Highly Selective MIF Ketonase Inhibitor KRP-6 Diminishes M1 Macrophage Polarization and Metabolic Reprogramming
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Compound
2.2. Leukocyte Isolation
2.3. Chemotaxis Assay
2.4. Apoptosis Assay
2.5. Cell Culture and Treatments
2.6. Measurements of Free Radical Scavenging Activity
2.7. Determination of ROS
2.8. Nitrite Measurement
2.9. TNF-α Production
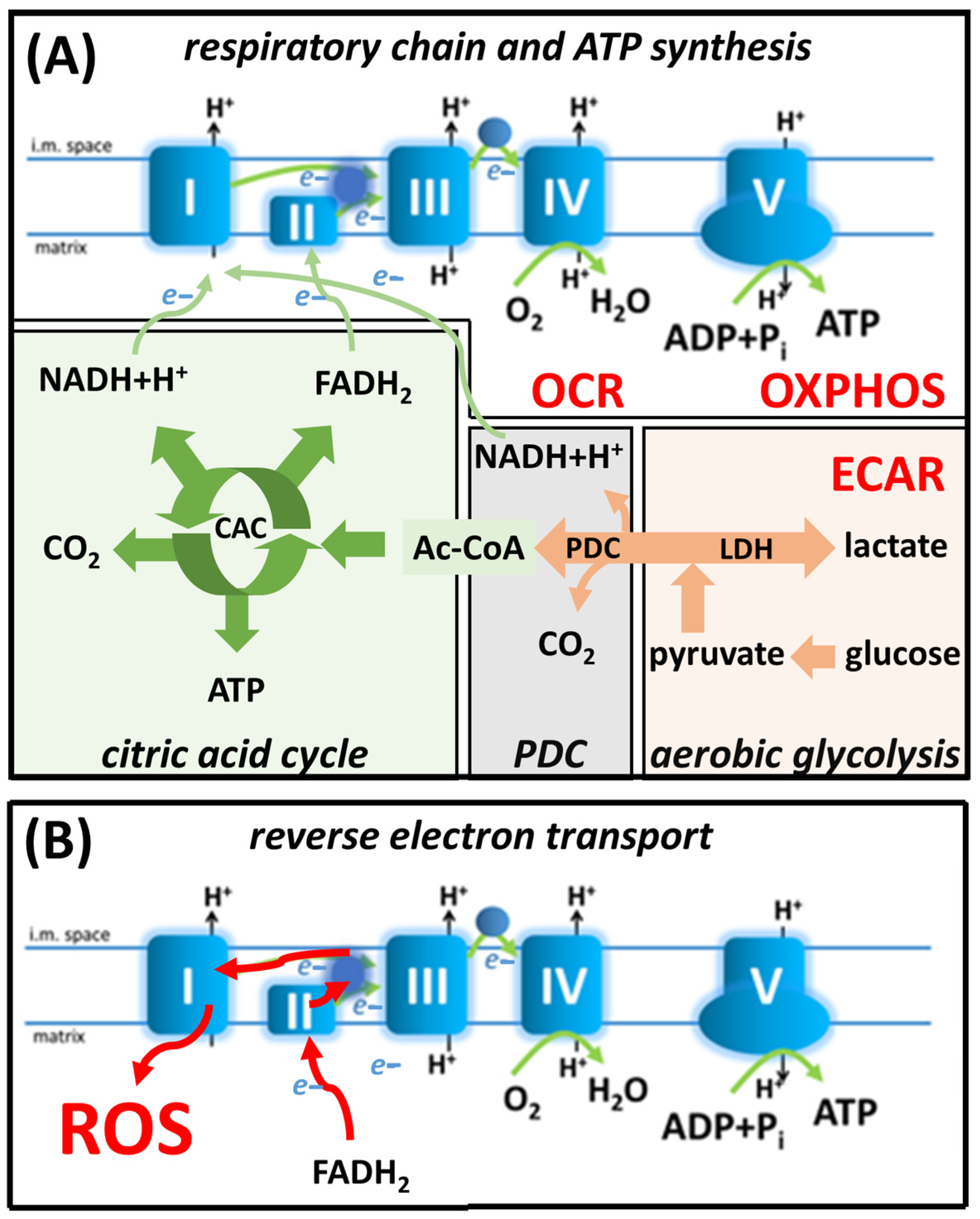
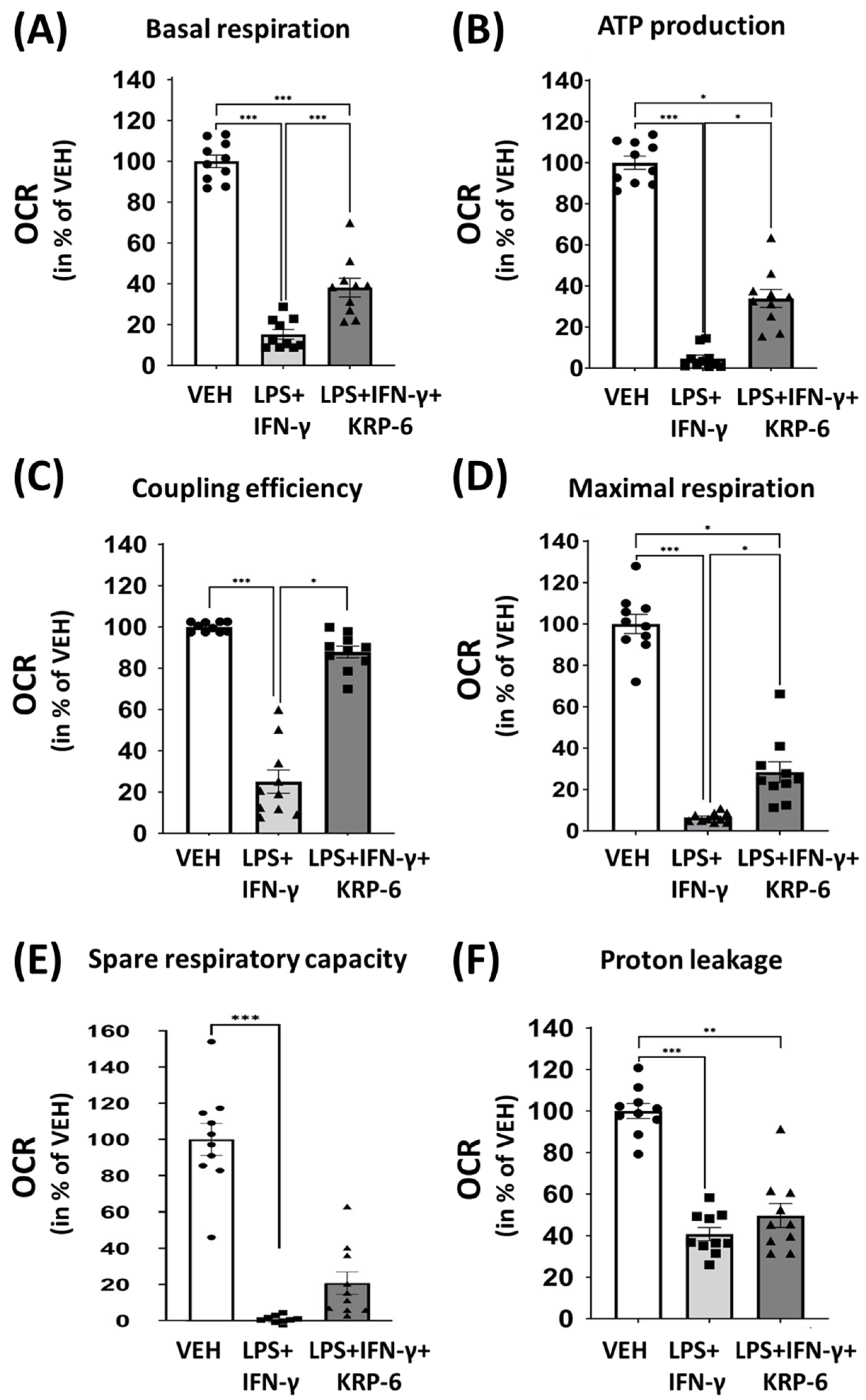
2.10. Measurements of OCR, ECAR and Mitochondrial Bioenergetics Parameters
2.11. RNA Isolation and qPCR
2.12. Statistical Analyses
3. Results
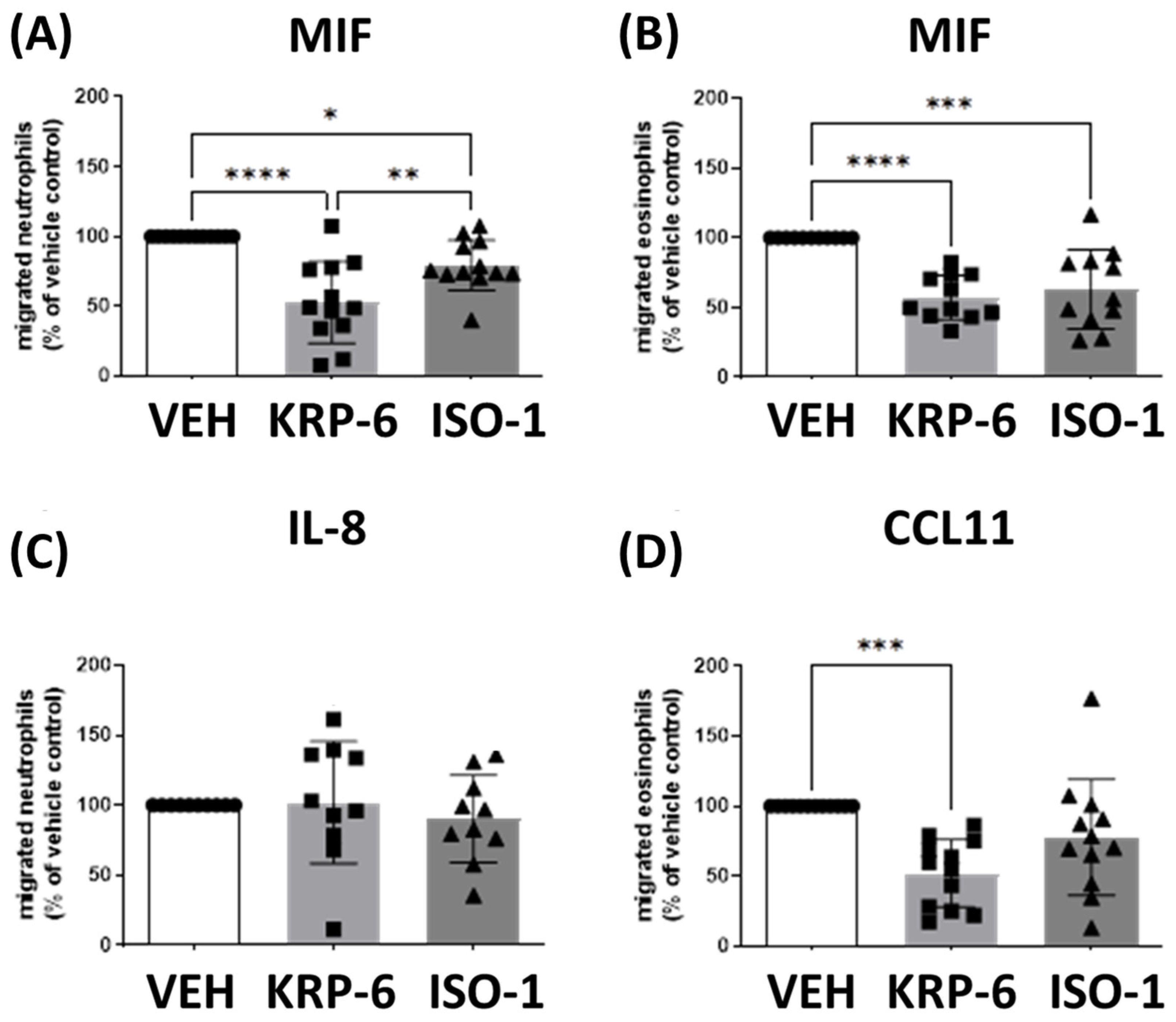
3.1. KRP-6 Inhibited Leukocyte Migration Similarly to ISO-1
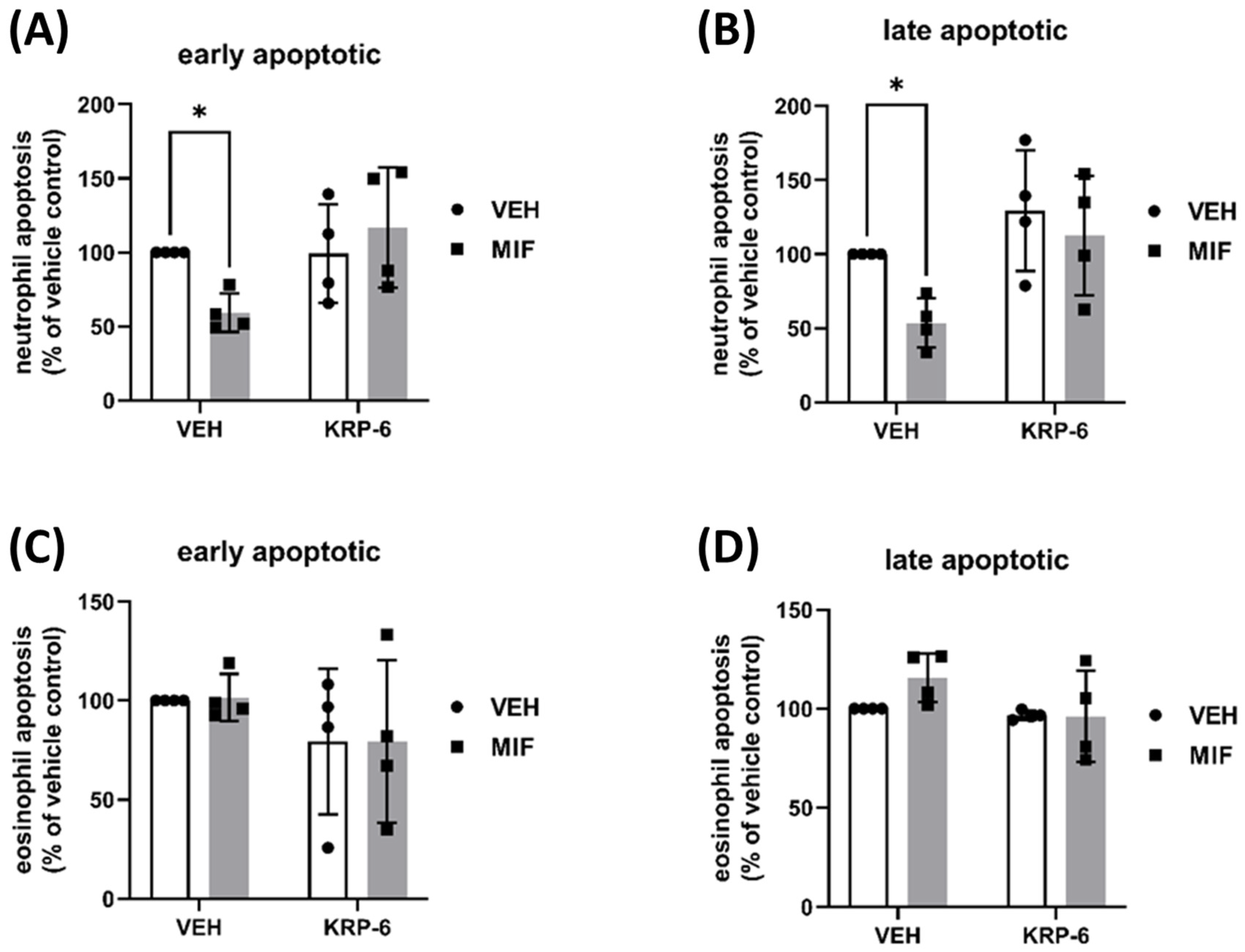
3.2. KRP-6 Counteracts the Anti-Apoptotic Effect of MIF in Human Neutrophils and Eosinophils
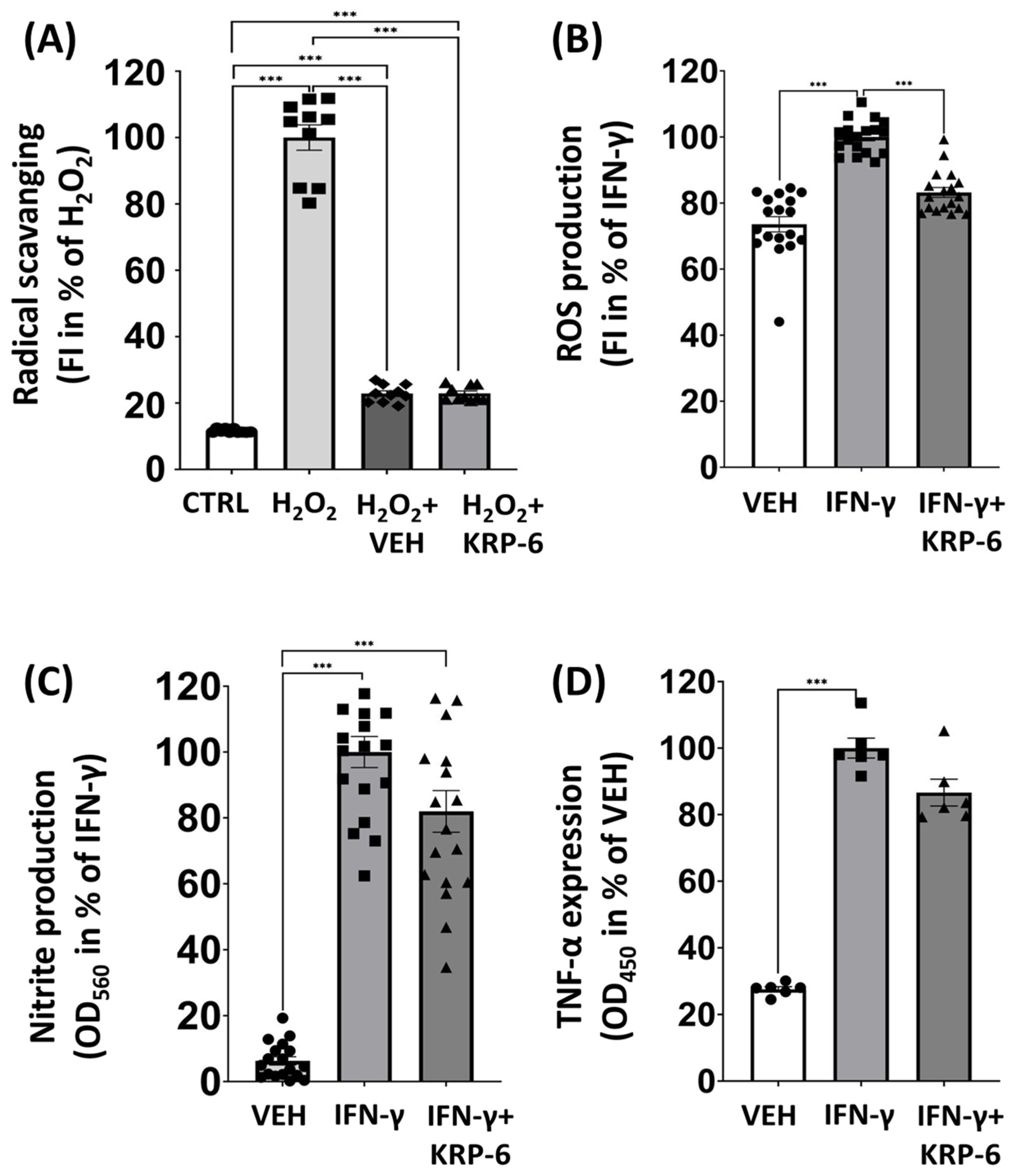
3.3. KRP-6 Reduces ROS in Macrophages without a Direct Antioxidant Effect
3.4. KRP-6 Reduces Glycolytic Flux in Activated Macrophages
3.5. KRP-6 Improves Mitochondrial Respiration of Macrophages
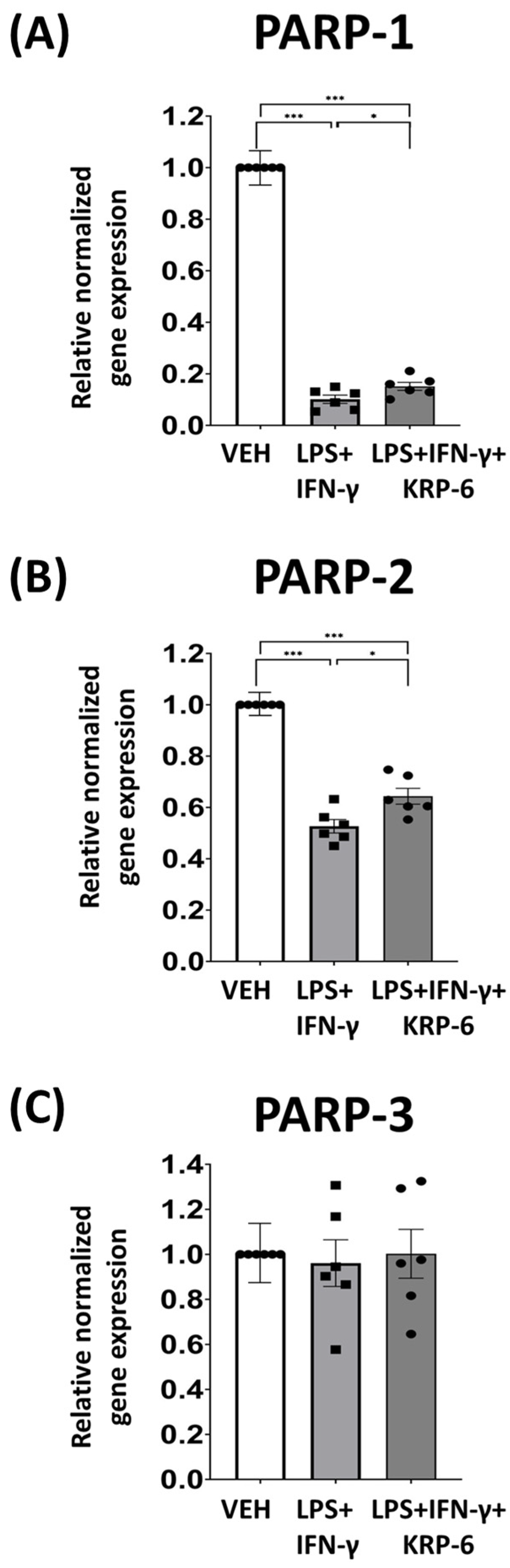
3.6. KRP-6 Diminishes M1 Macrophage Polarization-Associated PARP-1 and PARP-2 mRNA Downregulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holborough-Kerkvliet, M.D.; Kroos, S.; van de Wetering, R.; Toes, R.E.M. Addressing the key issue: Antigen-specific targeting of B cells in autoimmune diseases. Immunol. Lett. 2023, 259, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ye, F.; Xu, X.; Xu, P.; Wang, P.; Zheng, G.; Ye, G.; Yu, W.; Su, Z.; Lin, J.; et al. Targeting macrophage M1 polarization suppression through PCAF inhibition alleviates autoimmune arthritis via synergistic NF-κB and H3K9Ac blockade. J. Nanobiotechnol. 2023, 21, 280. [Google Scholar]
- Qu, R.; Zhou, M.; Qiu, Y.; Peng, Y.; Yin, X.; Liu, B.; Liu, B.; Bi, H.; Gao, Y.; Guo, D. Glucocorticoids improve the balance of M1/M2 macrophage polarization in experimental autoimmune uveitis through the P38MAPK-MEF2C axis. Int. Immunopharmacol. 2023, 120, 110392. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Jiang, L.; Hu, L.; Du, P.; Zhu, M.; Wu, H.; Zhao, M.; Lu, Q. Mivebresib alleviates systemic lupus erythematosus-associated diffuse alveolar hemorrhage via inhibiting infiltration of monocytes and M1 polarization of macrophages. Int. Immunopharmacol. 2023, 120, 110305. [Google Scholar] [CrossRef]
- Peng, Y.; Zhou, M.; Yang, H.; Qu, R.; Qiu, Y.; Hao, J.; Bi, H.; Guo, D. Regulatory Mechanism of M1/M2 Macrophage Polarization in the Development of Autoimmune Diseases. Mediat. Inflamm. 2023, 2023, 8821610. [Google Scholar] [CrossRef]
- Funes, S.C.; Rios, M.; Escobar-Vera, J.; Kalergis, A.M. Implications of macrophage polarization in autoimmunity. Immunology 2018, 154, 186–195. [Google Scholar] [CrossRef]
- Parisi, L.; Gini, E.; Baci, D.; Tremolati, M.; Fanuli, M.; Bassani, B.; Farronato, G.; Bruno, A.; Mortara, L. Macrophage Polarization in Chronic Inflammatory Diseases: Killers or Builders? J. Immunol. Res. 2018, 2018, 8917804. [Google Scholar] [CrossRef]
- Kim, H.; Park, H.-J.; Chang, H.W.; Back, J.H.; Lee, S.J.; Park, Y.E.; Kim, E.H.; Hong, Y.; Kwak, G.; Kwon, I.C.; et al. Exosome-guided direct reprogramming of tumor-associated macrophages from protumorigenic to antitumorigenic to fight cancer. Bioact. Mater. 2023, 25, 527–540. [Google Scholar] [CrossRef]
- Song, J.; Zeng, J.; Zheng, S.; Jiang, N.; Wu, A.; Guo, S.; Ye, R.; Hu, L.; Huang, F.; Wang, L.; et al. Sanguisorba officinalis L. promotes diabetic wound healing in rats through inflammation response mediated by macrophage. Phytother. Res. 2023, 37, 4265–4281. [Google Scholar] [CrossRef]
