Pleiotropic Roles of a KEAP1-Associated Deubiquitinase, OTUD1
Abstract
1. Introduction
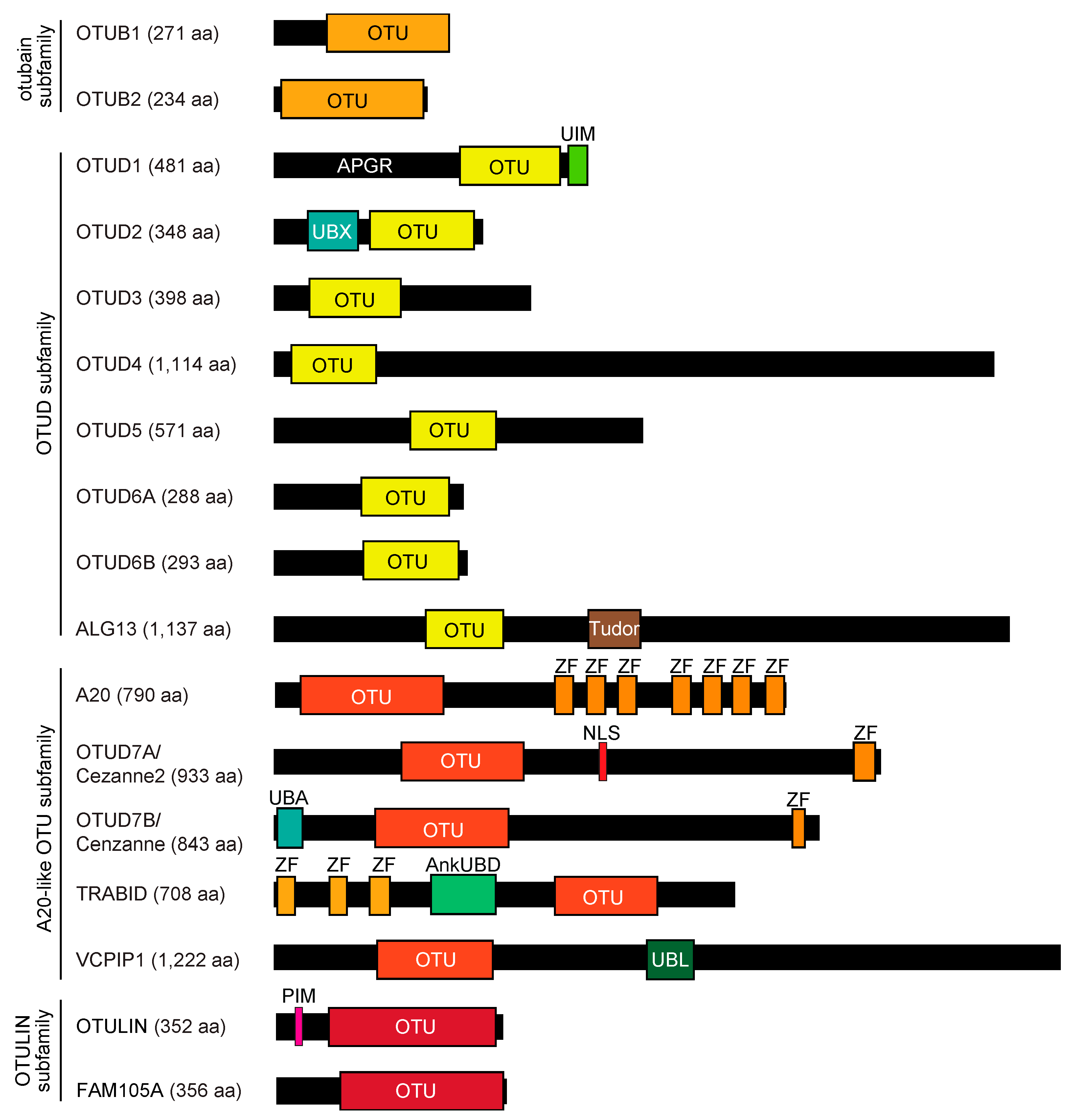
2. OTU Family DUBs
3. Pleiotropic Cellular Functions of OTUD1
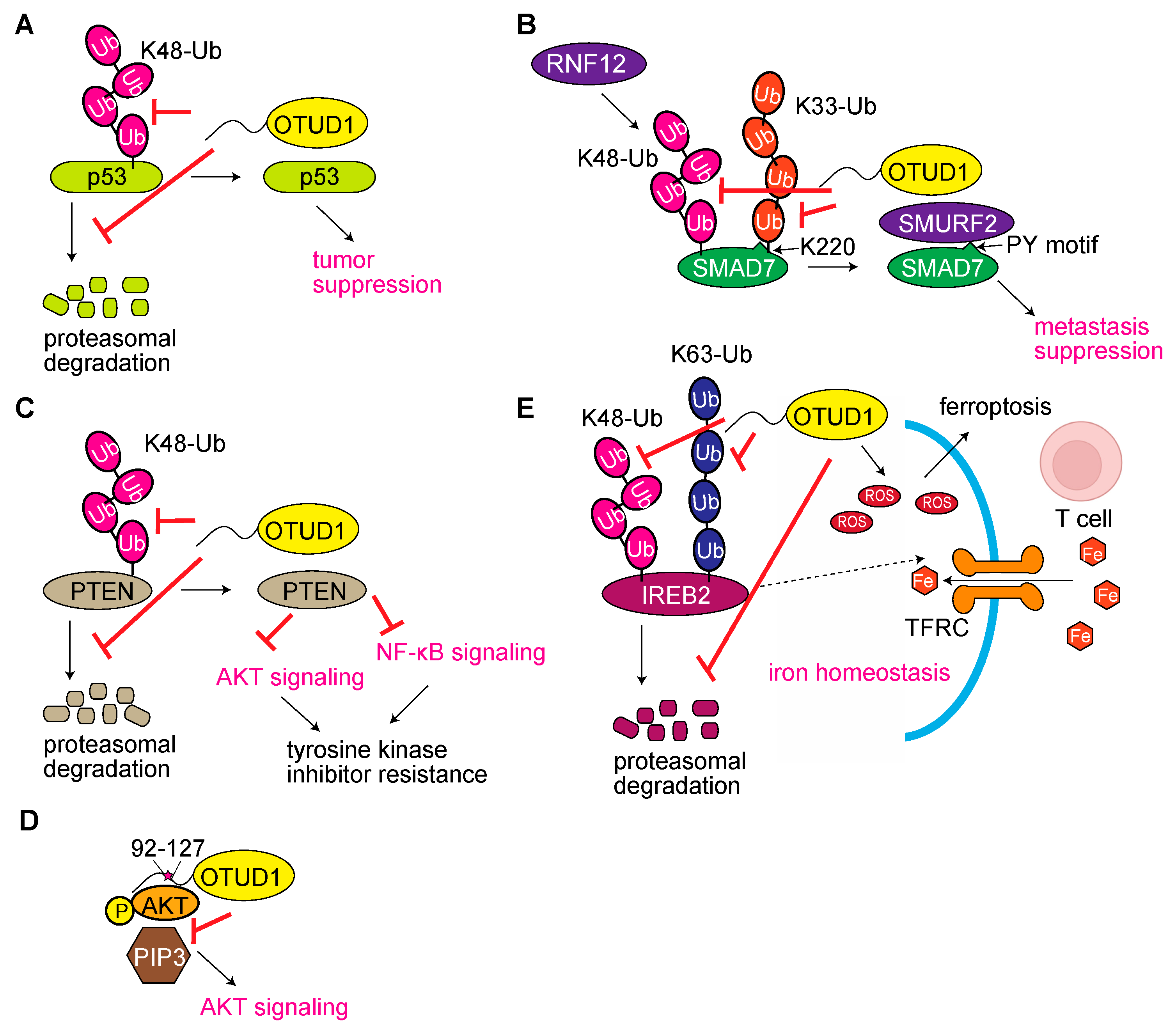
3.1. OTUD1 Functions as a Tumor Regulator
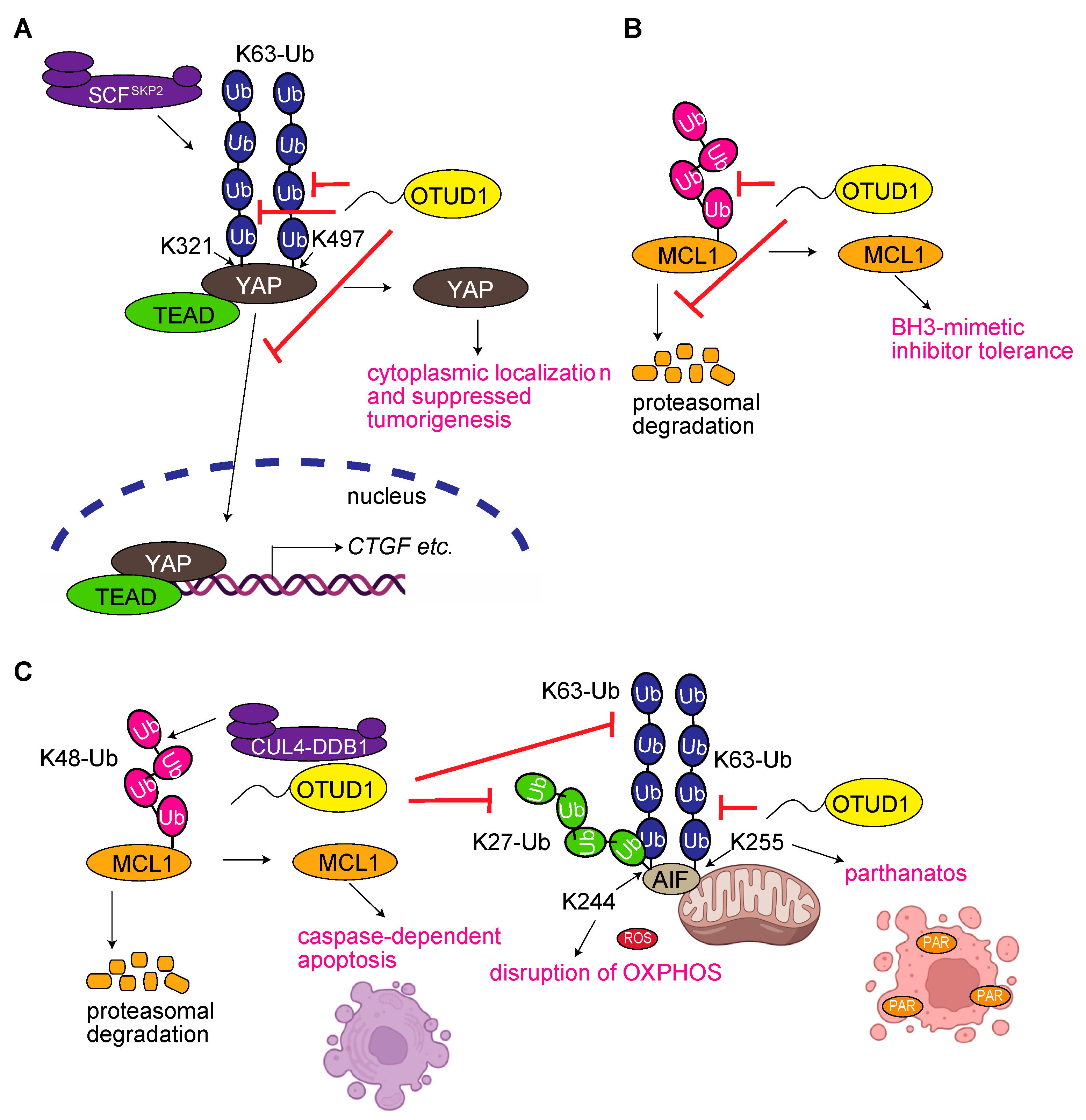
3.2. OTUD1 Regulates Yes-Associated Protein (YAP) Signaling
3.3. OTUD1 Regulates the Degradation of Myeloid Cell Leukemia 1 (MCL1)
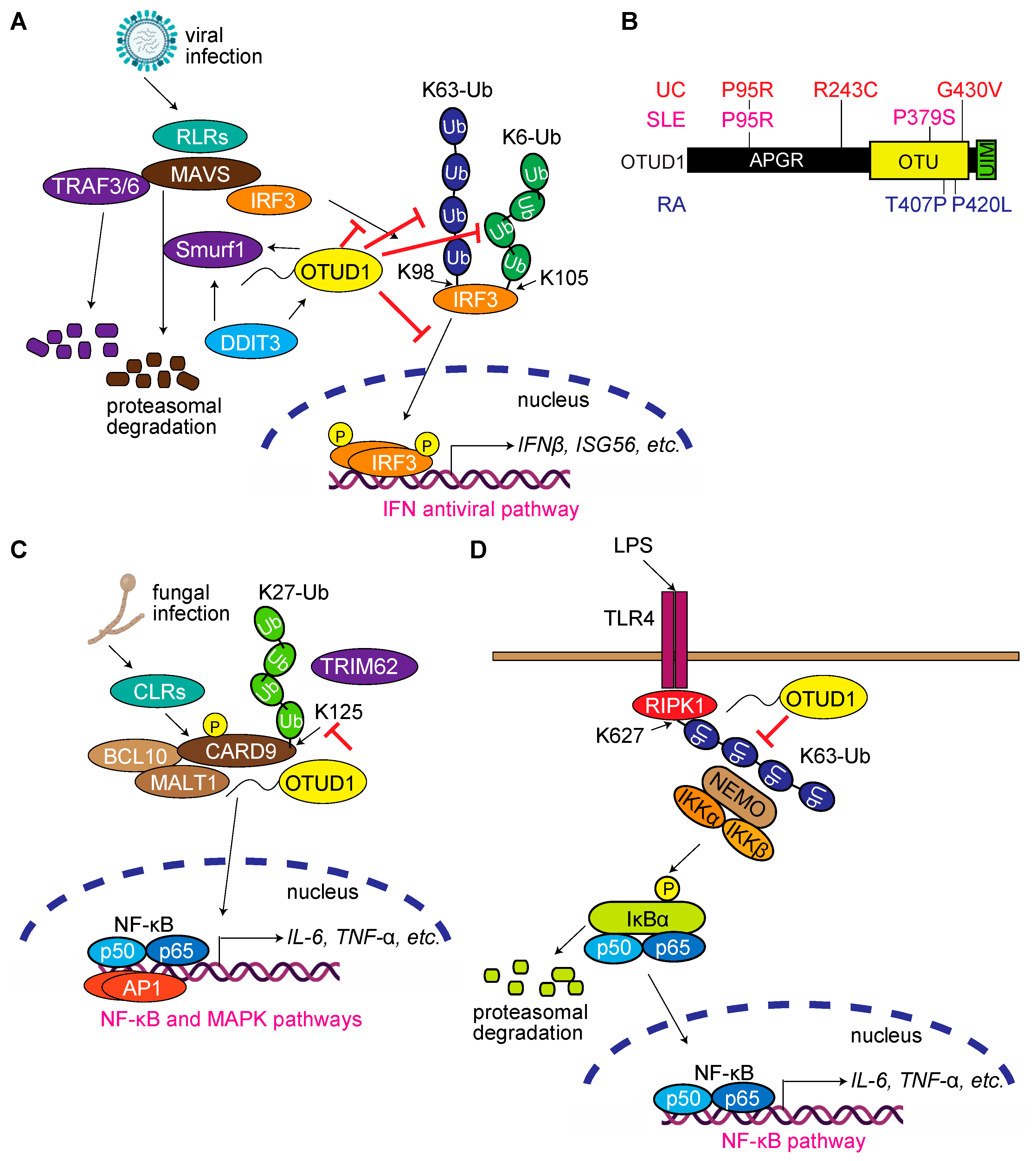
3.4. OTUD1 Regulates Antiviral and Innate Immune Responses, and Its Dysfunction Is Associated with Autoimmune and Inflammatory Diseases
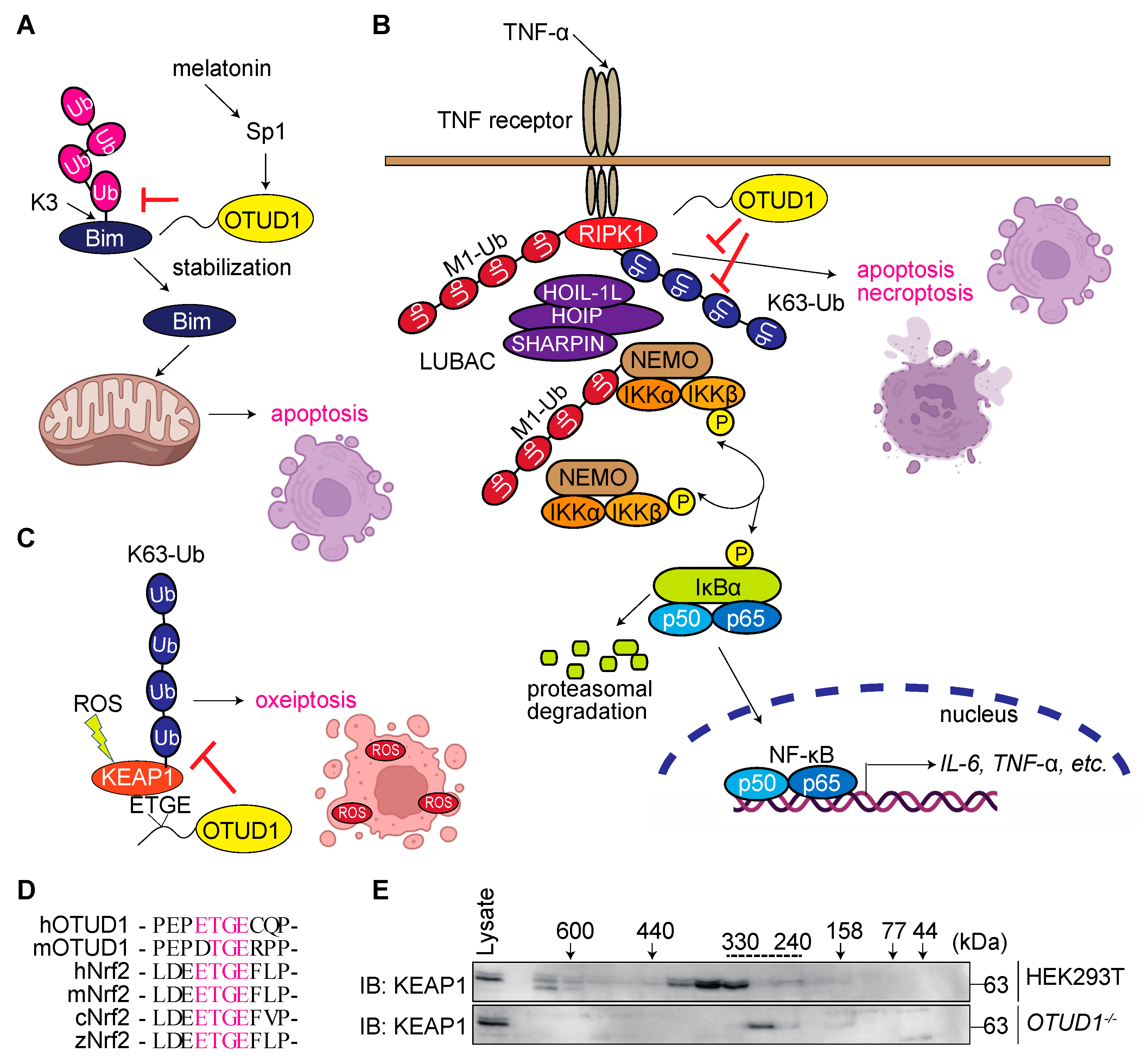
3.5. OTUD1 Regulates Cell Death Pathways
3.6. OTUD1 Regulates Acquired Immunity and Hematologic Disorders
3.7. OTUD1 Regulates Translation and RNA Metabolism
3.8. OTUD1 in Vascular Remodeling
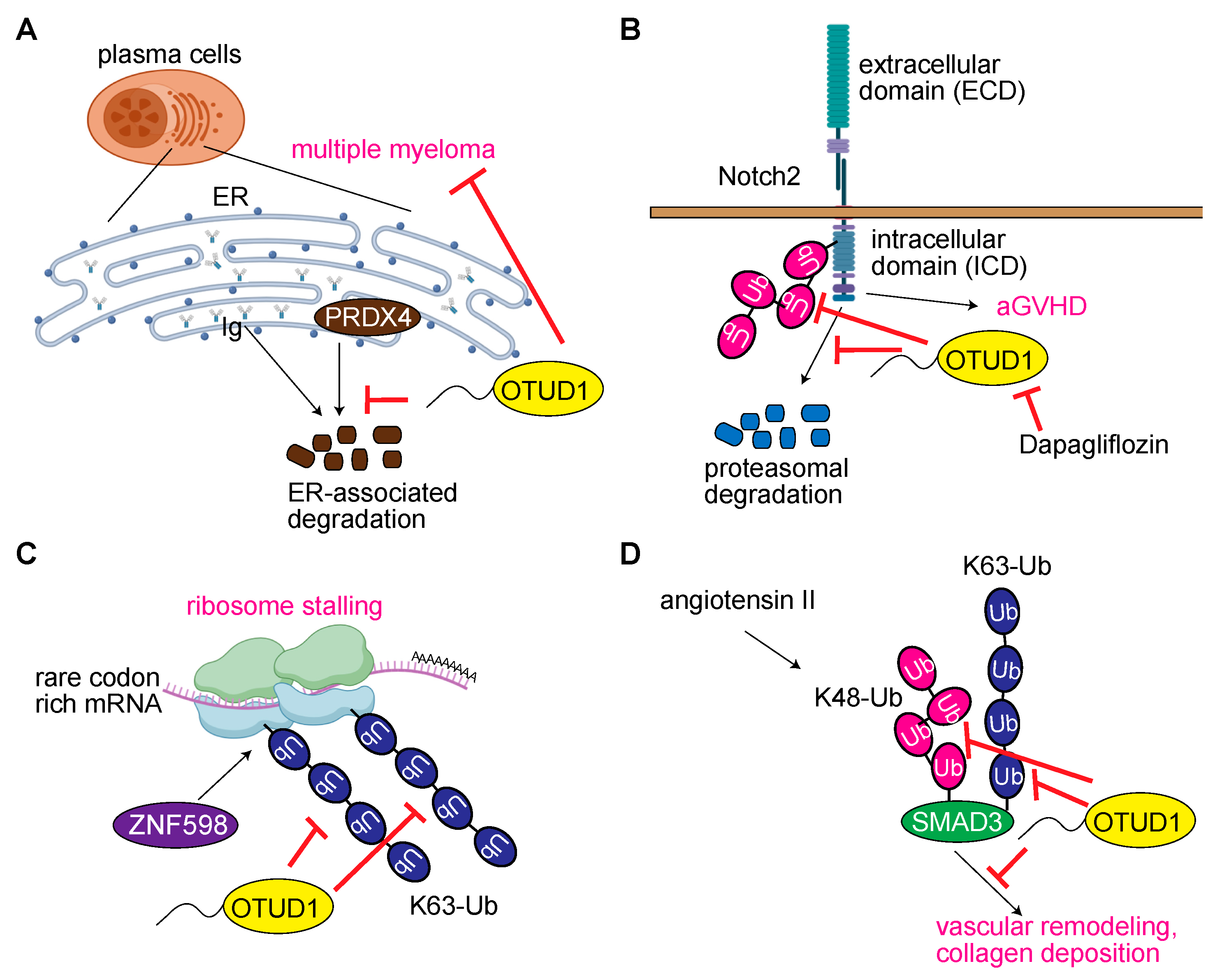
3.9. OTUD1 Is Involved in Stress Responses in a Wide Range of Species
3.10. Miscellaneous Functions of OTUD1
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Swatek, K.N.; Usher, J.L.; Kueck, A.F.; Gladkova, C.; Mevissen, T.E.T.; Pruneda, J.N.; Skern, T.; Komander, D. Insights into ubiquitin chain architecture using Ub-clipping. Nature 2019, 572, 533–537. [Google Scholar] [CrossRef] [PubMed]
- Kelsall, I.R. Non-lysine ubiquitylation: Doing things differently. Front. Mol. Biosci. 2022, 9, 1008175. [Google Scholar] [CrossRef] [PubMed]
- Sakamaki, J.-I.; Ode, K.L.; Kurikawa, Y.; Ueda, H.R.; Yamamoto, H.; Mizushima, N. Ubiquitination of phosphatidylethanolamine in organellar membranes. Mol. Cell 2022, 82, 3677–3692.e11. [Google Scholar] [CrossRef]
- Dikic, I.; Schulman, B.A. An expanded lexicon for the ubiquitin code. Nat. Rev. Mol. Cell Biol. 2023, in press. [Google Scholar] [CrossRef]
- Rehman, S.A.A.; Kristariyanto, Y.A.; Choi, S.-Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar] [CrossRef]
