Hypoxia Affects the Antioxidant Activity of Glutaredoxin 3 in Scylla paramamosain through Hypoxia Response Elements
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Hypoxia
2.2. Gene Cloning and Sequence Analysis
2.3. RNA Interference Assay
2.4. Real-Time PCR
2.5. Subcellular Localization of SpGrx3
2.6. Overexpression Assay
2.7. Dual-Luciferase Reporter Assays
2.8. Statistical Analyses
3. Results
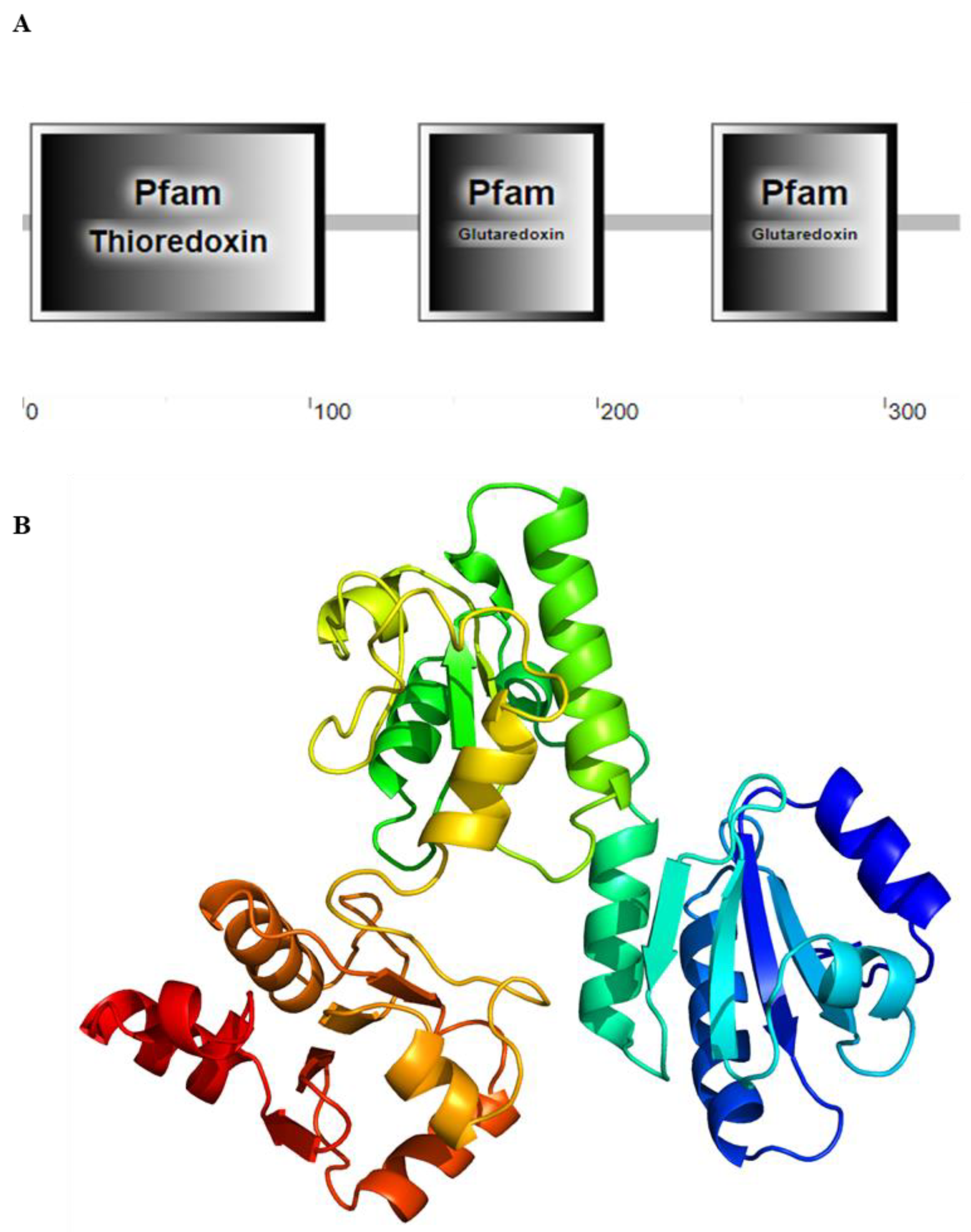
3.1. cDNA Cloning and Characterization of SpGrx3
3.2. Expression Profiles of SpGrx3
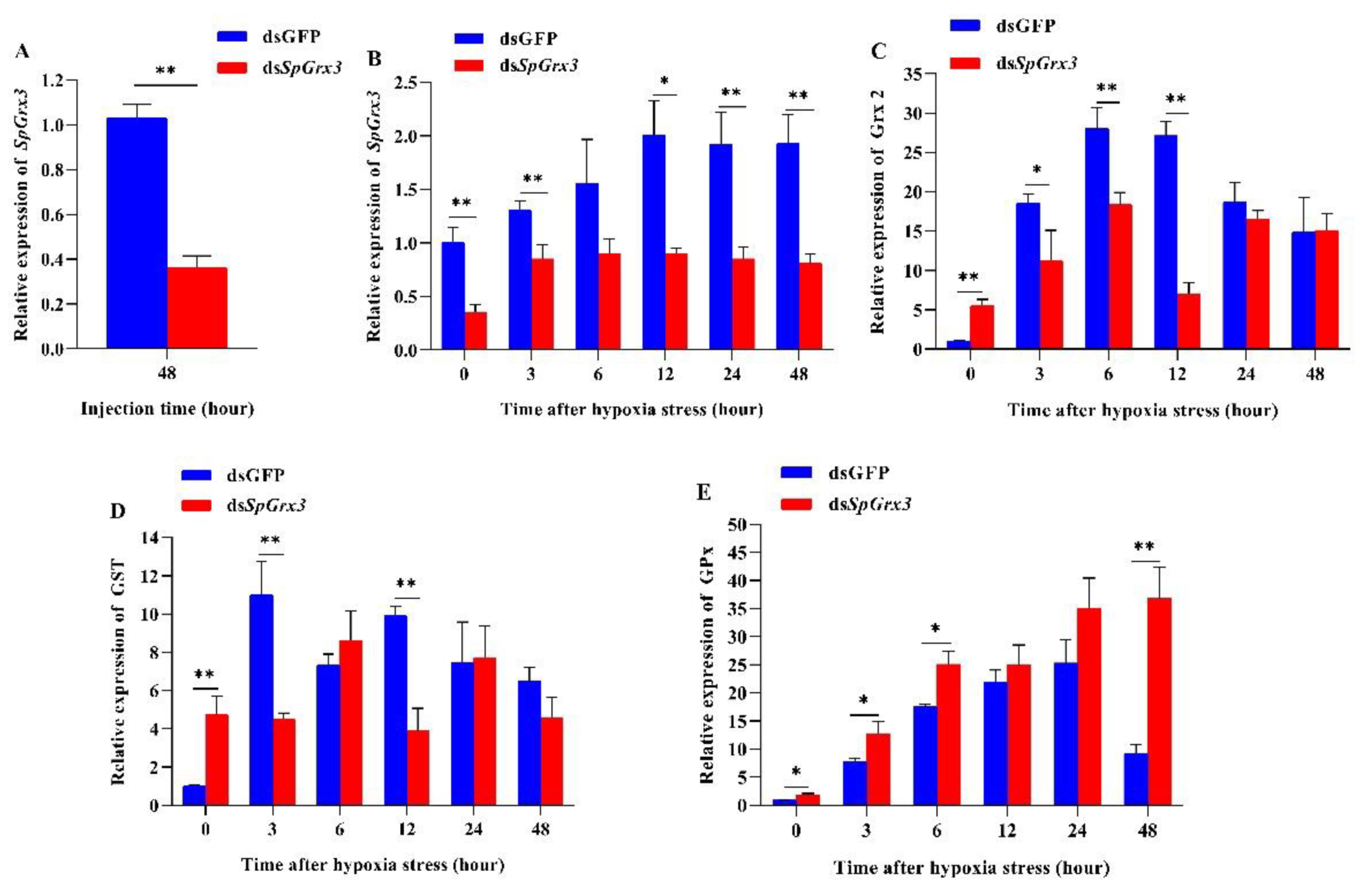
3.3. Expression Patterns of Antioxidant Genes in SpGrx3-Interfered Mud Crabs following Hypoxia
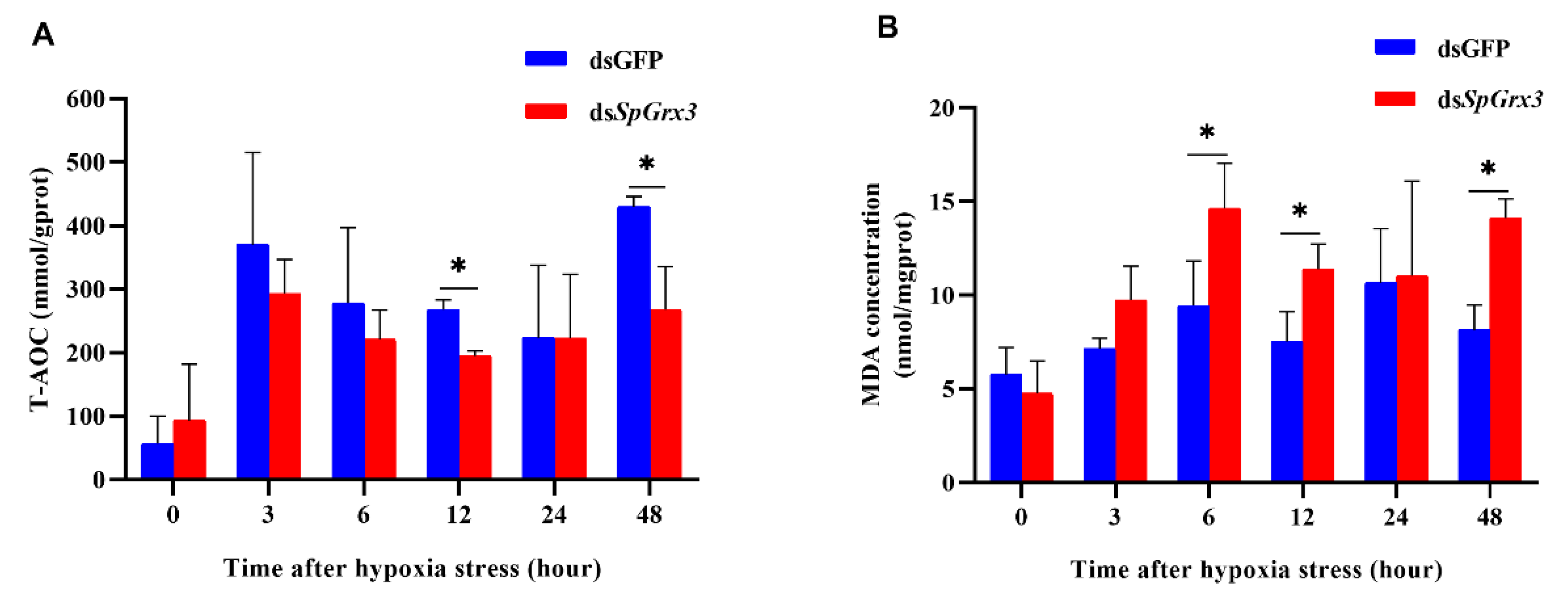
3.4. T-AOC and MDA Contents in SpGrx3-Interfered Mud Crabs after Hypoxia
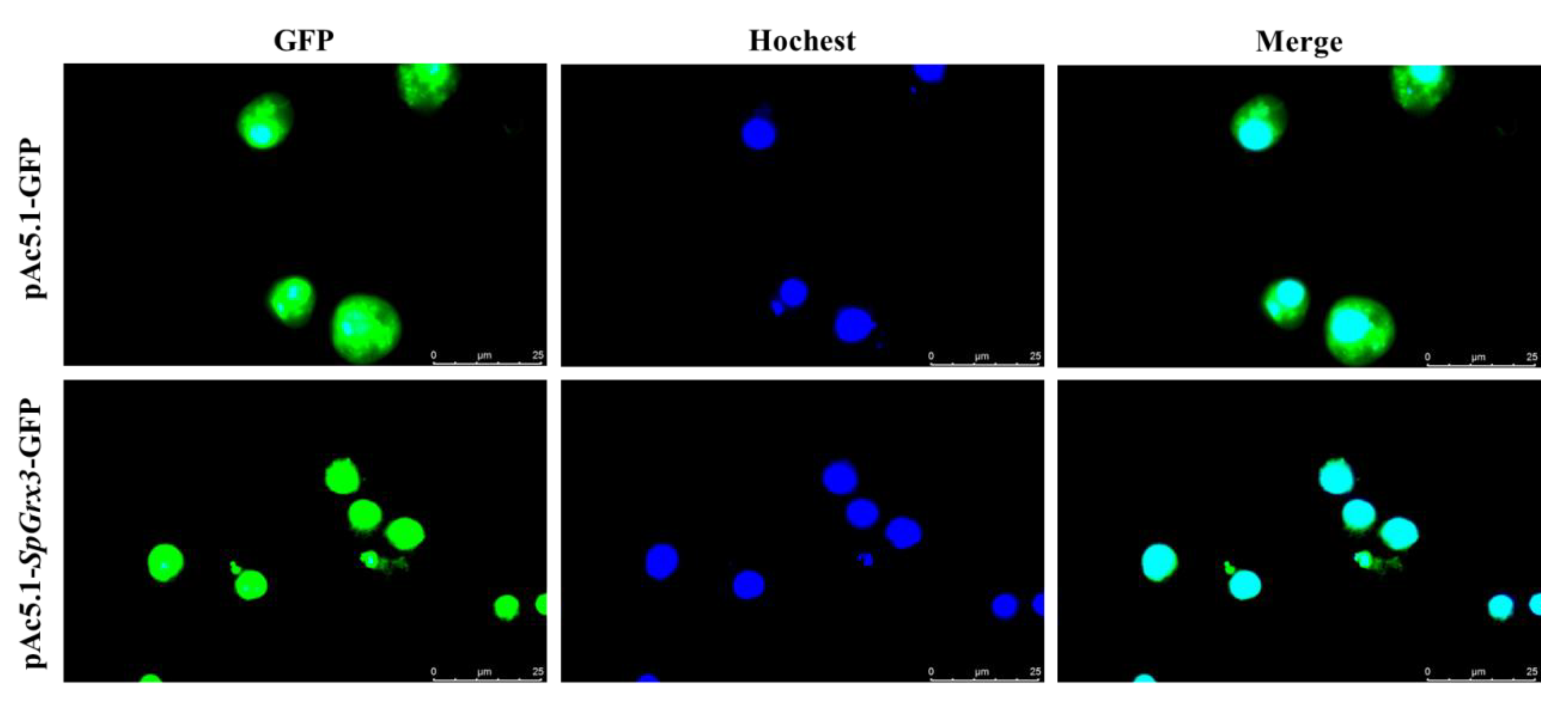
3.5. The Subcellular Localization of SpGrx3
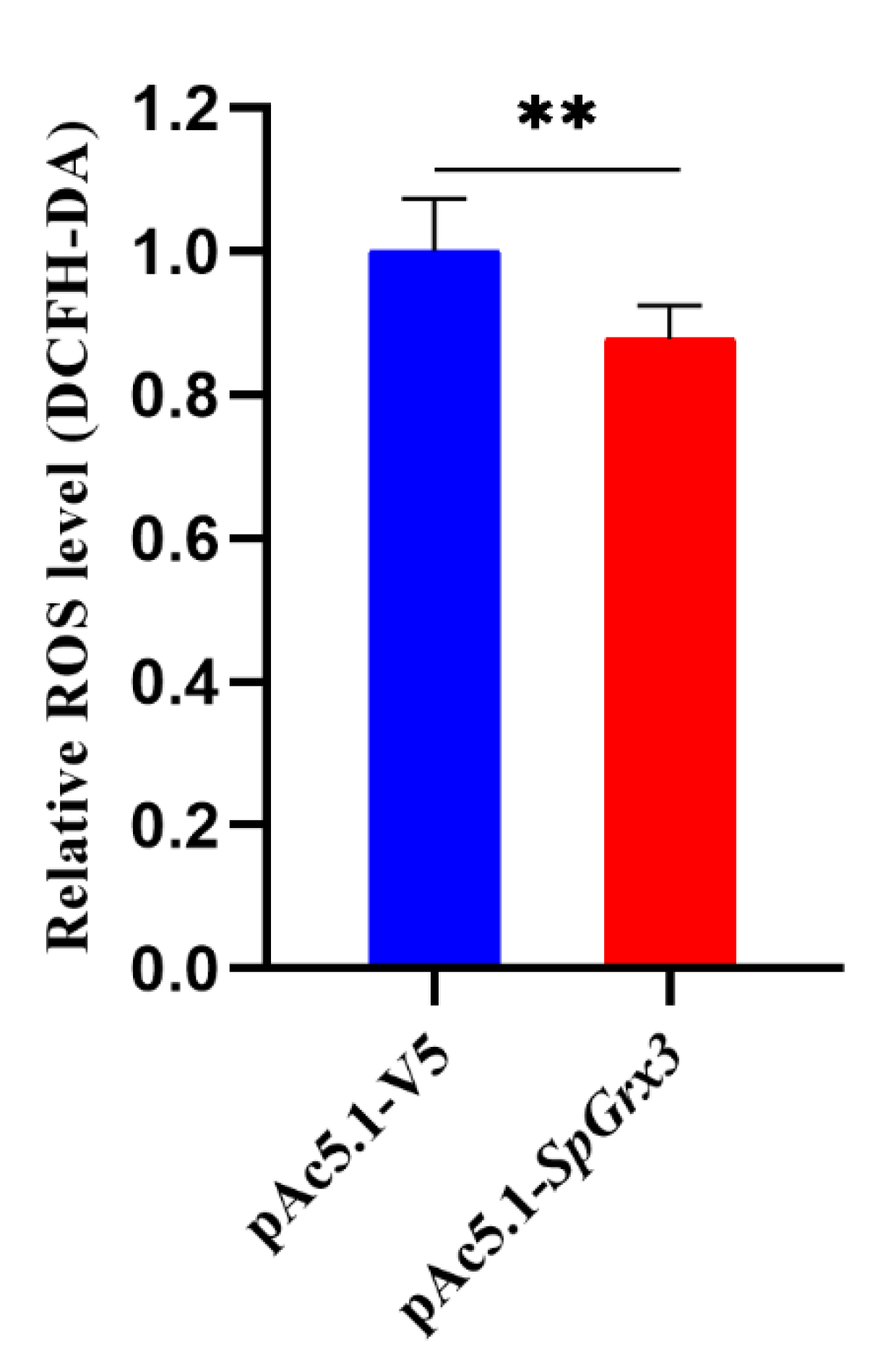
3.6. Effect of SpGrx3 Overexpression on Intracellular ROS Content after Hypoxia
3.7. Analysis of the SpGrx3 Promoter
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Portner, H.O.; Peck, M.A. Climate change effects on fishes and fisheries: Towards a cause-and-effect understanding. J. Fish Biol. 2010, 77, 1745–1779. [Google Scholar] [CrossRef]
- Ferreira, N.C.; Bonetti, C.; Seiffert, W.Q. Hydrological and Water Quality Indices as management tools in marine shrimp culture. Aquaculture 2011, 318, 425–433. [Google Scholar] [CrossRef]
- Jie, Y.K.; Cheng, C.H.; Wang, L.C.; Ma, H.L.; Deng, Y.Q.; Liu, G.X.; Feng, J.; Guo, Z.X.; Ye, L.T. Hypoxia-induced oxidative stress and transcriptome changes in the mud crab (Scylla paramamosain). Comp. Biochem. Physiol. Toxicol. Pharmacol. CBP 2021, 245, 109039. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wei, L.; Cao, J.; Qiu, L.; Jiang, X.; Li, P.; Song, Q.; Zhou, H.; Han, Q.; Diao, X. Oxidative stress, DNA damage and antioxidant enzyme activities in the pacific white shrimp (Litopenaeus vannamei) when exposed to hypoxia and reoxygenation. Chemosphere 2016, 144, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Guo, Z.; Fu, H.; Zhu, J.; Ge, X. Integrated metabolomic and transcriptomic analysis of brain energy metabolism in the male Oriental river prawn (Macrobrachium nipponense) in response to hypoxia and reoxygenation. Environ. Pollut. 2018, 243, 1154–1165. [Google Scholar] [CrossRef] [PubMed]