- Zhang, C.; Cheng, W.; Yang, T.; Fang, H.; Zhang, R. Lactate secreted by esophageal cancer cells induces M2 macrophage polarization via the AKT/ERK pathway. Thorac. Cancer 2023, 14, 2139–2148. [Google Scholar] [CrossRef]
- Chen, S.; Saeed, A.F.; Liu, Q.; Jiang, Q.; Xu, H.; Xiao, G.G.; Rao, L.; Duo, Y. Macrophages in immunoregulation and therapeutics. Signal Transduct. Target. Ther. 2023, 8, 207. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, A.J.; Elsawa, S.F. Macrophage Polarization States in the Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 6995. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Sheng, Y.; Wang, J.; Zhou, X.; Li, W.; Zhang, C.; Guo, L.; Yang, Y. NOX4 promotes mucosal barrier injury in inflammatory bowel disease by mediating macrophages M1 polarization through ROS. Int. Immunopharmacol. 2022, 104, 108361. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, D.; Zhang, T.; Ma, Y.; Tong, X.; Fan, H. The role of immunometabolism in macrophage polarization and its impact on acute lung injury/acute respiratory distress syndrome. Front. Immunol. 2023, 14, 1117548. [Google Scholar] [CrossRef]
- Hughey, C.C.; Puchalska, P.; Crawford, P.A. Integrating the contributions of mitochondrial oxidative metabolism to lipotoxicity and inflammation in NAFLD pathogenesis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2022, 1867, 159209. [Google Scholar] [CrossRef]
- Guan, S.; Zhao, L.; Peng, R. Mitochondrial Respiratory Chain Supercomplexes: From Structure to Function. Int. J. Mol. Sci. 2022, 23, 13880. [Google Scholar] [CrossRef]
- Greene, J.; Segaran, A.; Lord, S. Targeting OXPHOS and the electron transport chain in cancer; Molecular and therapeutic implications. Semin. Cancer Biol. 2022, 86 Pt 2, 851–859. [Google Scholar] [CrossRef]
- Zhao, W.; Xu, D.; Hong, W.; Zhang, L.; Wu, Q.; Gao, M.; Wang, J. Grossamide attenuates inflammation by balancing macrophage polarization through metabolic reprogramming of macrophages in mice. Int. Immunopharmacol. 2022, 112, 109190. [Google Scholar] [CrossRef]
- Mortezaee, K.; Majidpoor, J. Dysregulated metabolism: A friend-to-foe skewer of macrophages. Int. Rev. Immunol. 2023, 42, 287–303. [Google Scholar] [CrossRef]
- Bai, R.; Meng, Y.; Cui, J. Therapeutic strategies targeting metabolic characteristics of cancer cells. Crit. Rev. Oncol. Hematol. 2023, 187, 104037. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Li, X.; Chen, X.; Zhang, Y.; Liu, X.; Wu, S.; Li, Y.; Li, B. 2-DG Re-Normalized IFN-γ Production in T Cells Excluding TEMRA Cells from Patients with Aplastic Anemia. Immunol. Investig. 2023, 52, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Benmoussa, K.; Garaude, J.; Acín-Pérez, R. How Mitochondrial Metabolism Contributes to Macrophage Phenotype and Functions. J. Mol. Biol. 2018, 430, 3906–3921. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, F.; Fan, N.; Zhou, C.; Li, D.; Macvicar, T.; Dong, Q.; Bruns, C.J.; Zhao, Y. Targeting Glutaminolysis: New Perspectives to Understand Cancer Development and Novel Strategies for Potential Target Therapies. Front. Oncol. 2020, 10, 589508. [Google Scholar] [CrossRef] [PubMed]
- Loenarz, C.; Schofield, C.J. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem. Sci. 2011, 36, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.L.; Kelly, B.; Logan, A.; Costa, A.S.; Varma, M.; Bryant, C.; Tourlomousis, P.; Däbritz, J.H.M.; Gottlieb, E.; Latorre, I.; et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 2016, 167, 457–470.e13. [Google Scholar] [CrossRef]
- Murphy, M.P.; O’Neill, L.A.J. Krebs Cycle Reimagined: The Emerging Roles of Succinate and Itaconate as Signal Transducers. Cell 2018, 174, 780–784. [Google Scholar] [CrossRef]
- Ponath, V.; Kaina, B. Death of Monocytes through Oxidative Burst of Macrophages and Neutrophils: Killing in Trans. PLoS ONE 2017, 12, e0170347. [Google Scholar] [CrossRef]