- Haahr, P.; Borgermann, N.; Guo, X.; Typas, D.; Achuthankutty, D.; Hoffmann, S.; Shearer, R.; Sixma, T.K.; Mailand, N. ZUFSP Deubiquitylates K63-Linked Polyubiquitin Chains to Promote Genome Stability. Mol. Cell 2018, 70, 165–174.e6. [Google Scholar] [CrossRef]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef]
- Mevissen, T.E.; Komander, D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 2017, 86, 159–192. [Google Scholar] [CrossRef]
- Bello, A.I.; Goswami, R.; Brown, S.L.; Costanzo, K.; Shores, T.; Allan, S.; Odah, R.; Mohan, R.D. Deubiquitinases in Neurodegeneration. Cells 2022, 11, 556. [Google Scholar] [CrossRef]
- Makarova, K.S.; Aravind, L.; Koonin, E.V. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci. 2000, 25, 50–52. [Google Scholar] [CrossRef] [PubMed]
- Freitas, F.B.; Frouco, G.; Martins, C.; Ferreira, F. African swine fever virus encodes for an E2-ubiquitin conjugating enzyme that is mono- and di-ubiquitinated and required for viral replication cycle. Sci. Rep. 2018, 8, 3471. [Google Scholar] [CrossRef] [PubMed]
- Mevissen, T.E.T.; Hospenthal, M.K.; Geurink, P.P.; Elliott, P.R.; Akutsu, M.; Arnaudo, N.; Ekkebus, R.; Kulathu, Y.; Wauer, T.; El Oualid, F.; et al. OTU Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis. Cell 2013, 154, 169–184. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Fu, L.; Sui, Y.; Zhang, L. The function and regulation of OTU deubiquitinases. Front. Med. 2020, 14, 542–563. [Google Scholar] [CrossRef]
- De Cesare, V.; Lopez, D.C.; Mabbitt, P.D.; Fletcher, A.J.; Soetens, M.; Antico, O.; Wood, N.T.; Virdee, S. Deubiquitinating enzyme amino acid profiling reveals a class of ubiquitin esterases. Proc. Natl. Acad. Sci. USA 2021, 118, e2006947118. [Google Scholar] [CrossRef]
- Ceccarelli, D.F.; Ivantsiv, S.; Mullin, A.A.; Coyaud, E.; Manczyk, N.; Maisonneuve, P.; Kurinov, I.; Zhao, L.; Go, C.; Gingras, A.-C.; et al. FAM105A/OTULINL Is a Pseudodeubiquitinase of the OTU-Class that Localizes to the ER Membrane. Structure 2019, 27, 1000–1012.e6. [Google Scholar] [CrossRef]
