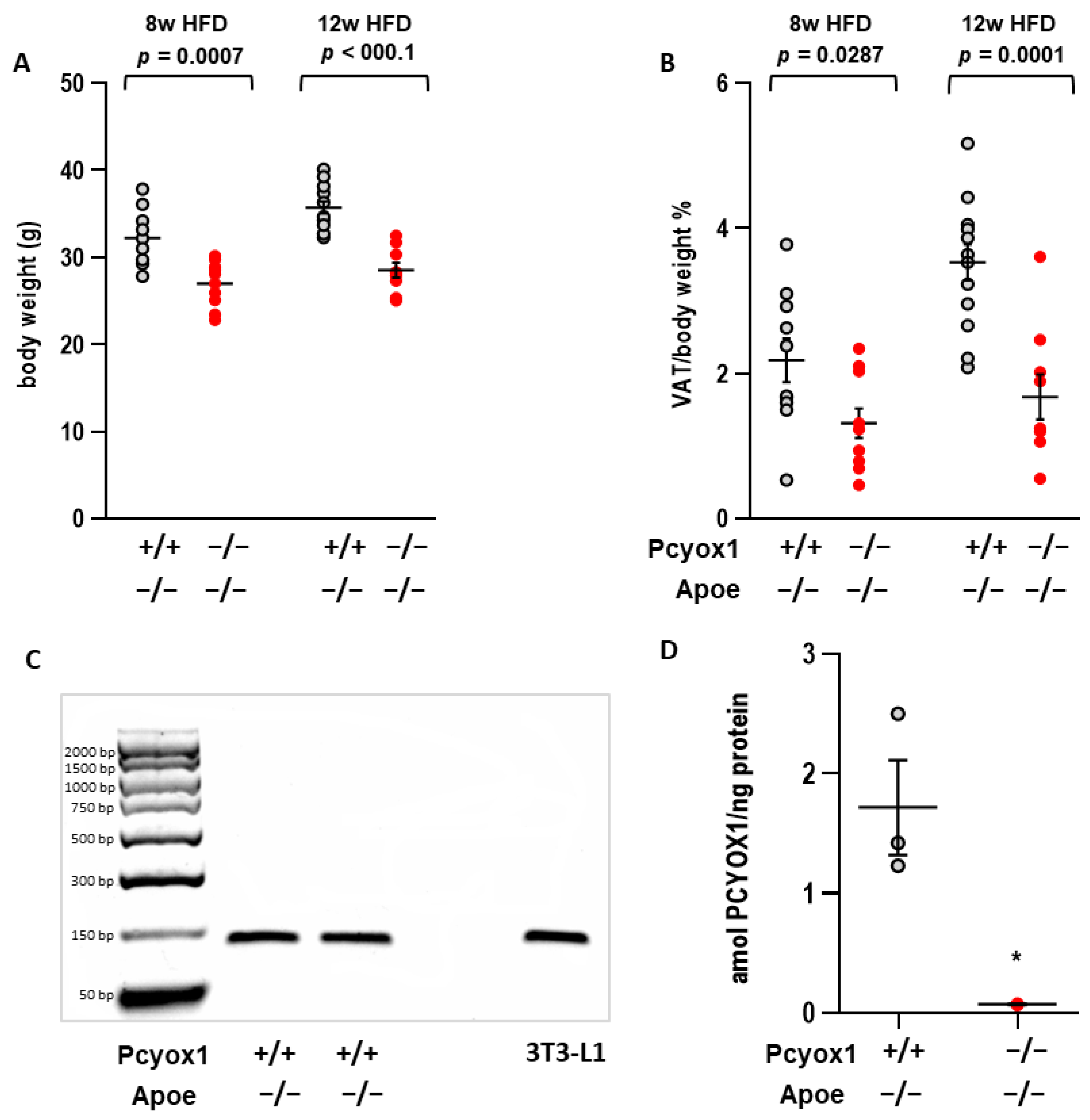
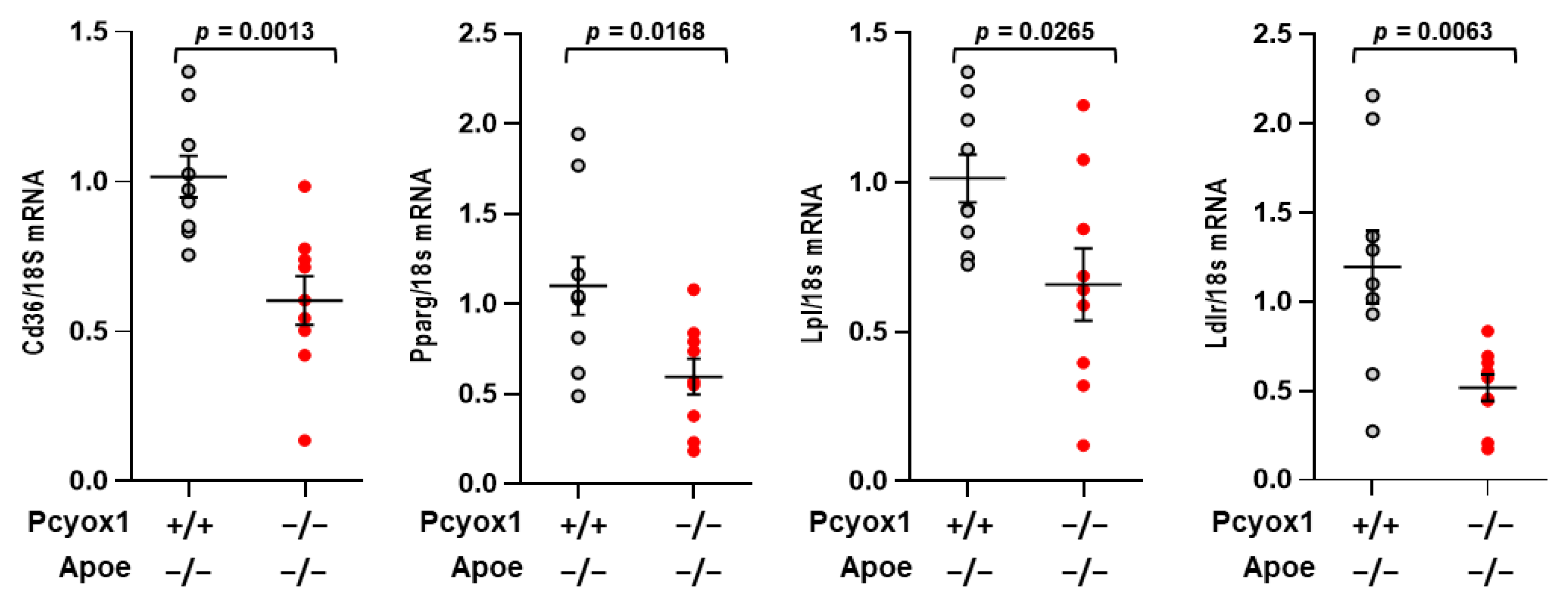
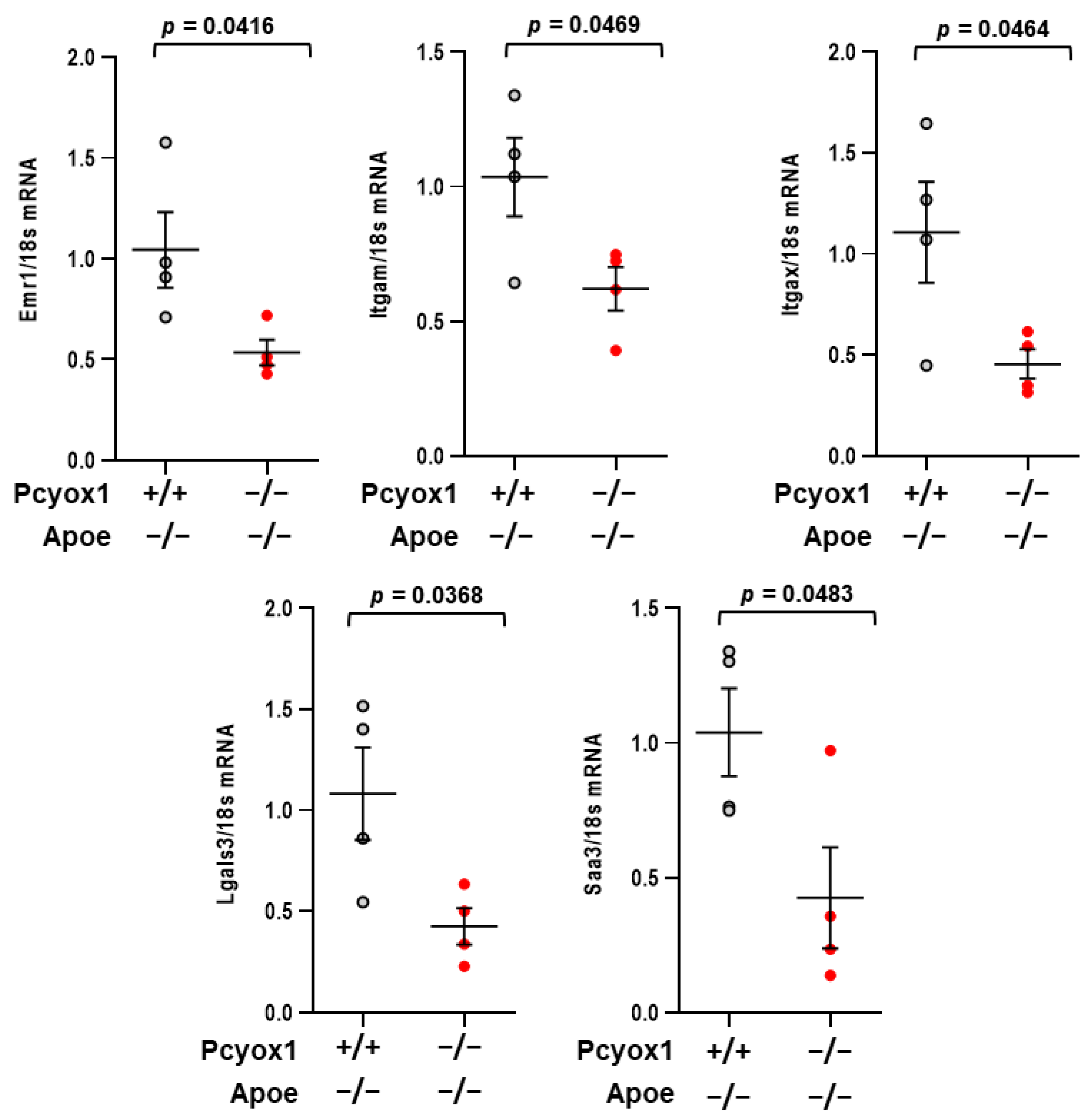
3.1. Pcyox1−/− Mice Have Decreased Adiposity
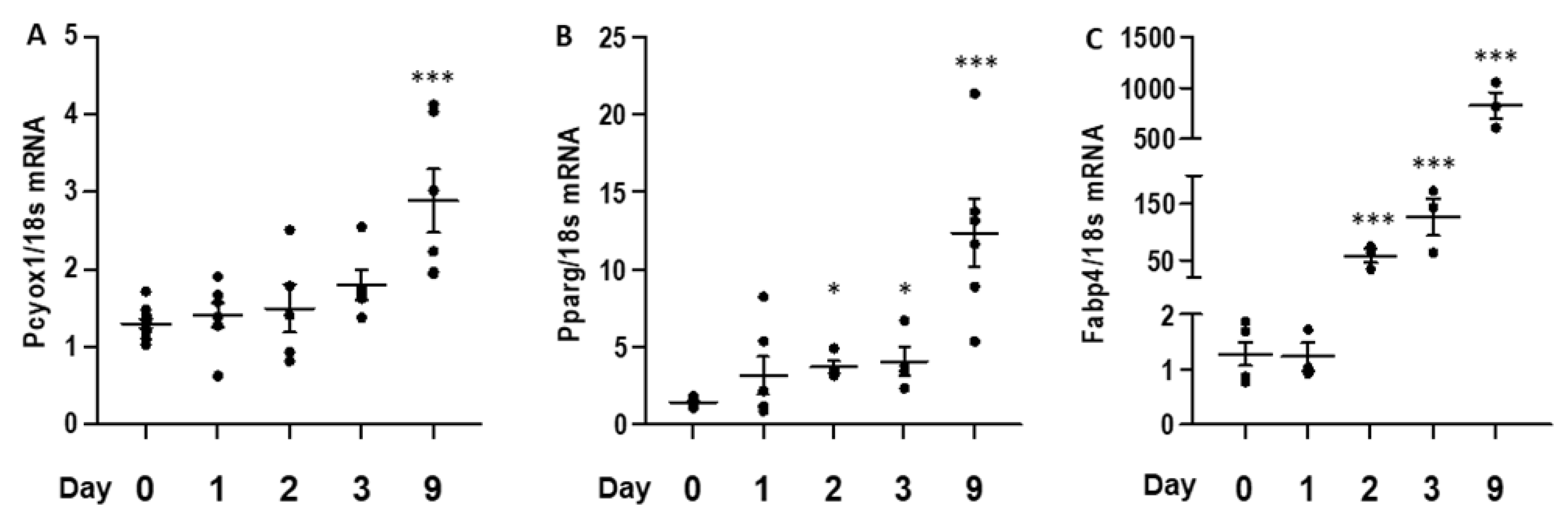
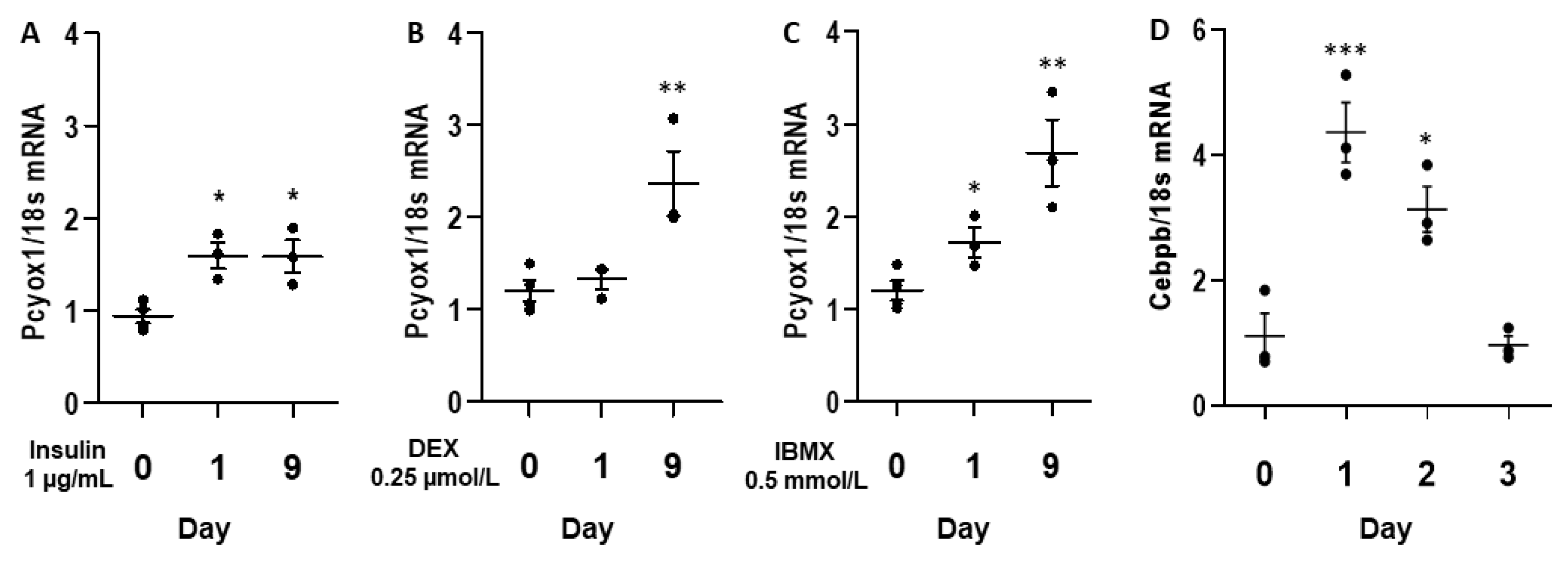
3.2. PCYOX1 Expression Is Induced during Adipocyte Differentiation In Vitro
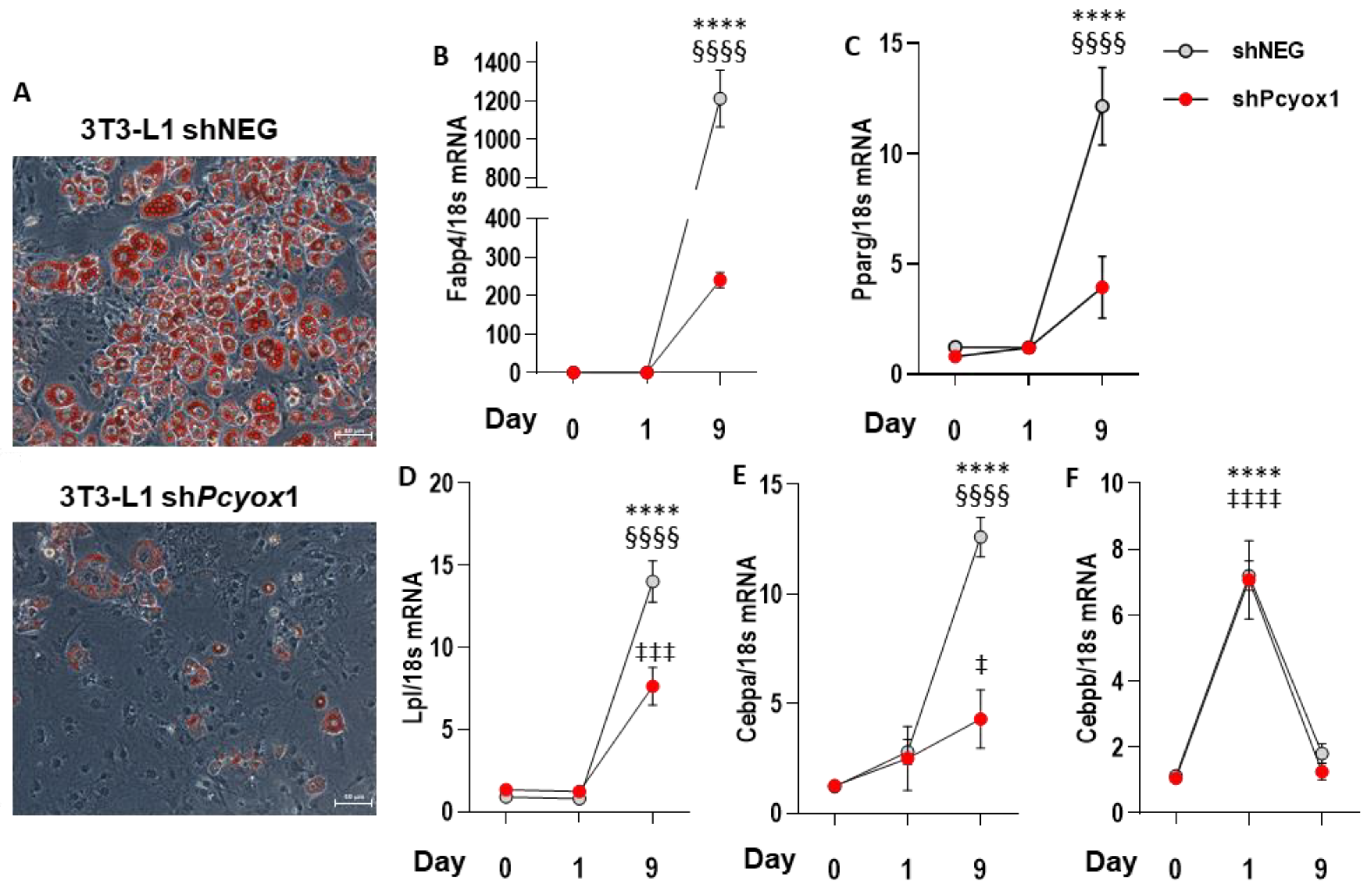
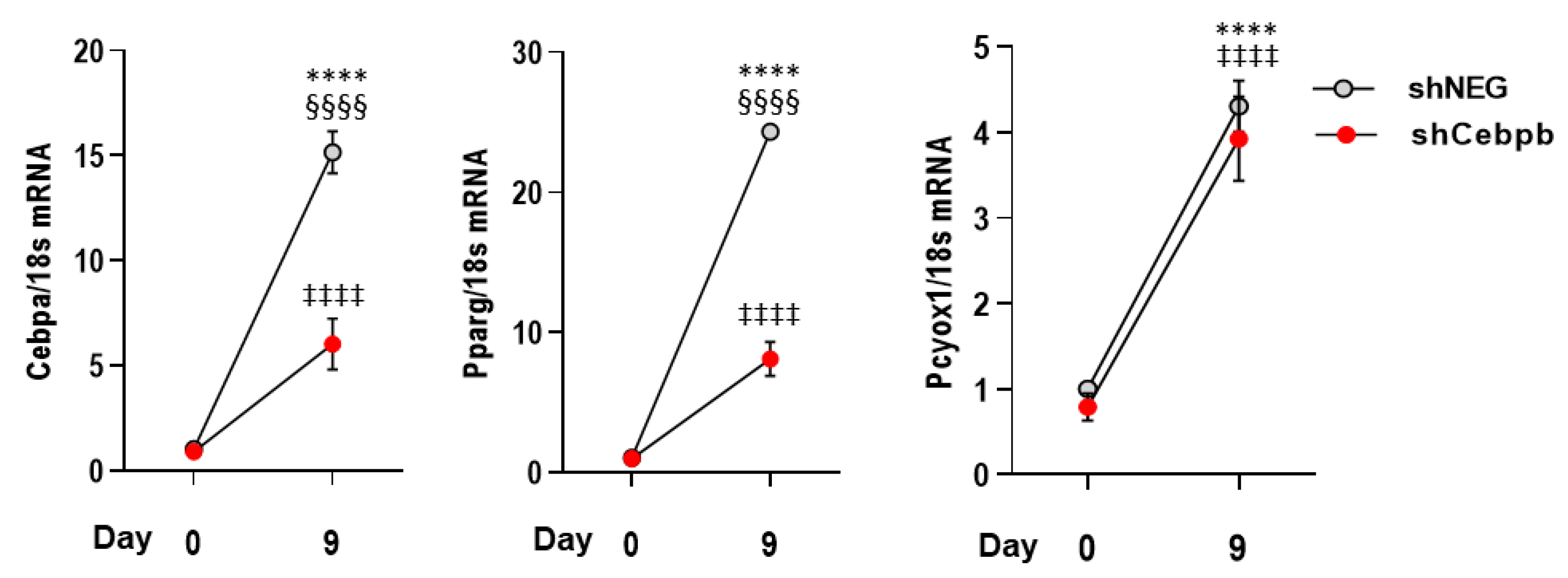
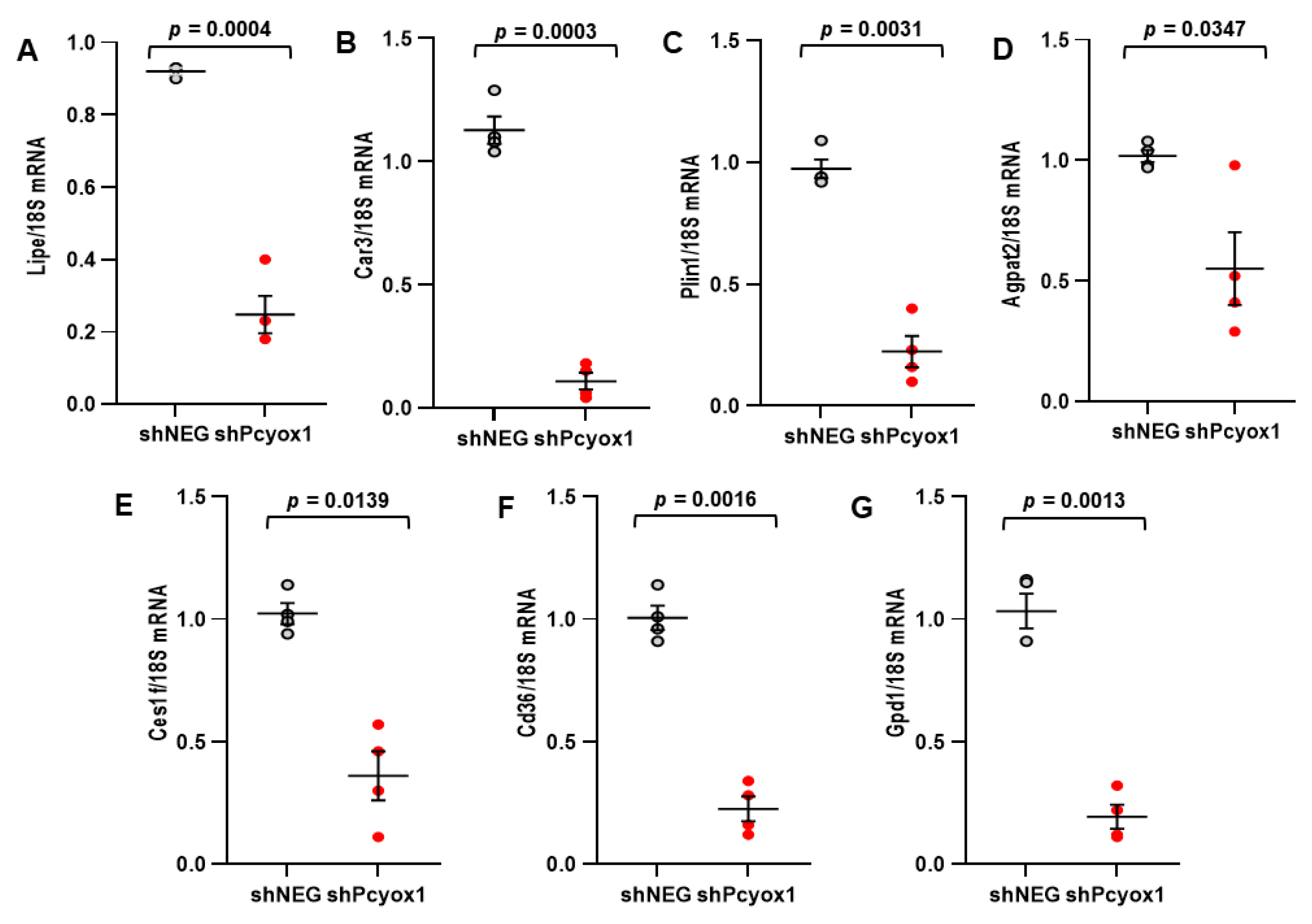
3.3. PCYOX1 Is Critical for Adipogenesis In Vitro
3.4. The Effects of PCYOX1 in Adipogenesis Are Independent of Its Pro-Oxidant Activity
3.5. PCYOX1 Significantly Affects the Cell Proteome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spiegelman, B.M.; Flier, J.S. Obesity and the regulation of energy balance. Cell 2001, 104, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Koenen, M.; Hill, M.A.; Cohen, P.; Sowers, J.R. Obesity, Adipose Tissue and Vascular Dysfunction. Circ. Res. 2021, 128, 951–968. [Google Scholar] [CrossRef] [PubMed]
- Moseti, D.; Regassa, A.; Kim, W.K. Molecular Regulation of Adipogenesis and Potential Anti-Adipogenic Bioactive Molecules. Int. J. Mol. Sci. 2016, 17, 124. [Google Scholar] [CrossRef] [PubMed]
- Banfi, C.; Baetta, R.; Barbieri, S.S.; Brioschi, M.; Guarino, A.; Ghilardi, S.; Sandrini, L.; Eligini, S.; Polvani, G.; Bergman, O.; et al. Prenylcysteine oxidase 1, an emerging player in atherosclerosis. Commun. Biol. 2021, 4, 1109. [Google Scholar] [CrossRef]
- Tschantz, W.R.; Digits, J.A.; Pyun, H.J.; Coates, R.M.; Casey, P.J. Lysosomal prenylcysteine lyase is a FAD-dependent thioether oxidase. J. Biol. Chem. 2001, 276, 2321–2324. [Google Scholar] [CrossRef]
- Zhang, L.; Tschantz, W.R.; Casey, P.J. Isolation and characterization of a prenylcysteine lyase from bovine brain. J. Biol. Chem. 1997, 272, 23354–23359. [Google Scholar] [CrossRef]
- Banfi, C.; Brioschi, M.; Barcella, S.; Pignieri, A.; Parolari, A.; Biglioli, P.; Tremoli, E.; Mussoni, L. Tissue factor induction by protease-activated receptor 1 requires intact caveolin-enriched membrane microdomains in human endothelial cells. J. Thromb. Haemost. 2007, 5, 2437–2444. [Google Scholar] [CrossRef]