- Herb, M.; Schramm, M. Functions of ROS in Macrophages and Antimicrobial Immunity. Antioxidants 2021, 10, 313. [Google Scholar] [CrossRef]
- Regdon, Z.; Robaszkiewicz, A.; Kovács, K.; Rygielska, Ż.; Hegedűs, C.; Bodoor, K.; Szabó, É.; Virág, L. LPS protects macrophages from AIF-independent parthanatos by downregulation of PARP1 expression, induction of SOD2 expression, and a metabolic shift to aerobic glycolysis. Free. Radic. Biol. Med. 2019, 131, 184–196. [Google Scholar] [CrossRef]
- Zhu, T.; Zheng, J.Y.; Huang, L.L.; Wang, Y.H.; Yao, D.F.; Dai, H.B. Human PARP1 substrates and regulators of its catalytic activity: An updated overview. Front. Pharmacol. 2023, 14, 1137151. [Google Scholar] [CrossRef]
- Kadam, A.; Jubin, T.; Roychowdhury, R.; Begum, R. Role of PARP-1 in mitochondrial homeostasis. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129669. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Yoon, S.P.; Kim, J. Poly(ADP-ribose) polymerase regulates glycolytic activity in kidney proximal tubule epithelial cells. Anat. Cell Biol. 2016, 49, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Andrabi, S.A.; Umanah, G.K.E.; Chang, C.; Stevens, D.A.; Karuppagounder, S.S.; Gagné, J.-P.; Poirier, G.G.; Dawson, V.L.; Dawson, T.M. Poly(ADP-ribose) polymerase-dependent energy depletion occurs through inhibition of glycolysis. Proc. Natl. Acad. Sci. USA 2014, 111, 10209–10214. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; An, R.; Umanah, G.K.; Park, H.; Nambiar, K.; Eacker, S.M.; Kim, B.; Bao, L.; Harraz, M.M.; Chang, C.; et al. A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science 2016, 354, aad6872. [Google Scholar] [CrossRef] [PubMed]
- Kleemann, R.; Kapurniotu, A.; Frank, R.W.; Gessner, A.; Mischke, R.; Flieger, O.; Jüttner, S.; Brunner, H.; Bernhagen, J. Disulfide analysis reveals a role for macrophage migration inhibitory factor (MIF) as thiol-protein oxidoreductase. J. Mol. Biol. 1998, 280, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Allam, V.S.R.R.; Pavlidis, S.; Liu, G.; Kermani, N.Z.; Simpson, J.; To, J.; Donnelly, S.; Guo, Y.-K.; Hansbro, P.M.; Phipps, S.; et al. Macrophage migration inhibitory factor promotes glucocorticoid resistance of neutrophilic inflammation in a murine model of severe asthma. Thorax 2023, 78, 661–673. [Google Scholar] [CrossRef]
- Lintomen, L.; Kluppel, L.M.; Kitoko, J.Z.; Montes-Cobos, E.; Vidal, V.M.; Tan, L.B.; de Farias, J.N.; de Souza, H.S.; Olsen, P.C.; Bozza, M.T. MIF is essential to the establishment of house dust mite-induced airway inflammation and tissue remodeling in mice. Eur. J. Immunol. 2023, 53, e2250016. [Google Scholar] [CrossRef]
- Bilsborrow, J.B.; Doherty, E.; Tilstam, P.V.; Bucala, R. Macrophage migration inhibitory factor (MIF) as a therapeutic target for rheumatoid arthritis and systemic lupus erythematosus. Expert Opin. Ther. Targets 2019, 23, 733–744. [Google Scholar] [CrossRef]
- Hao, H.; Hou, Y.; Li, A.; Niu, L.; Li, S.; He, B.; Zhang, X.; Song, H.; Cai, R.; Zhou, Y.; et al. HIF-1α promotes astrocytic production of macrophage migration inhibitory factor following spinal cord injury. CNS Neurosci. Ther. 2023, 1–13. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Z.; Yan, G.; Lin, C.; Luo, Y.; Ye, Y.; Zeng, X.; Yao, J. MIF promotes Th17 cell differentiation in Hashimoto’s thyroiditis by binding HVEM and activating NF-κB signaling pathway. Int. Immunopharmacol. 2023, 121, 110494. [Google Scholar] [CrossRef]