- Takahashi, H.; Yamanaka, S.; Kuwada, S.; Higaki, K.; Kido, K.; Sato, Y.; Fukai, S.; Tokunaga, F.; Sawasaki, T. A Human DUB Protein Array for Clarification of Linkage Specificity of Polyubiquitin Chain and Application to Evaluation of Its Inhibitors. Biomedicines 2020, 8, 152. [Google Scholar] [CrossRef]
- Deng, J.; Hou, G.; Fang, Z.; Liu, J.; Lv, X.D. Distinct expression and prognostic value of OTU domain-containing proteins in non-small-cell lung cancer. Oncol. Lett. 2019, 18, 5417–5427. [Google Scholar] [CrossRef]
- Oikawa, D.; Gi, M.; Kosako, H.; Shimizu, K.; Takahashi, H.; Shiota, M.; Hosomi, S.; Komakura, K.; Wanibuchi, H.; Tsuruta, D.; et al. OTUD1 deubiquitinase regulates NF-κB- and KEAP1-mediated inflammatory responses and reactive oxygen species-associated cell death pathways. Cell Death Dis. 2022, 13, 694. [Google Scholar] [CrossRef]
- Carneiro, A.P.; Reis, C.F.; Morari, E.C.; Maia, Y.C.P.; Nascimento, R.; Bonatto, J.M.C.; de Souza, M.; Goulart, L.R.; Ward, L.S. A putative OTU domain-containing protein 1 deubiquitinating enzyme is differentially expressed in thyroid cancer and identifies less-aggressive tumours. Br. J. Cancer 2014, 111, 551–558. [Google Scholar] [CrossRef]
- Piao, S.; Pei, H.Z.; Huang, B.; Baek, S.-H. Ovarian tumor domain-containing protein 1 deubiquitinates and stabilizes p53. Cell. Signal. 2017, 33, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Fan, Y.; Xie, F.; Zhou, H.; Jin, K.; Shao, L.; Shi, W.; Fang, P.; Yang, B.; van Dam, H.; et al. Breast cancer metastasis suppressor OTUD1 deubiquitinates SMAD7. Nat. Commun. 2017, 8, 2116. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yan, B.; Yu, H.; Ren, J.; Peng, M.; Zhu, L.; Wang, Y.; Jin, X.; Yi, L. OTUD1 stabilizes PTEN to inhibit the PI3K/AKT and TNF-alpha/NF-kappaB signaling pathways and sensitize ccRCC to TKIs. Int. J. Biol. Sci. 2022, 18, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Wang, F.; Chen, Y.; Zheng, Q.; Xiong, J.; Lv, Q.; Wu, K.; Xiong, J.; Wei, L.; Li, D.; et al. The deubiquitinase OTUD1 noncanonically suppresses Akt activation through its N-terminal intrinsically disordered region. Cell Rep. 2023, 42, 111916. [Google Scholar] [CrossRef]