- Banfi, C.; Brioschi, M.; Vicentini, L.M.; Cattaneo, M.G. The Effects of Silencing PTX3 on the Proteome of Human Endothelial Cells. Int. J. Mol. Sci. 2022, 23, 13487. [Google Scholar] [CrossRef]
- Brioschi, M.; Eligini, S.; Crisci, M.; Fiorelli, S.; Tremoli, E.; Colli, S.; Banfi, C. A mass spectrometry-based workflow for the proteomic analysis of in vitro cultured cell subsets isolated by means of laser capture microdissection. Anal. Bioanal. Chem. 2014, 406, 2817–2825. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef]
- Pontremoli, M.; Brioschi, M.; Baetta, R.; Ghilardi, S.; Banfi, C. Identification of DKK-1 as a novel mediator of statin effects in human endothelial cells. Sci. Rep. 2018, 8, 16671. [Google Scholar] [CrossRef]
- Banfi, C.; Amadio, P.; Zara, M.; Brioschi, M.; Sandrini, L.; Barbieri, S.S. Prenylcysteine Oxidase 1 (PCYOX1), a New Player in Thrombosis. Int. J. Mol. Sci. 2022, 23, 2831. [Google Scholar] [CrossRef]
- Merrett, J.E.; Bo, T.; Psaltis, P.J.; Proud, C.G. Identification of DNA response elements regulating expression of CCAAT/enhancer-binding protein (C/EBP) beta and delta and MAP kinase-interacting kinases during early adipogenesis. Adipocyte 2020, 9, 427–442. [Google Scholar] [CrossRef]
- Nagy, L.; Tontonoz, P.; Alvarez, J.G.; Chen, H.; Evans, R.M. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell 1998, 93, 229–240. [Google Scholar] [CrossRef]
- Lu, J.Y.; Hofmann, S.L. Thematic review series: Lipid posttranslational modifications. Lysosomal metabolism of lipid-modified proteins. J. Lipid Res. 2006, 47, 1352–1357. [Google Scholar] [CrossRef]
- Banfi, C.; Brioschi, M.; Barcella, S.; Wait, R.; Begum, S.; Galli, S.; Rizzi, A.; Tremoli, E. Proteomic analysis of human low-density lipoprotein reveals the presence of prenylcysteine lyase, a hydrogen peroxide-generating enzyme. Proteomics 2009, 9, 1344–1352. [Google Scholar] [CrossRef]
- Beigneux, A.; Withycombe, S.K.; Digits, J.A.; Tschantz, W.R.; Weinbaum, C.A.; Griffey, S.M.; Bergo, M.; Casey, P.J.; Young, S.G. Prenylcysteine lyase deficiency in mice results in the accumulation of farnesylcysteine and geranylgeranylcysteine in brain and liver. J. Biol. Chem. 2002, 277, 38358–38363. [Google Scholar] [CrossRef]