- Kondo, N.; Nakamura, H.; Masutani, H.; Yodoi, J. Redox regulation of human thioredoxin network. Antioxid. Redox Signal. 2006, 8, 1881–1890. [Google Scholar] [CrossRef] [PubMed]
- Lillig, C.H.; Berndt, C.; Holmgren, A. Glutaredoxin systems. Biochim. Biophys. Acta 2008, 1780, 1304–1317. [Google Scholar] [CrossRef] [PubMed]
- Holmgren, A.; Johansson, C.; Berndt, C.; Lonn, M.E.; Hudemann, C.; Lillig, C.H. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem. Soc. Trans. 2005, 33, 1375–1377. [Google Scholar] [CrossRef]
- Kalinina, E.V.; Chernov, N.N.; Novichkova, M.D. Role of glutathione, glutathione transferase, and glutaredoxin in regulation of redox-dependent processes. Biochemistry (Moscow) 2014, 79, 1562–1583. [Google Scholar] [CrossRef]
- Lillig, C.H.; Berndt, C.; Vergnolle, O.; Lönn, M.E.; Hudemann, C.; Bill, E.; Holmgren, A. Characterization of human glutaredoxin 2 as iron-sulfur protein: A possible role as redox sensor. Proc. Natl. Acad. Sci. USA 2005, 102, 8168–8173. [Google Scholar] [CrossRef]
- Vlamis-Gardikas, A.; Aslund, F.; Spyrou, G.; Bergman, T.; Holmgren, A. Cloning, overexpression, and characterization of glutaredoxin 2, an atypical glutaredoxin from Escherichia coli. J. Biol. Chem. 1997, 272, 11236–11243. [Google Scholar] [CrossRef] [PubMed]
- Herrero, E.; de la Torre-Ruiz, M.A. Monothiol glutaredoxins: A common domain for multiple functions. Cell Mol. Life Sci. 2007, 64, 1518–1530. [Google Scholar] [CrossRef] [PubMed]
- Berndt, C.; Christ, L.; Rouhier, N.; Mühlenhoff, U. Glutaredoxins with iron-sulphur clusters in eukaryotes—Structure, function and impact on disease. Biochim. Biophys. Acta Bioenergy 2021, 1862, 148317. [Google Scholar] [CrossRef]