- Katsarou, A.; Pantopoulos, K. Basics and principles of cellular and systemic iron homeostasis. Mol. Asp. Med. 2020, 75, 100866. [Google Scholar] [CrossRef]
- Song, J.; Liu, T.; Yin, Y.; Zhao, W.; Lin, Z.; Yin, Y.; Lu, D.; You, F. The deubiquitinase OTUD1 enhances iron transport and potentiates host antitumor immunity. EMBO Rep. 2021, 22, e51162. [Google Scholar] [CrossRef]
- Ma, X.; Wang, L.; Shi, G.; Sun, S. The deubiquitinase OTUD1 inhibits non-small cell lung cancer progression by deubiquitinating and stabilizing KLF4. Thorac. Cancer 2022, 13, 761–770. [Google Scholar] [CrossRef]
- Qadir, F.; Aziz, M.A.; Sari, C.P.; Ma, H.; Dai, H.; Wang, X.; Raithatha, D.; Da Silva, L.G.L.; Hussain, M.; Poorkasreiy, S.P.; et al. Transcriptome reprogramming by cancer exosomes: Identification of novel molecular targets in matrix and immune modulation. Mol. Cancer 2018, 17, 97. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Y.; Zhang, C.; Zhang, C.; Guan, X.; Jia, W. Differences of macrophages in the tumor microenvironment as an underlying key factor in glioma patients. Front. Immunol. 2022, 13, 1028937. [Google Scholar] [CrossRef]
- Yu, F.-X.; Zhao, B.; Guan, K.L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015, 163, 811–828. [Google Scholar] [CrossRef]
- Yao, F.; Zhou, Z.; Kim, J.; Hang, Q.; Xiao, Z.; Ton, B.N.; Chang, L.; Liu, N.; Zeng, L.; Wang, W.; et al. SKP2- and OTUD1-regulated non-proteolytic ubiquitination of YAP promotes YAP nuclear localization and activity. Nat. Commun. 2018, 9, 2269. [Google Scholar] [CrossRef] [PubMed]
- Grattarola, M.; Cucci, M.A.; Roetto, A.; Dianzani, C.; Barrera, G.; Pizzimenti, S. Post-translational down-regulation of Nrf2 and YAP proteins, by targeting deubiquitinases, reduces growth and chemoresistance in pancreatic cancer cells. Free. Radic. Biol. Med. 2021, 174, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhong, L.; Lu, Y.; Liu, X.; Wei, J.; Ding, Y.; Huang, H.; Nie, Q.; Liao, X. Deubiquitylase OTUD1 confers Erlotinib sensitivity in non-small cell lung cancer through inhibition of nuclear translocation of YAP1. Cell Death Discov. 2022, 8, 403. [Google Scholar] [CrossRef] [PubMed]