- King, V.L.; Hatch, N.W.; Chan, H.W.; de Beer, M.C.; de Beer, F.C.; Tannock, L.R. A murine model of obesity with accelerated atherosclerosis. Obesity 2010, 18, 35–41. [Google Scholar] [CrossRef]
- Rosen, E.D.; Hsu, C.H.; Wang, X.; Sakai, S.; Freeman, M.W.; Gonzalez, F.J.; Spiegelman, B.M. C/EBPalpha induces adipogenesis through PPARgamma: A unified pathway. Genes Dev. 2002, 16, 22–26. [Google Scholar] [CrossRef]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Yoshida, N.; Kishimoto, T.; Akira, S. Defective adipocyte differentiation in mice lacking the C/EBPbeta and/or C/EBPdelta gene. EMBO J. 1997, 16, 7432–7443. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Sun, T.; Bookout, A.L.; Downes, M.; Yu, R.T.; Evans, R.M.; Mangelsdorf, D.J. A Nuclear Receptor Atlas: 3T3-L1 adipogenesis. Mol. Endocrinol. 2005, 19, 2437–2450. [Google Scholar] [CrossRef] [PubMed]
- Soukas, A.; Socci, N.D.; Saatkamp, B.D.; Novelli, S.; Friedman, J.M. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J. Biol. Chem. 2001, 276, 34167–34174. [Google Scholar] [CrossRef]
- Ambele, M.A.; Dhanraj, P.; Giles, R.; Pepper, M.S. Adipogenesis: A Complex Interplay of Multiple Molecular Determinants and Pathways. Int. J. Mol. Sci. 2020, 21, 4283. [Google Scholar] [CrossRef]
- Martinez-Botas, J.; Anderson, J.B.; Tessier, D.; Lapillonne, A.; Chang, B.H.; Quast, M.J.; Gorenstein, D.; Chen, K.H.; Chan, L. Absence of perilipin results in leanness and reverses obesity in Lepr(db/db) mice. Nat. Genet. 2000, 26, 474–479. [Google Scholar] [CrossRef]
- Tansey, J.T.; Sztalryd, C.; Gruia-Gray, J.; Roush, D.L.; Zee, J.V.; Gavrilova, O.; Reitman, M.L.; Deng, C.X.; Li, C.; Kimmel, A.R.; et al. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. USA 2001, 98, 6494–6499. [Google Scholar] [CrossRef]
- Itabe, H.; Yamaguchi, T.; Nimura, S.; Sasabe, N. Perilipins: A diversity of intracellular lipid droplet proteins. Lipids Health Dis. 2017, 16, 83. [Google Scholar] [CrossRef]
- Subauste, A.R.; Das, A.K.; Li, X.; Elliott, B.G.; Evans, C.; El Azzouny, M.; Treutelaar, M.; Oral, E.; Leff, T.; Burant, C.F. Alterations in lipid signaling underlie lipodystrophy secondary to AGPAT2 mutations. Diabetes 2012, 61, 2922–2931. [Google Scholar] [CrossRef]