- de Souza, H.S.; Tortori, C.A.; Lintomen, L.; Figueiredo, R.T.; Bernardazzi, C.; Leng, L.; Bucala, R.; Madi, K.; Buongusto, F.; Elia, C.C.S.; et al. Macrophage migration inhibitory factor promotes eosinophil accumulation and tissue remodeling in eosinophilic esophagitis. Mucosal Immunol. 2015, 8, 1154–1165. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.L.; Fan, H.; Hall, P.; Ngo, D.; Mackay, C.R.; Fingerle-Rowson, G.; Bucala, R.; Hickey, M.J.; Morand, E.F. Macrophage migration inhibitory factor regulates neutrophil chemotactic responses in inflammatory arthritis in mice. Arthritis Rheum. 2011, 63, 960–970. [Google Scholar] [CrossRef]
- Cotzomi-Ortega, I.; Nieto-Yañez, O.; Juárez-Avelar, I.; Rojas-Sanchez, G.; Montes-Alvarado, J.B.; Reyes-Leyva, J.; Aguilar-Alonso, P.; Rodriguez-Sosa, M.; Maycotte, P. Autophagy inhibition in breast cancer cells induces ROS-mediated MIF expression and M1 macrophage polarization. Cell. Signal. 2021, 86, 110075. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, P.T.M.; Oliveira-Scussel, A.C.M.; Sousa, R.A.P.; Gomes, B.Q.; Félix, J.E.; Silva, R.J.; Millian, I.B.; Assunção, T.S.F.; Teixeira, S.C.; Gomes, M.d.L.M.; et al. Macrophage Migration Inhibitory Factor contributes to drive phenotypic and functional macrophages activation in response to Toxoplasma gondii infection. Immunobiology 2023, 228, 152357. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Li, D.; Wang, H.; Chen, K.; Wang, S.; Xu, J.; Ping, J. Exosomes from adipose-derived stem cells regulate M1/M2 macrophage phenotypic polarization to promote bone healing via miR-451a/MIF. Stem Cell Res. Ther. 2022, 13, 149. [Google Scholar] [CrossRef]
- Caltabiano, R.; De Pasquale, R.; Piombino, E.; Campo, G.; Nicoletti, F.; Cavalli, E.; Mangano, K.; Fagone, P. Macrophage Migration Inhibitory Factor (MIF) and Its Homologue d-Dopachrome Tautomerase (DDT) Inversely Correlate with Inflammation in Discoid Lupus Erythematosus. Molecules 2021, 26, 184. [Google Scholar] [CrossRef]
- Song, S.; Xiao, Z.; Dekker, F.J.; Poelarends, G.J.; Melgert, B.N. Macrophage migration inhibitory factor family proteins are multitasking cytokines in tissue injury. Cell. Mol. Life Sci. 2022, 79, 105. [Google Scholar] [CrossRef]
- Lubetsky, J.B.; Dios, A.; Han, J.; Aljabari, B.; Ruzsicska, B.; Mitchell, R.; Lolis, E.; Al-Abed, Y. The Tautomerase Active Site of Macrophage Migration Inhibitory Factor Is a Potential Target for Discovery of Novel Anti-inflammatory Agents. J. Biol. Chem. 2002, 277, 24976–24982. [Google Scholar] [CrossRef]
- Garai, J.; Radnai, B.; Vámos, E.; Kovács, D.; Vántus, V.B.; Rumbus, Z.; Pákai, E.; Garami, A.; Gulyás-Fekete, G.; Agócs, A.; et al. Synthesis and evaluation of a new class of MIF-inhibitors in activated macrophage cells and in experimental septic shock in mice. Eur. J. Med. Chem. 2023, 247, 115050. [Google Scholar] [CrossRef]
- Garai, J.; Lóránd, T.; Molnár, V. Ketone bodies affect the enzymatic activity of macrophage migration inhibitory factor. Life Sci. 2005, 77, 1375–1380. [Google Scholar] [CrossRef]
- Garai, J.; Krekó, M.; Őrfi, L.; Jakus, P.B.; Rumbus, Z.; Kéringer, P.; Garami, A.; Vámos, E.; Kovács, D.; Vántus, V.B.; et al. Tetralone derivatives are MIF tautomerase inhibitors and attenuate macrophage activation and amplify the hypothermic response in endotoxemic mice. J. Enzym. Inhib. Med. Chem. 2021, 36, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Das, U.; Lorand, T.; Dimmock, S.G.; Perjesi, P.; Dimmock, J.R. 3-benzylidene-4-chromanones: A novel cluster of anti-tubercular agents. J. Enzym. Inhib. Med. Chem. 2015, 30, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Roula, D.; Theiler, A.; Luschnig, P.; Sturm, G.J.; Tomazic, P.V.; Marsche, G.; Heinemann, A.; Sturm, E.M. Apolipoprotein A-IV acts as an endogenous anti-inflammatory protein and is reduced in treatment-naïve allergic patients and allergen-challenged mice. Allergy 2020, 75, 392–402. [Google Scholar] [CrossRef]