- Winder, M.L.; Campbell, K.J. MCL-1 is a clinically targetable vulnerability in breast cancer. Cell Cycle 2022, 21, 1439–1455. [Google Scholar] [CrossRef]
- Wu, L.; Lin, Y.; Feng, J.; Qi, Y.; Wang, X.; Lin, Q.; Shi, W.; Zheng, E.; Wang, W.; Hou, Z.; et al. The deubiquitinating enzyme OTUD1 antagonizes BH3-mimetic inhibitor induced cell death through regulating the stability of the MCL1 protein. Cancer Cell Int. 2019, 19, 222. [Google Scholar] [CrossRef]
- Luo, Q.; Wu, X.; Zhao, P.; Nan, Y.; Chang, W.; Zhu, X.; Su, D.; Liu, Z. OTUD1 Activates Caspase-Independent and Caspase-Dependent Apoptosis by Promoting AIF Nuclear Translocation and MCL1 Degradation. Adv. Sci. 2021, 8, 2002874. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, J.; Qian, L.; Feng, Q.; Wang, X.; Yuan, Y.; Zuo, Y.; Cheng, Q.; Miao, Y.; Guo, T.; et al. Induction of OTUD1 by RNA viruses potently inhibits innate immune responses by promoting degradation of the MAVS/TRAF3/TRAF6 signalosome. PLoS Pathog. 2018, 14, e1007067. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, D.; Wang, P.; Zhao, Y.; You, F. OTUD1 Negatively Regulates Type I IFN Induction by Disrupting Noncanonical Ubiquitination of IRF3. J. Immunol. 2020, 204, 1904–1918. [Google Scholar] [CrossRef]
- Wang, S.; Hou, P.; Pan, W.; He, W.; He, D.C.; Wang, H.; He, H. DDIT3 Targets Innate Immunity via the DDIT3-OTUD1-MAVS Pathway to Promote Bovine Viral Diarrhea Virus Replication. J. Virol. 2021, 95, e02351-20. [Google Scholar] [CrossRef]
- Zhang, H.-G.; Bin Wang, B.; Yang, Y.; Liu, X.; Wang, J.; Xin, N.; Li, S.; Miao, Y.; Wu, Q.; Guo, T.; et al. Depression compromises antiviral innate immunity via the AVP-AHI1-Tyk2 axis. Cell Res. 2022, 32, 897–913. [Google Scholar] [CrossRef]