- Fernandes, A.P.; Fladvad, M.; Berndt, C.; Andrésen, C.; Lillig, C.H.; Neubauer, P.; Sunnerhagen, M.; Holmgren, A.; Vlamis-Gardikas, A. A novel monothiol glutaredoxin (Grx4) from Escherichia coli can serve as a substrate for thioredoxin reductase. J. Biol. Chem. 2005, 280, 24544–24552. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.M. Role of the glutathione/glutaredoxin and thioredoxin systems in yeast growth and response to stress conditions. Mol. Microbiol. 2001, 39, 533–541. [Google Scholar] [CrossRef]
- Hanschmann, E.M.; Berndt, C.; Hecker, C.; Garn, H.; Bertrams, W.; Lillig, C.H.; Hudemann, C. Glutaredoxin 2 Reduces Asthma-Like Acute Airway Inflammation in Mice. Front. Immunol. 2020, 11, 561724. [Google Scholar] [CrossRef]
- Lee, J.; You, J.H.; Shin, D.; Roh, J.L. Inhibition of Glutaredoxin 5 predisposes Cisplatin-resistant Head and Neck Cancer Cells to Ferroptosis. Theranostics 2020, 10, 7775–7786. [Google Scholar] [CrossRef]
- Fan, R.; Li, Y.; Jiang, S.; Jiang, S.; Yang, Q.; Yang, L.; Huang, J.; Zhou, F. cDNA cloning and expression analysis of glutaredoxin 3 in black tiger shrimp Penaeus monodon. Aquac. Int. 2021, 29, 2661–2679. [Google Scholar] [CrossRef]
- Fan, R.; Jiang, S.; Li, Y.; Yang, Q.; Jiang, S.; Huang, J.; Yang, L.; Chen, X.; Zhou, F. Molecular Characterization and Expression Analysis of Glutaredoxin 5 in Black Tiger Shrimp (Penaeus monodon) and Correlation Analysis Between the SNPs of PmGrx5 and Ammonia-N Stress Tolerance Trait. Front. Mar. Sci. 2022, 9, 909827. [Google Scholar] [CrossRef]
- Berndt, C.; Poschmann, G.; Stühler, K.; Holmgren, A.; Bräutigam, L. Zebrafish heart development is regulated via glutaredoxin 2 dependent migration and survival of neural crest cells. Redox Biol. 2014, 2, 673–678. [Google Scholar] [CrossRef]
- Omeka, W.K.M.; Liyanage, D.S.; Yang, H.; Lee, J. Glutaredoxin 2 from big belly seahorse (Hippocampus abdominalis) and its potential involvement in cellular redox homeostasis and host immune responses. Fish Shellfish Immunol. 2019, 95, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Omeka, W.K.M.; Liyanage, D.S.; Priyathilaka, T.T.; Godahewa, G.I.; Lee, S.; Lee, S.; Lee, J. Glutaredoxin 1 from big-belly seahorse (Hippocampus abdominalis): Molecular, transcriptional, and functional evidence in teleost immune responses. Fish Shellfish Immunol. 