- Luschnig, P.; Kienzl, M.; Roula, D.; Pilic, J.; Atallah, R.; Heinemann, A.; Sturm, E.M. The JAK1/2 inhibitor baricitinib suppresses eosinophil effector function and restricts allergen-induced airway eosinophilia. Biochem. Pharmacol. 2021, 192, 114690. [Google Scholar] [CrossRef]
- Apak, R.; Calokerinos, A.; Gorinstein, S.; Segundo, M.A.; Hibbert, D.B.; Gülçin, I.; Çekiç, S.D.; Güçlü, K.; Özyürek, M.; Çelik, S.E.; et al. Methods to evaluate the scavenging activity of antioxidants toward reactive oxygen and nitrogen species (IUPAC Technical Report). Pure Appl. Chem. 2022, 94, 87–144. [Google Scholar] [CrossRef]
- Kang, K.S.; Kim, H.Y.; Pyo, J.S.; Yokozawa, T. Increase in the free radical scavenging activity of ginseng by heat-processing. Biol. Pharm. Bull. 2006, 29, 750–754. [Google Scholar] [CrossRef]
- Henderson, L.M.; Chappell, J.B. Dihydrorhodamine 123: A fluorescent probe for superoxide generation? Eur. J. Biochem. 1993, 217, 973–980. [Google Scholar] [CrossRef]
- Váradi, L.; Breedon, M.; Chen, F.F.; Trinchi, A.; Cole, I.S.; Wei, G. Evaluation of novel Griess-reagent candidates for nitrite sensing in aqueous media identified via molecular fingerprint searching. RSC Adv. 2019, 9, 3994–4000. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, W. Anti-Inflammatory Effect of Wogonin on RAW 264.7 Mouse Macrophages Induced with Polyinosinic-Polycytidylic Acid. Molecules 2015, 20, 6888–6900. [Google Scholar] [CrossRef]
- Desousa, B.R.; Kim, K.K.; Jones, A.E.; Ball, A.B.; Hsieh, W.Y.; Swain, P.; Morrow, D.H.; Brownstein, A.J.; Ferrick, D.A.; Shirihai, O.S.; et al. Calculation of ATP production rates using the Seahorse XF Analyzer. EMBO Rep. 2023, 8, 1154–1165. [Google Scholar] [CrossRef]
- Kovács, D.; Vántus, V.B.; Vámos, E.; Kálmán, N.; Schicho, R.; Gallyas, F.; Radnai, B. Olaparib: A Clinically Applied PARP Inhibitor Protects from Experimental Crohn’s Disease and Maintains Barrier Integrity by Improving Bioenergetics through Rescuing Glycolysis in Colonic Epithelial Cells. Oxidative Med. Cell. Longev. 2021, 2021, 7308897. [Google Scholar] [CrossRef] [PubMed]
- Noguera, N.I.; Tavian, D.; Angelini, C.; Cortese, F.; Filosto, M.; Garibaldi, M.; Missaglia, S.; Smigliani, A.; Zaza, A.; Pennisi, E.M. Effects of Triheptanoin on Mitochondrial Respiration and Glycolysis in Cultured Fibroblasts from Neutral Lipid Storage Disease Type M (NLSD-M) Patients. Biomolecules 2023, 13, 452. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Hall, P.; Santos, L.L.; Gregory, J.L.; Fingerle-Rowson, G.; Bucala, R.; Morand, E.F.; Hickey, M.J. Macrophage migration inhibitory factor and CD74 regulate macrophage chemotactic responses via mapk and rho gtpase. J. Immunol. 2011, 186, 4915–4924. [Google Scholar] [CrossRef] [PubMed]
- Dhayni, K.; Zibara, K.; Issa, H.; Kamel, S.; Bennis, Y. Targeting CXCR1 and CXCR2 receptors in cardiovascular diseases. Pharmacol. Ther. 2022, 237, 108257. [Google Scholar] [CrossRef]
- Nazarinia, D.; Behzadifard, M.; Gholampour, J.; Karimi, R.; Gholampour, M. Eotaxin-1 (CCL11) in neuroinflammatory disorders and possible role in COVID-19 neurologic complications. Acta Neurol. Belg. 2022, 122, 865–869. [Google Scholar] [CrossRef]
- Baumann, R.; Casaulta, C.; Simon, D.; Conus, S.; Yousefi, S.; Simon, H.U. Macrophage migration inhibitory factor delays apoptosis in neutrophils by inhibiting the mitochondria-dependent death pathway. FASEB J. 2003, 17, 2221–2230. [Google Scholar] [CrossRef]
- Schindler, L.; Zwissler, L.; Krammer, C.; Hendgen-Cotta, U.; Rassaf, T.; Hampton, M.B.; Dickerhof, N.; Bernhagen, J. Macrophage migration inhibitory factor inhibits neutrophil apoptosis by inducing cytokine release from mononuclear cells. J. Leukoc. Biol. 2021, 110, 893–905. [Google Scholar] [CrossRef]