- Lu, D.; Song, J.; Sun, Y.; Qi, F.; Liu, L.; Jin, Y.; McNutt, M.A.; Yin, Y. Mutations of deubiquitinase OTUD1 are associated with autoimmune disorders. J. Autoimmun. 2018, 94, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiang, B.; Hao, H.; Liu, Z. CARD9 Signaling, Inflammation, and Diseases. Front. Immunol. 2022, 13, 880879. [Google Scholar] [CrossRef]
- Cao, Z.; Conway, K.L.; Heath, R.J.; Rush, J.S.; Leshchiner, E.S.; Ramirez-Ortiz, Z.G.; Nedelsky, N.B.; Huang, H.; Ng, A.; Gardet, A.; et al. Ubiquitin Ligase TRIM62 Regulates CARD9-Mediated Anti-fungal Immunity and Intestinal Inflammation. Immunity 2015, 43, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, H.; Wang, X.; Shao, Z.; Li, Y.; Zhao, G.; Liu, F.; Liu, B.; Zheng, Y.; Chen, T.; et al. OTUD1 Regulates Antifungal Innate Immunity through Deubiquitination of CARD9. J. Immunol. 2021, 206, 1832–1843. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Qiang, L.; Zhang, Y.; Fu, Y.; Zhao, M.; Lei, Z.; Lu, Z.; Wei, Y.-G.; Dai, H.; Ge, Y.; et al. The deubiquitinase OTUD1 inhibits colonic inflammation by suppressing RIPK1-mediated NF-κB signaling. Cell Mol. Immunol. 2022, 19, 276–289. [Google Scholar] [CrossRef]
- Xie, J.; Li, H.; Li, S.; Li, J.; Li, Y. Molecular Mechanism of Sevoflurane Preconditioning Based on Whole-transcriptome Sequencing of Lipopolysaccharide-induced Cardiac Dysfunction in Mice. J. Cardiovasc. Pharmacol. 2022, 79, 846–857. [Google Scholar] [CrossRef] [PubMed]
- Buzzatto-Leite, I.; Afonso, J.; Silva-Vignato, B.; Coutinho, L.; Alvares, L. Differential gene co-expression network analyses reveal novel molecules associated with transcriptional dysregulation of key biological processes in osteoarthritis knee cartilage. Osteoarthr. Cartil. Open 2022, 4, 100316. [Google Scholar] [CrossRef] [PubMed]
- Sionov, R.V.; Vlahopoulos, S.A.; Granot, Z. Regulation of Bim in Health and Disease. Oncotarget 2015, 6, 23058–23134. [Google Scholar] [CrossRef]
- Woo, S.M.; Seo, S.U.; Min, K.; Kwon, T.K. Melatonin induces apoptotic cell death through Bim stabilization by Sp1-mediated OTUD1 upregulation. J. Pineal Res. 2022, 72, e12781. [Google Scholar] [CrossRef]