- Gale, S.E.; Frolov, A.; Han, X.; Bickel, P.E.; Cao, L.; Bowcock, A.; Schaffer, J.E.; Ory, D.S. A regulatory role for 1-acylglycerol-3-phosphate-O-acyltransferase 2 in adipocyte differentiation. J. Biol. Chem. 2006, 281, 11082–11089. [Google Scholar] [CrossRef]
- McIntyre, T.M.; Pontsler, A.V.; Silva, A.R.; St Hilaire, A.; Xu, Y.; Hinshaw, J.C.; Zimmerman, G.A.; Hama, K.; Aoki, J.; Arai, H.; et al. Identification of an intracellular receptor for lysophosphatidic acid (LPA): LPA is a transcellular PPARgamma agonist. Proc. Natl. Acad. Sci. USA 2003, 100, 131–136. [Google Scholar] [CrossRef]
- Lian, J.; Nelson, R.; Lehner, R. Carboxylesterases in lipid metabolism: From mouse to human. Protein Cell 2018, 9, 178–195. [Google Scholar] [CrossRef]
- Dolinsky, V.W.; Gilham, D.; Hatch, G.M.; Agellon, L.B.; Lehner, R.; Vance, D.E. Regulation of triacylglycerol hydrolase expression by dietary fatty acids and peroxisomal proliferator-activated receptors. Biochim. Biophys. Acta 2003, 1635, 20–28. [Google Scholar] [CrossRef]
- Dominguez, E.; Galmozzi, A.; Chang, J.W.; Hsu, K.L.; Pawlak, J.; Li, W.; Godio, C.; Thomas, J.; Partida, D.; Niessen, S.; et al. Integrated phenotypic and activity-based profiling links Ces3 to obesity and diabetes. Nat. Chem. Biol. 2014, 10, 113–121. [Google Scholar] [CrossRef]
- Jernas, M.; Olsson, B.; Arner, P.; Jacobson, P.; Sjostrom, L.; Walley, A.; Froguel, P.; McTernan, P.G.; Hoffstedt, J.; Carlsson, L.M. Regulation of carboxylesterase 1 (CES1) in human adipose tissue. Biochem. Biophys. Res. Commun. 2009, 383, 63–67. [Google Scholar] [CrossRef]
- Friedrichsen, M.; Poulsen, P.; Wojtaszewski, J.; Hansen, P.R.; Vaag, A.; Rasmussen, H.B. Carboxylesterase 1 gene duplication and mRNA expression in adipose tissue are linked to obesity and metabolic function. PLoS ONE 2013, 8, e56861. [Google Scholar] [CrossRef]
- Shen, W.J.; Yu, Z.; Patel, S.; Jue, D.; Liu, L.F.; Kraemer, F.B. Hormone-sensitive lipase modulates adipose metabolism through PPARgamma. Biochim. Biophys. Acta 2011, 1811, 9–16. [Google Scholar] [CrossRef]