- Kang, K.; Bachu, M.; Park, S.H.; Kang, K.; Bae, S.; Park-Min, K.-H.; Ivashkiv, L.B. IFN-γ selectively suppresses a subset of TLR4-activated genes and enhancers to potentiate macrophage activation. Nat. Commun. 2019, 10, 3320. [Google Scholar] [CrossRef]
- Viola, A.; Munari, F.; Sánchez-Rodríguez, R.; Scolaro, T.; Castegna, A. The Metabolic Signature of Macrophage Responses. Front. Immunol. 2019, 10, 1462. [Google Scholar] [CrossRef]
- Shapouri-Moghaddam, A.; Mohammadian, S.; Vazini, H.; Taghadosi, M.; Esmaeili, S.A.; Mardani, F.; Seifi, B.; Mohammadi, A.; Afshari, J.T. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018, 233, 6425–6440. [Google Scholar] [CrossRef]
- Sharma, N.; Akkoyunlu, M.; Rabin, R.L. Macrophages-common culprit in obesity and asthma. Allergy 2018, 73, 1196–1205. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.L.; Zhi, Y.K.; Yi, L.; Luo, J.F.; Li, J.; Bai, S.S.; Liu, L.; Wang, P.X.; Zhou, H.; Dong, Y. Sinomenine regulates CD14/TLR4, JAK2/STAT3 pathway and calcium signal via α7nAChR to inhibit inflammation in LPS-stimulated macrophages. Immunopharmacol. Immunotoxicol. 2019, 41, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.W.; Su, M.X.; Zhang, W.; Wang, L.; Qian, C.Y. Atorvastatin increases lipopolysaccharide-induced expression of tumour necrosis factor-α-induced protein 8-like 2 in RAW264.7 cells. Exp. Ther. Med. 2014, 8, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Leng, L.; Metz, C.N.; Fang, Y.; Xu, J.; Donnelly, S.; Baugh, J.; Delohery, T.; Chen, Y.; Mitchell, R.A.; Bucala, R. MIF signal transduction initiated by binding to CD74. J. Exp. Med. 2003, 197, 1467–1476. [Google Scholar] [CrossRef]
- Shi, X.; Leng, L.; Wang, T.; Wang, W.; Du, X.; Li, J.; McDonald, C.; Chen, Z.; Murphy, J.W.; Lolis, E.; et al. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006, 25, 595–606. [Google Scholar] [CrossRef]
- Tilstam, P.V.; Schulte, W.; Holowka, T.; Kim, B.-S.; Nouws, J.; Sauler, M.; Piecychna, M.; Pantouris, G.; Lolis, E.; Leng, L.; et al. MIF but not MIF-2 recruits inflammatory macrophages in an experimental polymicrobial sepsis model. J. Clin. Investig. 2021, 131, e127171. [Google Scholar] [CrossRef]
- Senter, P.D.; Al-Abed, Y.; Metz, C.N.; Benigni, F.; Mitchell, R.A.; Chesney, J.; Han, J.; Gartner, C.G.; Nelson, S.D.; Todaro, G.J.; et al. Inhibition of macrophage migration inhibitory factor (MIF) tautomerase and biological activities by acetaminophen metabolites. Proc. Natl. Acad. Sci. USA 2002, 99, 144–149. [Google Scholar] [CrossRef]
- Fingerle-Rowson, G.; Kaleswarapu, D.R.; Schlander, C.; Kabgani, N.; Brocks, T.; Reinart, N.; Busch, R.; Schütz, A.; Lue, H.; Du, X.; et al. A tautomerase-null macrophage migration-inhibitory factor (MIF) gene knock-in mouse model reveals that protein interactions and not enzymatic activity mediate MIF-dependent growth regulation. Mol. Cell. Biol. 2009, 29, 1922–1932. [Google Scholar] [CrossRef]
- Li, Y.H.; Wen, K.; Zhu, L.L.; Lv, S.K.; Cao, Q.; Li, Q.; Deng, L.; Chen, T.; Wang, X.; Deng, K.Y.; et al. Tautomerase Activity-Lacking of the Macrophage Migration Inhibitory Factor Alleviates the Inflammation and Insulin Tolerance in High Fat Diet-Induced Obese Mice. Front. Endocrinol. 2020, 11, 134. [Google Scholar] [CrossRef]
- Hermanowski-Vosatka, A.; Mundt, S.S.; Ayala, J.M.; Goyal, S.; Hanlon, W.A.; Czerwinski, R.M.; Wright, S.D.; Whitman, C.P. Enzymatically inactive macrophage migration inhibitory factor inhibits monocyte chemotaxis and random migration. Biochemistry 1999, 38, 12841–12849. [Google Scholar] [CrossRef]