- Oikawa, D.; Sato, Y.; Ito, H.; Tokunaga, F. Linear Ubiquitin Code: Its Writer, Erasers, Decoders, Inhibitors, and Implications in Disorders. Int. J. Mol. Sci. 2020, 21, 3381. [Google Scholar] [CrossRef]
- Vdovin, A.; Jelinek, T.; Zihala, D.; Sevcikova, T.; Durech, M.; Sahinbegovic, H.; Snaurova, R.; Radhakrishnan, D.; Turi, M.; Chyra, Z.; et al. The deubiquitinase OTUD1 regulates immunoglobulin production and proteasome inhibitor sensitivity in multiple myeloma. Nat. Commun. 2022, 13, 6820. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Wang, D.; Lai, X.; Liu, Y.; Zuo, Y.; Zhang, W.; Lei, L.; Chen, J.; Liu, H.; Wang, Y.; et al. The OTUD1-Notch2-ICD axis orchestrates allogeneic T cell-mediated graft-versus-host disease. Blood 2023, in press. [Google Scholar] [CrossRef]
- Dougherty, S.E.; Maduka, A.O.; Inada, T.; Silva, G.M. Expanding Role of Ubiquitin in Translational Control. Int. J. Mol. Sci. 2020, 21, 1151. [Google Scholar] [CrossRef] [PubMed]
- Narita, M.; Denk, T.; Matsuo, Y.; Sugiyama, T.; Kikuguchi, C.; Ito, S.; Sato, N.; Suzuki, T.; Hashimoto, S.; Machová, I.; et al. A distinct mammalian disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation. Nat. Commun. 2022, 13, 6411. [Google Scholar] [CrossRef]
- Garshott, D.M.; Sundaramoorthy, E.; Leonard, M.; Bennett, E.J. Distinct regulatory ribosomal ubiquitylation events are reversible and hierarchically organized. Elife 2020, 9, e54023. [Google Scholar] [CrossRef]
- Snaurova, R.; Vdovin, A.; Durech, M.; Nezval, J.; Zihala, D.; Jelinek, T.; Hajek, R.; Simicek, M. Deubiquitinase OTUD1 Resolves Stalled Translation on polyA and Rare Codon Rich mRNAs. Mol. Cell. Biol. 2022, 42, e0026522. [Google Scholar] [CrossRef]
- Huang, Z.; Shen, S.; Wang, M.; Li, W.; Wu, G.; Huang, W.; Luo, W.; Liang, G. Mouse endothelial OTUD1 promotes angiotensin II-induced vascular remodeling by deubiquitinating SMAD3. EMBO Rep. 2023, in press. [Google Scholar] [CrossRef]