2019, 90, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Godoy, J.R.; Oesteritz, S.; Hanschmann, E.M.; Ockenga, W.; Ackermann, W.; Lillig, C.H. Segment-specific overexpression of redoxins after renal ischemia and reperfusion: Protective roles of glutaredoxin 2, peroxiredoxin 3, and peroxiredoxin 6. Free Radic. Biol. Med. 2011, 51, 552–561. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xin, H.; Shi, Y.; Mu, J. Glutaredoxin 2 protects cardiomyocytes from hypoxia/reoxygenation-induced injury by suppressing apoptosis, oxidative stress, and inflammation via enhancing Nrf2 signaling. Int. Immunopharmacol. 2021, 94, 107428. [Google Scholar] [CrossRef]
- Petry, S.F.; Sun, L.M.; Knapp, A.; Reinl, S.; Linn, T. Distinct Shift in Beta-Cell Glutaredoxin 5 Expression Is Mediated by Hypoxia and Lipotoxicity Both In Vivo and In Vitro. Front. Endocrinol. (Lausanne) 2018, 9, 84. [Google Scholar] [CrossRef]
- Watanabe, Y.; Murdoch, C.E.; Sano, S.; Ido, Y.; Bachschmid, M.M.; Cohen, R.A.; Matsui, R. Glutathione adducts induced by ischemia and deletion of glutaredoxin-1 stabilize HIF-1α and improve limb revascularization. Proc. Natl. Acad. Sci. USA 2016, 113, 6011–6016. [Google Scholar] [CrossRef]
- Jaakkola, P.; Mole, D.R.; Tian, Y.M.; Wilson, M.I.; Gielbert, J.; Gaskell, S.J.; von Kriegsheim, A.; Hebestreit, H.F.; Mukherji, M.; Schofield, C.J.; et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001, 292, 468–472. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia-inducible factor 1 and cardiovascular disease. Annu. Rev. Physiol. 2014, 76, 39–56. [Google Scholar] [CrossRef]
- Cheng, C.H.; Ma, H.L.; Su, Y.L.; Deng, Y.Q.; Feng, J.; Xie, J.W.; Chen, X.L.; Guo, Z.X. Ammonia toxicity in the mud crab (Scylla paramamosain): The mechanistic insight from physiology to transcriptome analysis. Ecotoxicol. Environ. Saf. 2019, 179, 9–16. [Google Scholar] [CrossRef]
- Xie, J.; Cheng, C.; Jie, Y.; Ma, H.; Feng, J.; Su, Y.; Deng, Y.; Xu, H.; Guo, Z. Expression of lactate dehydrogenase is induced during hypoxia via HIF-1 in the mud crab Scylla paramamosain. Comp. Biochem. Physiol. Toxicol. Pharmacol. CBP 2019, 225, 108563. [Google Scholar] [CrossRef]