- Kawamura, M.; Jensen, D.F.; Wancewicz, E.V.; Joy, L.L.; Khoo, J.C.; Steinberg, D. Hormone-sensitive lipase in differentiated 3T3-L1 cells and its activation by cyclic AMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 1981, 78, 732–736. [Google Scholar] [CrossRef]
- Takahata, T.; Kumano, T.; Ookawa, K.; Hayakari, M.; Kakizaki, I.; Tsuchida, S. Inhibition of 3T3-L1 adipocyte differentiation by 6-ethoxyzolamide: Repressed peroxisome proliferator-activated receptor gamma mRNA and enhanced CCAAT/enhancer binding protein beta mRNA levels. Biochem. Pharmacol. 2004, 67, 1667–1675. [Google Scholar] [CrossRef]
- Lynch, C.J.; Brennan, W.A., Jr.; Vary, T.C.; Carter, N.; Dodgson, S.J. Carbonic anhydrase III in obese Zucker rats. Am. J. Physiol. 1993, 264, E621–E630. [Google Scholar] [CrossRef]
- Yamamoto, H.; Uramaru, N.; Kawashima, A.; Higuchi, T. Carbonic anhydrase 3 increases during liver adipogenesis even in pre-obesity, and its inhibitors reduce liver adipose accumulation. FEBS Open Bio. 2022, 12, 827–834. [Google Scholar] [CrossRef]
- Van Schothorst, E.M.; Franssen-van Hal, N.; Schaap, M.M.; Pennings, J.; Hoebee, B.; Keijer, J. Adipose gene expression patterns of weight gain suggest counteracting steroid hormone synthesis. Obes. Res. 2005, 13, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Gabrielsson, B.L.; Carlsson, B.; Carlsson, L.M. Partial genome scale analysis of gene expression in human adipose tissue using DNA array. Obes. Res. 2000, 8, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, C.; Larose, M.; Lafond, N.; Yoshioka, M.; Rodrigue, M.A.; Morissette, J.; Labrie, C.; Raymond, V.; St-Amand, J. Adipose tissue transcriptome by serial analysis of gene expression. Obes. Res. 2004, 12, 750–757. [Google Scholar] [CrossRef] [PubMed]
- Hames, K.C.; Vella, A.; Kemp, B.J.; Jensen, M.D. Free fatty acid uptake in humans with CD36 deficiency. Diabetes 2014, 63, 3606–3614. [Google Scholar] [CrossRef]
- Cai, L.; Wang, Z.; Ji, A.; Meyer, J.M.; van der Westhuyzen, D.R. Scavenger receptor CD36 expression contributes to adipose tissue inflammation and cell death in diet-induced obesity. PLoS ONE 2012, 7, e36785. [Google Scholar] [CrossRef]
- Kennedy, D.J.; Kuchibhotla, S.; Westfall, K.M.; Silverstein, R.L.; Morton, R.E.; Febbraio, M. A CD36-dependent pathway enhances macrophage and adipose tissue inflammation and impairs insulin signalling. Cardiovasc. Res. 2011, 89, 604–613. [Google Scholar] [CrossRef]