- Dagia, N.M.; Kamath, D.V.; Bhatt, P.; Gupte, R.D.; Dadarkar, S.S.; Fonseca, L.; Agarwal, G.; Chetrapal-Kunwar, A.; Balachandran, S.; Srinivasan, S.; et al. A fluorinated analog of ISO-1 blocks the recognition and biological function of MIF and is orally efficacious in a murine model of colitis. Eur. J. Pharmacol. 2009, 607, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Kok, T.; Wapenaar, H.; Wang, K.; Neochoritis, C.G.; Zarganes-Tzitzikas, T.; Proietti, G.; Eleftheriadis, N.; Kurpiewska, K.; Kalinowska-Tłuścik, J.; Cool, R.H.; et al. Discovery of chromenes as inhibitors of macrophage migration inhibitory factor. Bioorg. Med. Chem. 2018, 26, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yang, C.; Wang, L.; Yang, Z.; Guo, D.; Fan, C. Repurposing old drugs as novel inhibitors of human MIF from structural and functional analysis. Bioorg. Med. Chem. Lett. 2022, 55, 128445. [Google Scholar] [CrossRef] [PubMed]
- Ohkawara, T.; Okubo, N.; Maehara, O.; Nishihira, J.; Takeda, H. Protective effect of ISO-1 with inhibition of RIPK3 up-regulation and neutrophilic accumulation on acetaminophen-induced liver injury in mice. Toxicol. Lett. 2021, 339, 51–59. [Google Scholar] [CrossRef]
- Vieira-De-Abreu, A.; Calheiros, A.S.; Mesquita-Santos, F.P.; Magalhães, E.S.; Mourão-Sá, D.; Castro-Faria-Neto, H.C.; Bozza, M.T.; Bandeira-Melo, C.; Bozza, P.T. Cross-talk between macrophage migration inhibitory factor and eotaxin in allergic eosinophil activation forms leukotriene C₄-synthesizing lipid bodies. Am. J. Respir. Cell Mol. Biol. 2011, 44, 509–516. [Google Scholar] [CrossRef]
- Weber, C.; Kraemer, S.; Drechsler, M.; Lue, H.; Koenen, R.R.; Kapurniotu, A.; Zernecke, A.; Bernhagen, J. Structural determinants of MIF functions in CXCR2-mediated inflammatory and atherogenic leukocyte recruitment. Proc. Natl. Acad. Sci. USA 2008, 105, 16278–16283. [Google Scholar] [CrossRef]
- Hébert, C.A.; Vitangcol, R.V.; Baker, J.B. Scanning mutagenesis of interleukin-8 identifies a cluster of residues required for receptor binding. J. Biol. Chem. 1991, 266, 18989–18994. [Google Scholar] [CrossRef]
- Stamps, S.L.; Taylor, A.B.; Wang, S.C.; Hackert, M.L.; Whitman, C.P. Mechanism of the Phenylpyruvate Tautomerase Activity of Macrophage Migration Inhibitory Factor: Properties of the P1G, P1A, Y95F, and N97A Mutants. Biochemistry 2000, 39, 9671–9678. [Google Scholar] [CrossRef]
- Onodera, S.; Kaneda, K.; Mizue, Y.; Koyama, Y.; Fujinaga, M.; Nishihira, J. Macrophage Migration Inhibitory Factor Up-regulates Expression of Matrix Metalloproteinases in Synovial Fibroblasts of Rheumatoid Arthritis. J. Biol. Chem. 2000, 275, 444–450. [Google Scholar] [CrossRef]
- Calandra, T.; Bernhagen, J.; Mitchell, R.A.; Bucala, R. The macrophage is an important and previously unrecognized source of macrophage migration inhibitory factor. J. Exp. Med. 1994, 179, 1895–1902. [Google Scholar] [CrossRef]
- Kleemann, R.; Hausser, A.; Geiger, G.; Mischke, R.; Burger-Kentischer, A.; Flieger, O.; Franz-Josef, J.; Roger, T.; Calandra, T.; Kapurniotu, A.; et al. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature 2000, 408, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Gui, Z.; Zhang, X.; Han, Q.; Hang, Z.; Tan, R.; Gu, M.; Wang, Z. Macrophage polarization induces endothelium-to-myofibroblast transition in chronic allograft dysfunction. Ren. Fail. 2023, 45, 2220418. [Google Scholar] [CrossRef]
- Liu, M.H.; Lin, X.L.; Xiao, L.L. Hydrogen sulfide attenuates TMAO-induced macrophage inflammation through increased SIRT1 sulfhydration. Mol. Med. Rep. 2023, 28, 129. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, L.; Liu, M.; Du, A.; Qiu, M.; Shu, H.; Li, L.; Kong, X.; Sun, W. ROS inhibition increases KDM6A-mediated NOX2 transcription and promotes macrophages oxidative stress and M1 polarization. Cell Stress Chaperones 2023, 28, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Benigni, F.; Atsumi, T.; Calandra, T.; Metz, C.; Echtenacher, B.; Peng, T.; Bucala, R. The proinflammatory mediator macrophage migration inhibitory factor induces glucose catabolism in muscle. J. Clin. Investig. 2000, 106, 1291–1300. [Google Scholar] [CrossRef]