- Ma, J.; Liu, Y.; Niu, D.; Li, X. Effects of chlorpyrifos on the transcription of CYP3A cDNA, activity of acetylcholinesterase, and oxidative stress response of goldfish (Carassius auratus). Environ. Toxicol. 2015, 30, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, H.; Ohishi, N.; Yagi, K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 1979, 95, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.W.; Cheng, C.H.; Ma, H.L.; Feng, J.; Su, Y.L.; Deng, Y.Q.; Guo, Z.X. Molecular characterization, expression and antimicrobial activities of a c-type lysozyme from the mud crab, Scylla paramamosain. Dev. Comp. Immunol. 2019, 98, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Haunhorst, P.; Hanschmann, E.M.; Bräutigam, L.; Stehling, O.; Hoffmann, B.; Mühlenhoff, U.; Lill, R.; Berndt, C.; Lillig, C.H. Crucial function of vertebrate glutaredoxin 3 (PICOT) in iron homeostasis and hemoglobin maturation. Mol. Biol. Cell 2013, 24, 1895–1903. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Zhang, X.; Wang, D.; Li, J.; Zhang, Z.; Lu, Y.; Xian, J.; Wang, A.; Wang, L. Molecular Characterization and Expression Analysis of a Novel Glutaredoxin 3 Gene in Pacific White Shrimp (Litopenaeus vannamei). Front. Mar. Sci. 2021, 8, 687377. [Google Scholar] [CrossRef]
- Romano, N.; Zeng, C.S. Survival, osmoregulation and ammonia-N excretion of blue swimmer crab, Portunus pelagicus, juveniles exposed to different ammonia-N and salinity combinations. Comp. Biochem. Physiol. C-Toxicol. Pharmacol. 2010, 151, 222–228. [Google Scholar] [CrossRef]
- Borowiec, B.G.; Scott, G.R. Hypoxia acclimation alters reactive oxygen species homeostasis and oxidative status in estuarine killifish (Fundulus heteroclitus). J. Exp. Biol. 2020, 223. [Google Scholar] [CrossRef]
- Li, S.Y.; Fu, Z.J.; Lo, A.C. Hypoxia-induced oxidative stress in ischemic retinopathy. Oxidative Med. Cell. Longev. 2012, 2012, 426769. [Google Scholar] [CrossRef]