- Sanada, Y.; Yamamoto, T.; Satake, R.; Yamashita, A.; Kanai, S.; Kato, N.; van de Loo, F.A.; Nishimura, F.; Scherer, P.E.; Yanaka, N. Serum Amyloid A3 Gene Expression in Adipocytes is an Indicator of the Interaction with Macrophages. Sci. Rep. 2016, 6, 38697. [Google Scholar] [CrossRef]
- Herrera-Marcos, L.V.; Lou-Bonafonte, J.M.; Martinez-Gracia, M.V.; Arnal, C.; Navarro, M.A.; Osada, J. Prenylcysteine oxidase 1, a pro-oxidant enzyme of low density lipoproteins. Front. Biosci. 2018, 23, 1020–1037. [Google Scholar] [CrossRef]
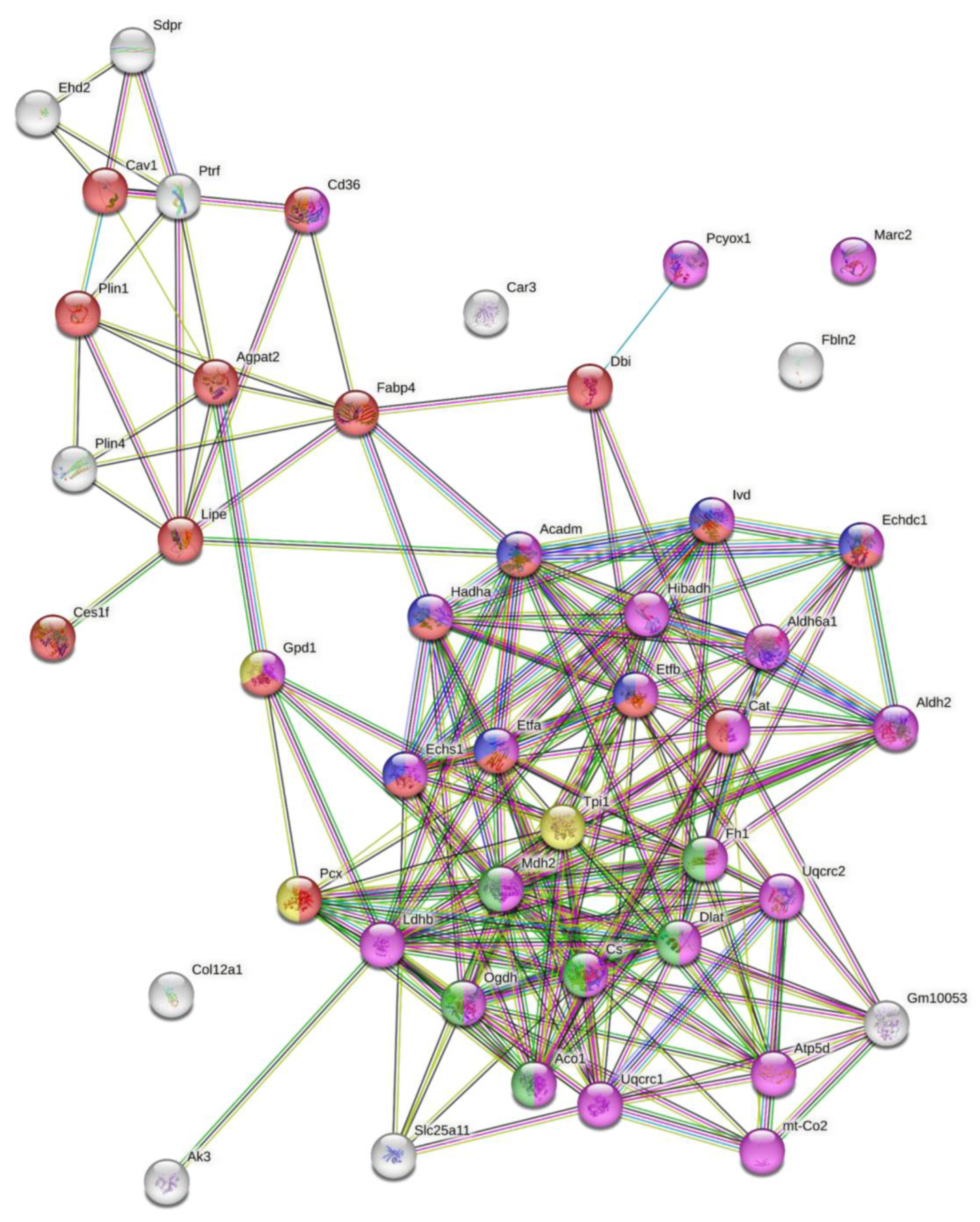
| Accession | Unique Peptides | q Value | Max Fold Change | Highest Mean Condition | Description |
|---|---|---|---|---|---|
| Reduced in Pcyox1 silenced cells | |||||
| P54310 | 3 | 0.003 | 7.07 | NEG | Hormone-sensitive lipase OS = Mus musculus GN = Lipe |
| Q9CQF9 | 2 | 0.001 | 4.84 | NEG | Prenylcysteine oxidase OS = Mus musculus GN = Pcyox1 |
| P16015 | 5 | 0.001 | 4.73 | NEG | Carbonic anhydrase 3 OS = Mus musculus GN = Car3 |
| P04117 | 5 | 0.003 | 3.51 | NEG | Fatty acid-binding protein_ adipocyte OS = Mus musculus GN = Fabp4 |
| Q8CGN5 | 5 | 0.001 | 3.34 | NEG | Perilipin-1 OS = Mus musculus GN = Plin1 |
| Q8K3K7 | 3 | 0.003 | 2.91 | NEG | 1-acyl-sn-glycerol-3-phosphate acyltransferase beta OS = Mus musculus GN = Agpat2 |
| Q91WU0 | 2 | 0.006 | 2.77 | NEG | Carboxylesterase 1F OS = Mus musculus GN = Ces1f |
| Q08857 | 2 | 0.012 | 2.32 | NEG | Platelet glycoprotein 4 OS = Mus musculus GN = Cd36 |
| P13707 | 9 | 0.006 | 2.32 | NEG | Glycerol-3-phosphate dehydrogenase [NAD(+)]_ cytoplasmic OS = Mus musculus GN = Gpd1 |
| P28271 | 3 | 0.004 | 2.18 | NEG | Cytoplasmic aconitate hydratase OS = Mus musculus GN = Aco1 |
| P16125 | 2 | 0.011 | 2.03 | NEG | L-lactate dehydrogenase B chain OS = Mus musculus GN = Ldhb P |
| P24270 | 2 | 0.004 | 1.92 | NEG | Catalase OS = Mus musculus GN = Cat |
| Q63918 | 4 | 0.001 | 1.92 | NEG | Serum deprivation-response protein OS = Mus musculus GN = Sdpr |
| Q05920 | 18 | 0.001 | 1.88 | NEG | Pyruvate carboxylase_ mitochondrial OS = Mus musculus GN = Pc |