- Skugor, A.; Kjos, N.P.; Sundaram, A.Y.M.; Mydland, L.T.; Ånestad, R.; Tauson, A.-H.; Øverland, M. Effects of long-term feeding of rapeseed meal on skeletal muscle transcriptome, production efficiency and meat quality traits in Norwegian Landrace growing-finishing pigs. PLoS ONE 2019, 14, e0220441. [Google Scholar] [CrossRef]
- Gondret, F.; Le Floc’H, N.; Batonon-Alavo, D.; Perruchot, M.-H.; Mercier, Y.; Lebret, B. Flash dietary methionine supply over growth requirements in pigs: Multi-facetted effects on skeletal muscle metabolism. Animal 2021, 15, 100268. [Google Scholar] [CrossRef]
- Maharjan, P.; Beitia, A.; Weil, J.; Suesuttajit, N.; Hilton, K.; Caldas, J.; Umberson, C.; Martinez, D.; Kong, B.; Owens, C.M.; et al. Woody breast myopathy broiler show age-dependent adaptive differential gene expression in Pectoralis major and altered in-vivo triglyceride kinetics in adipogenic tissues. Poult. Sci. 2021, 100, 101092. [Google Scholar] [CrossRef]
- Ren, J.; Long, Y.; Liu, R.; Song, G.; Li, Q.; Cui, Z. Characterization of Biological Pathways Regulating Acute Cold Resistance of Zebrafish. Int. J. Mol. Sci. 2021, 22, 3028. [Google Scholar] [CrossRef] [PubMed]
- Cozier, Y.C.; Ruiz-Narvaez, E.; McKinnon, C.J.; Berman, J.S.; Rosenberg, L.; Palmer, J.R. Fine-mapping in African-American women confirms the importance of the 10p12 locus to sarcoidosis. Genes Immun. 2012, 13, 573–578. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oikawa, D.; Shimizu, K.; Tokunaga, F. Pleiotropic Roles of a KEAP1-Associated Deubiquitinase, OTUD1. Antioxidants 2023, 12, 350. https://doi.org/10.3390/antiox12020350
Oikawa D, Shimizu K, Tokunaga F. Pleiotropic Roles of a KEAP1-Associated Deubiquitinase, OTUD1. Antioxidants. 2023; 12(2):350. https://doi.org/10.3390/antiox12020350
Chicago/Turabian StyleOikawa, Daisuke, Kouhei Shimizu, and Fuminori Tokunaga. 2023. "Pleiotropic Roles of a KEAP1-Associated Deubiquitinase, OTUD1" Antioxidants 12, no. 2: 350. https://doi.org/10.3390/antiox12020350
APA StyleOikawa, D., Shimizu, K., & Tokunaga, F. (2023). Pleiotropic Roles of a KEAP1-Associated Deubiquitinase, OTUD1. Antioxidants, 12(2), 350. https://doi.org/10.3390/antiox12020350