- Pujol-Carrion, N.; de la Torre-Ruiz, M.A. Glutaredoxins Grx4 and Grx3 of Saccharomyces cerevisiae play a role in actin dynamics through their Trx domains, which contributes to oxidative stress resistance. Appl. Environ. Microbiol. 2010, 76, 7826–7835. [Google Scholar] [CrossRef]
- Yu, H.J.; Liu, J.Q.; Bock, A.; Li, J.; Luo, G.M.; Shen, J.C. Engineering glutathione transferase to a novel glutathione peroxidase mimic with high catalytic efficiency—Incorporation of selenocysteine into a glutathione-binding scaffold using an auxotrophic expression system. J. Biol. Chem. 2005, 280, 11930–11935. [Google Scholar] [CrossRef]
- Takebe, G.; Yarimizu, J.; Saito, Y.; Hayashi, T.; Nakamura, H.; Yodoi, J.; Nagasawa, S.; Takahashi, K. A comparative study on the hydroperoxide and thiol specificity of the glutathione peroxidase family and selenoprotein P. J. Biol. Chem. 2002, 277, 41254–41258. [Google Scholar] [CrossRef]
- Ge, Y.; Qi, Z.; Wang, Y.; Liu, X.; Li, J.; Xu, J.; Liu, J.; Shen, J. Engineered selenium-containing glutaredoxin displays strong glutathione peroxidase activity rivaling natural enzyme. Int. J. Biochem. Cell Biol. 2009, 41, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Lönn, M.E.; Hudemann, C.; Berndt, C.; Cherkasov, V.; Capani, F.; Holmgren, A.; Lillig, C.H. Expression pattern of human glutaredoxin 2 isoforms: Identification and characterization of two testis/cancer cell-specific isoforms. Antioxid. Redox Signal. 2008, 10, 547–557. [Google Scholar] [CrossRef] [PubMed]
- Pujol-Carrion, N.; Belli, G.; Herrero, E.; Nogues, A.; de la Torre-Ruiz, M.A. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J. Cell Sci. 2006, 119, 4554–4564. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Peng, Y.; Li, J.; Holmgren, A.; Lu, J. Modulation of thiol-dependent redox system by metal ions via thioredoxin and glutaredoxin systems. Metallomics 2018, 10, 218–228. [Google Scholar] [CrossRef]
- Lopreiato, R.; Facchin, S.; Sartori, G.; Arrigoni, G.; Casonato, S.; Ruzzene, M.; Pinna, L.A.; Carignani, G. Analysis of the interaction between piD261/Bud32, an evolutionarily conserved protein kinase of Saccharomyces cerevisiae, and the Grx4 glutaredoxin. Biochem. J. 2004, 377, 395–405. [Google Scholar] [CrossRef]