| P31786 | 2 | 0.021 | 1.87 | NEG | Acyl-CoA-binding protein OS = Mus musculus GN = Dbi |
| O88492 | 3 | 0.019 | 1.86 | NEG | Perilipin-4 OS = Mus musculus GN = Plin4 |
| P45952 | 3 | 0.006 | 1.81 | NEG | Medium-chain specific acyl-CoA dehydrogenase_ mitochondrial OS = Mus musculus GN = Acadm |
| Q8BMS1 | 13 | 0.007 | 1.80 | NEG | Trifunctional enzyme subunit alpha_ mitochondrial OS = Mus musculus GN = Hadha |
| P97807 | 6 | 0.004 | 1.79 | NEG | Fumarate hydratase_ mitochondrial OS = Mus musculus GN = Fh |
| Q8BH95 | 3 | 0.037 | 1.70 | NEG | Enoyl-CoA hydratase_ mitochondrial OS = Mus musculus GN = Echs1 |
| Q9WTP7 | 2 | 0.007 | 1.68 | NEG | GTP:AMP phosphotransferase AK3_ mitochondrial OS = Mus musculus GN = Ak3 |
| P49817 | 2 | 0.016 | 1.53 | NEG | Caveolin-1 OS = Mus musculus GN = Cav1 |
| Q9CZU6 | 4 | 0.017 | 1.51 | NEG | Citrate synthase_ mitochondrial OS = Mus musculus GN = Cs |
| P62897 | 2 | 0.005 | 1.49 | NEG | Cytochrome c_ somatic OS = Mus musculus GN = Cycs |
| Q9EQ20 | 3 | 0.001 | 1.48 | NEG | Methylmalonate-semialdehyde dehydrogenase [acylating]_ mitochondrial OS = Mus musculus GN = Aldh6a1 |
| Q9JHI5 | 2 | 0.007 | 1.48 | NEG | Isovaleryl-CoA dehydrogenase_ mitochondrial OS = Mus musculus GN = Ivd |
| Q9CZ13 | 3 | 0.009 | 1.47 | NEG | Cytochrome b-c1 complex subunit 1_ mitochondrial OS = Mus musculus GN = Uqcrc1 |
| O54724 | 6 | 0.001 | 1.47 | NEG | Polymerase I and transcript release factor OS = Mus musculus GN = Ptrf |
| Q8BH64 | 8 | 0.003 | 1.46 | NEG | EH domain-containing protein 2 OS = Mus musculus GN = Ehd2 |
| P17751 | 9 | 0.001 | 1.46 | NEG | Triosephosphate isomerase OS = Mus musculus GN = Tpi1 |
| Q9D3D9 | 2 | 0.010 | 1.46 | NEG | ATP synthase subunit delta_ mitochondrial OS = Mus musculus GN = Atp5d |
| Q8BMF4 | 5 | 0.011 | 1.45 | NEG | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex_ mitochondrial OS = Mus musculus GN = Dlat |
| Q60597 | 4 | 0.022 | 1.44 | NEG | 2-oxoglutarate dehydrogenase_ mitochondrial OS = Mus musculus GN = Ogdh |
| Q9DB77 | 7 | 0.014 | 1.43 | NEG | Cytochrome b-c1 complex subunit 2_ mitochondrial OS = Mus musculus GN = Uqcrc2 |
| Q99L13 | 3 | 0.003 | 1.42 | NEG | 3-hydroxyisobutyrate dehydrogenase_ mitochondrial OS = Mus musculus GN = Hibadh |