- Marsin, A.S.; Bouzin, C.; Bertrand, L.; Hue, L. The stimulation of glycolysis by hypoxia in activated monocytes is mediated by AMP-activated protein kinase and inducible 6-phosphofructo-2-kinase. J. Biol. Chem. 2002, 277, 30778–30783. [Google Scholar] [CrossRef]
- Liu, B.; Ding, C.; Tang, W.; Zhang, C.; Gu, Y.; Wang, Z.; Yu, T.; Li, Z. Hepatic ROS Mediated Macrophage Activation Is Responsible for Irinotecan Induced Liver Injury. Cells 2022, 11, 3791. [Google Scholar] [CrossRef]
- Zhang, B.; Yang, Y.; Yi, J.; Zhao, Z.; Ye, R. Hyperglycemia modulates M1/M2 macrophage polarization via reactive oxygen species overproduction in ligature-induced periodontitis. J. Periodontal Res. 2021, 56, 991–1005. [Google Scholar] [CrossRef]
- Molavian, H.R.; Kohandel, M.; Sivaloganathan, S. High Concentrations of H2O2 Make Aerobic Glycolysis Energetically More Favorable for Cellular Respiration. Front. Physiol. 2016, 7, 362. [Google Scholar] [CrossRef]
- Zhang, Y.; Marcillat, O.; Giulivi, C.; Ernster, L.; Davies, K.J. The oxidative inactivation of mitochondrial electron transport chain components and ATPase. J. Biol. Chem. 1990, 265, 16330–16336. [Google Scholar] [CrossRef]
- Lippe, G.; Comelli, M.; Mazzilis, D.; Sala, F.D.; Mavelli, I. The inactivation of mitochondrial F1 ATPase by H2O2 is mediated by iron ions not tightly bound in the protein. Biochem. Biophys. Res. Commun. 1991, 181, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Rai, Y.; Anita; Kumari, N.; Singh, S.; Kalra, N.; Soni, R.; Bhatt, A.N. Mild mitochondrial uncoupling protects from ionizing radiation induced cell death by attenuating oxidative stress and mitochondrial damage. Biochim. Biophys. Acta Bioenerg. 2021, 1862, 148325. [Google Scholar] [CrossRef] [PubMed]



| Accession NM | Name | Sequence (5′-3′) (from Integrated DNA Technologies, BVBA, Belgium) | Amplicon Size (bp) |
|---|---|---|---|
| NM_007415 | PARP1 | F-GAGTACAGTGCCAGTCAGC | 117 |
| R-CACCTCGTCACCTTTTCTCTT | |||
| NM_009632 | PARP2 | F-GTGGACCCAGAGTGTGCAGCC | 194 |
| R-CCCGTCTTTCCAACTCGGCCC | |||
| NM_145619 | PARP3 | F-TGCTGCTGGTGCTAGCGGAC | 281 |
| R-GCCCAGTTTGGAGTGGGCCTG | |||
| NM_008084 | GAPDH | F-AATGGTGAAGGTCGGTGTG | 150 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vámos, E.; Kálmán, N.; Sturm, E.M.; Nayak, B.B.; Teppan, J.; Vántus, V.B.; Kovács, D.; Makszin, L.; Loránd, T.; Gallyas, F., Jr.; et al. Highly Selective MIF Ketonase Inhibitor KRP-6 Diminishes M1 Macrophage Polarization and Metabolic Reprogramming. Antioxidants 2023, 12, 1790. https://doi.org/10.3390/antiox12101790
Vámos E, Kálmán N, Sturm EM, Nayak BB, Teppan J, Vántus VB, Kovács D, Makszin L, Loránd T, Gallyas F Jr., et al. Highly Selective MIF Ketonase Inhibitor KRP-6 Diminishes M1 Macrophage Polarization and Metabolic Reprogramming. Antioxidants. 2023; 12(10):1790. https://doi.org/10.3390/antiox12101790
Chicago/Turabian StyleVámos, Eszter, Nikoletta Kálmán, Eva Maria Sturm, Barsha Baisakhi Nayak, Julia Teppan, Viola Bagóné Vántus, Dominika Kovács, Lilla Makszin, Tamás Loránd, Ferenc Gallyas, Jr., and et al. 2023. "Highly Selective MIF Ketonase Inhibitor KRP-6 Diminishes M1 Macrophage Polarization and Metabolic Reprogramming" Antioxidants 12, no. 10: 1790. https://doi.org/10.3390/antiox12101790
APA StyleVámos, E., Kálmán, N., Sturm, E. M., Nayak, B. B., Teppan, J., Vántus, V. B., Kovács, D., Makszin, L., Loránd, T., Gallyas, F., Jr., & Radnai, B. (2023). Highly Selective MIF Ketonase Inhibitor KRP-6 Diminishes M1 Macrophage Polarization and Metabolic Reprogramming. Antioxidants, 12(10), 1790. https://doi.org/10.3390/antiox12101790