- Molina, M.M.; Bellí, G.; de la Torre, M.A.; Rodríguez-Manzaneque, M.T.; Herrero, E. Nuclear monothiol glutaredoxins of Saccharomyces cerevisiae can function as mitochondrial glutaredoxins. J. Biol. Chem. 2004, 279, 51923–51930. [Google Scholar] [CrossRef]
- Pham, K.; Pal, R.; Qu, Y.; Liu, X.; Yu, H.; Shiao, S.L.; Wang, X.; O’Brian Smith, E.; Cui, X.; Rodney, G.G.; et al. Nuclear glutaredoxin 3 is critical for protection against oxidative stress-induced cell death. Free Radic. Biol. Med. 2015, 85, 197–206. [Google Scholar] [CrossRef]
- Murata, H.; Ihara, Y.; Nakamura, H.; Yodoi, J.; Sumikawa, K.; Kondo, T. Glutaredoxin exerts an antiapoptotic effect by regulating the redox state of Akt. J. Biol. Chem. 2003, 278, 50226–50233. [Google Scholar] [CrossRef]
- Miyazaki, K.; Kawamoto, T.; Tanimoto, K.; Nishiyama, M.; Honda, H.; Kato, Y. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J. Biol. Chem. 2002, 277, 47014–47021. [Google Scholar] [CrossRef]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu. Rev. Pathol. 2014, 9, 47–71. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, C.K.; Mazumder, B.; Fox, P.L. Role of hypoxia-inducible factor-1 in transcriptional activation of ceruloplasmin by iron deficiency. J. Biol. Chem. 2000, 275, 21048–21054. [Google Scholar] [CrossRef]
- Semenza, G.L.; Nejfelt, M.K.; Chi, S.M.; Antonarakis, S.E. Hypoxia-inducible nuclear factors bind to an enhancer element located 3′ to the human erythropoietin gene. Proc. Natl. Acad. Sci. USA 1991, 88, 5680–5684. [Google Scholar] [CrossRef]
- Blanchard, K.L.; Acquaviva, A.M.; Galson, D.L.; Bunn, H.F. Hypoxic induction of the human erythropoietin gene: Cooperation between the promoter and enhancer, each of which contains steroid receptor response elements. Mol. Cell Biol. 1992, 12, 5373–5385. [Google Scholar] [CrossRef] [PubMed]
- Caro, J. Hypoxia regulation of gene transcription. High Alt. Med. Biol. 2001, 2, 145–154. [Google Scholar] [CrossRef]