| P08249 | 13 | 0.019 | 1.42 | NEG | Malate dehydrogenase_ mitochondrial OS = Mus musculus GN = Mdh2 |
| Q9DCW4 | 4 | 0.017 | 1.42 | NEG | Electron transfer flavoprotein subunit beta OS = Mus musculus GN = Etfb |
| Q99LC5 | 9 | 0.011 | 1.42 | NEG | Electron transfer flavoprotein subunit alpha_ mitochondrial OS = Mus musculus GN = Etfa |
| Q9CR62 | 2 | 0.003 | 1.41 | NEG | Mitochondrial 2-oxoglutarate/malate carrier protein OS = Mus musculus GN = Slc25a11 |
| P00405 | 4 | 0.046 | 1.41 | NEG | Cytochrome c oxidase subunit 2 OS = Mus musculus GN = Mtco2 |
| Q922Q1 | 2 | 0.034 | 1.41 | NEG | Mitochondrial amidoxime reducing component 2 OS = Mus musculus GN = Marc2 |
| P47738 | 8 | 0.016 | 1.40 | NEG | Aldehyde dehydrogenase_ mitochondrial OS = Mus musculus GN = Aldh2 |
| Q9D9V3 | 4 | 0.022 | 1.40 | NEG | Ethylmalonyl-CoA decarboxylase OS = Mus musculus GN = Echdc1 |
| Increased in Pcyox1 silenced cells | |||||
| Q60847 | 4 | 0.001 | 1.99 | shPCYOX1 | Collagen alpha-1(XII) chain OS = Mus musculus GN = Col12a1 |
| P37889 | 16 | 0.004 | 1.42 | shPCYOX1 | Fibulin-2 OS = Mus musculus GN = Fbln2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banfi, C.; Mallia, A.; Ghilardi, S.; Brioschi, M.; Gianazza, E.; Eligini, S.; Sahlén, P.; Baetta, R. Prenylcysteine Oxidase 1 Is a Key Regulator of Adipogenesis. Antioxidants 2023, 12, 542. https://doi.org/10.3390/antiox12030542
Banfi C, Mallia A, Ghilardi S, Brioschi M, Gianazza E, Eligini S, Sahlén P, Baetta R. Prenylcysteine Oxidase 1 Is a Key Regulator of Adipogenesis. Antioxidants. 2023; 12(3):542. https://doi.org/10.3390/antiox12030542
Chicago/Turabian StyleBanfi, Cristina, Alice Mallia, Stefania Ghilardi, Maura Brioschi, Erica Gianazza, Sonia Eligini, Pelin Sahlén, and Roberta Baetta. 2023. "Prenylcysteine Oxidase 1 Is a Key Regulator of Adipogenesis" Antioxidants 12, no. 3: 542. https://doi.org/10.3390/antiox12030542
APA StyleBanfi, C., Mallia, A., Ghilardi, S., Brioschi, M., Gianazza, E., Eligini, S., Sahlén, P., & Baetta, R. (2023). Prenylcysteine Oxidase 1 Is a Key Regulator of Adipogenesis. Antioxidants, 12(3), 542. https://doi.org/10.3390/antiox12030542