- Sun, S.; Fu, H.; Zhu, J.; Ge, X.; Wu, X.; Qiao, H.; Jin, S.; Zhang, W. Molecular Cloning and Expression Analysis of Lactate Dehydrogenase from the Oriental River Prawn Macrobrachium nipponense in Response to Hypoxia. Int. J. Mol. Sci. 2018, 19, 1990. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Huang, C.H.; Chen, B.X.; Liu, H.; Wang, W.M.; Gul, Y.; Wang, H.L. Alternative splicing transcription of Megalobrama amblycephala HIF prolyl hydroxylase PHD3 and up-regulation of PHD3 by HIF-1α. Biochem. Biophys. Res. Commun. 2016, 469, 737–742. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zhou, J.; Zhao, Y.; Toselli, P.; Li, W. Hypoxia-response element (HRE)-directed transcriptional regulation of the rat lysyl oxidase gene in response to cobalt and cadmium. Toxicol. Sci. 2013, 132, 379–389. [Google Scholar] [CrossRef]
- Kaluz, S.; Kaluzová, M.; Stanbridge, E.J. Regulation of gene expression by hypoxia: Integration of the HIF-transduced hypoxic signal at the hypoxia-responsive element. Clin. Chim. Acta 2008, 395, 6–13. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jie, Y.-K.; Cheng, C.-H.; Ma, H.-L.; Liu, G.-X.; Fan, S.-G.; Jiang, J.-J.; Guo, Z.-X. Hypoxia Affects the Antioxidant Activity of Glutaredoxin 3 in Scylla paramamosain through Hypoxia Response Elements. Antioxidants 2023, 12, 76. https://doi.org/10.3390/antiox12010076
Jie Y-K, Cheng C-H, Ma H-L, Liu G-X, Fan S-G, Jiang J-J, Guo Z-X. Hypoxia Affects the Antioxidant Activity of Glutaredoxin 3 in Scylla paramamosain through Hypoxia Response Elements. Antioxidants. 2023; 12(1):76. https://doi.org/10.3390/antiox12010076
Chicago/Turabian StyleJie, Yu-Kun, Chang-Hong Cheng, Hong-Ling Ma, Guang-Xin Liu, Si-Gang Fan, Jian-Jun Jiang, and Zhi-Xun Guo. 2023. "Hypoxia Affects the Antioxidant Activity of Glutaredoxin 3 in Scylla paramamosain through Hypoxia Response Elements" Antioxidants 12, no. 1: 76. https://doi.org/10.3390/antiox12010076
APA StyleJie, Y.-K., Cheng, C.-H., Ma, H.-L., Liu, G.-X., Fan, S.-G., Jiang, J.-J., & Guo, Z.-X. (2023). Hypoxia Affects the Antioxidant Activity of Glutaredoxin 3 in Scylla paramamosain through Hypoxia Response Elements. Antioxidants, 12(1), 76. https://doi.org/10.3390/antiox12010076





