Abstract
Contact between SARS-CoV-2 and human lung cells involves the viral spike protein and the human angiotensin-converting enzyme 2 (ACE2) receptor on epithelial cells, the latter being strongly involved in the regulation of inflammation as well as blood pressure homeostasis. SARS-CoV-2 infection is characterized by a strong inflammatory response defined as a “cytokine storm”. Among recent therapeutic approaches against SARS-CoV-2 targeting the dramatic inflammatory reaction, some natural products are promising. Diatoms are microalgae able to produce bioactive secondary metabolites, such as the xanthophyll diatoxanthin (Dt). The aim of this study is to demonstrate the anti-inflammatory effects of Dt on the A549-hACE2 lung cell line, exploring its interaction with the ACE2 receptor, as well as depicting its role in inhibiting a cytokine storm induced by the SARS-CoV-2 spike glycoprotein. Results showed that Dt enhanced the cell metabolism, e.g., the percent of metabolically active cells, as well as the ACE2 enzymatic activity. Moreover, Dt strongly affected the response of the SARS-CoV-2 spike glycoprotein-exposed A549-hACE2 cells in decreasing the interleukin-6 production and increasing the interleukin-10 release. Moreover, Dt upregulated genes encoding for the interferon pathway related to antiviral defense and enhanced proteins belonging to the innate immunity response. The potential interest of Dt as a new therapeutic agent in the treatment and/or prevention of the severe inflammatory syndrome related to SARS-CoV-2 infection is postulated.
1. Introduction
The first interaction between SARS-CoV-2 and human lung cells corresponds to the recognition of the viral spike protein by the human angiotensin-converting enzyme 2 (ACE2) receptor on epithelial cells in the airways. This is followed by virus entry and priming of human transmembrane protease serine 2 (TMPRSS2), which cleaves the S protein and initiates viral fusion [1]. One of the most pronounced cellular responses to SARS-CoV-2 in lung cells is the cytokine storm [2], which can dramatically lead to acute respiratory distress syndrome (ARDS), with subsequent lung injury [3]. This storm results from the sudden acute increase in circulating levels of different pro-inflammatory cytokines, including the interleukins IL-6, IL-1, and the TNFα that are the most important pro-inflammatory mediators of the innate immune response [4,5,6]. This detrimental biological event is therefore a target for a therapeutic solution to SARS-CoV-2 infection; for instance, the tocilizumab drug that acts on the cytokine IL-6 [7]. Anti-inflammatory therapies fighting the cytokine responses are suggested to decrease both the morbidity and mortality in infected SARS-CoV-2 patients [8], such as nonsteroidal anti-inflammatory drugs [9]. Natural compounds/extracts, e.g., from the vegetal kingdom, exhibiting antimicrobial or antiviral properties are strongly attractive [10]. Natural compounds with anti-inflammatory activities are also appealing [11]; for instance, those displaying interests against SARS-CoV-2 [12,13,14,15,16,17]. Although some terrestrial plants are currently used as human health benefit resources [18], there is a growing interest in marine microalgae [19], thanks to the scientifically recognized strengths offered by those organisms [20,21]. Among microalgae, diverse families of bioactive compounds show antiviral properties, as reported for sulfated polysaccharides [22] or polyphenols [23]. Moreover, carotenoids, among the most bioactive compound families, do present some antiviral potential, such as the exclusively aquatic species, astaxanthin [24], fucoxanthin, or siphonaxanthin [25]. Nevertheless, in silico analysis carried out on nine xanthophylls demonstrated their ability to inhibit the main protease (Mpro) and the papain-like protease (PLpro) of SARS-CoV-2 [26]. It is noteworthy that microalgae present a high diversity of carotenoids, with some of them understudied [27]. This is the case of the xanthophyll diatoxanthin (Dt), belonging to diatoms and exerting a (photo-)protective role [28,29,30,31,32]. Recently, its great antioxidant property and in vitro bioactivity have been demonstrated ([27], unpublished data), satisfying chemopreventive and anti-inflammatory potential, e.g., inhibiting a cytokine storm [27,33]. These recent studies pave the way for further investigation on the human health-induced bioactivity of Dt. The aim of the present study is to underline in vitro the diatoxanthin effects on the expressing ACE2 carcinoma cell line, A549-hACE2. On the one hand, it focused on the interactions between Dt and the ACE2 receptor, performing a docking analysis, and on the ACE2 enzymatic activity. On the other hand, the study explores the Dt role in inhibiting a cytokine storm induced by the SARS-CoV-2 spike glycoprotein, investigating at the molecular and protein levels. The potential interest of using Dt as a new therapeutic agent in the treatment and/or prevention of the severe inflammatory syndrome related to SARS-CoV-2 infection is discussed.
2. Materials and Methods
2.1. The A549-hACE2 Cell Line Cultivation
The lung carcinoma cell line (A549-hACE2) was purchased from Invivogen (cat. a549-hace2, Invivogen, San Diego, CA, USA). The A549-hACE2 cells, generated from the A549 lung carcinoma cell line, were stably transfected to express the human ACE2 (hACE2) gene. The A549-hACE2 cell line is an in vitro model commonly used for SARS-CoV-2 investigations [34,35].
The A549-hACE2 cell line was grown in cultured DMEM/F12 (Dulbecco’s odified Eagle’s medium/nutrient mixture F-12 Ham), supplemented with 10% FBS, 100 units mL−1 penicillin and 100 units mL−1 streptomycin, in a 5% CO2 atmosphere at 37 °C. Cells were cultured until reaching an adequate confluence, and then seeded in multi-well plates for the experimental purposes described below.
2.2. Diatoxanthin
Pure diatoxanthin (C40H54O2, CAS No. 31063-73-7; Figure S1) was purchased from D.H.I. Water & Environment (Hørsholm, Denmark). The purity was then assessed through HPLC analysis, using the method described in Pistelli et al. [36].
2.3. MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide) Assay
The A549-hACE2 cells (2 × 103 cells well−1, 100 μL well−1) were seeded in 96-well plates (TPP Techno Plastic Products AG, Trasadingen, Switzerland) and kept overnight for attachment. Cells were incubated for 48 h with Dt in DMSO at four different concentrations, namely 0.1, 1, 10, and 100 ng mL−1. The final concentration of DMSO was 0.1% v/v for the highest concentration of Dt (100 ng). At the end of the 48 h-incubation, OranguTM (Cell Guidance Systems, Cambridge, UK) was used to measure cell viability according to the manufacturer’s instructions. The absorbance was recorded using a Microplate Reader: Infinite® M1000 PRO (TECAN, Männedorf, Switzerland) at a wavelength of 450 nm. The same analysis was conducted incubating cells with DMSO without Dt (untreated cells = control). The results were represented as a percent of metabolically active cells estimated as the ratio between the absorbance of each sample (A549-hACE2 cells treated with Dt) and the absorbance of the control (untreated cells).
2.4. Angiotensin-Converting Enzyme 2 (ACE2) Activity Assay
The A549-hACE2 cells (2 × 106 cells well−1, 2 mL well−1) were seeded in 6-well plates (TPP Techno Plastic Products AG, Trasadingen, Switzerland) and kept overnight for attachment. Cells were then incubated with Dt in DMSO at the concentration 0.1 ng mL−1. Control was setup with A549-hACE2 cells plus DMSO without Dt addition. After 24 h incubation, the medium was removed from each well and the attached cells were rapidly washed with phosphate buffered saline (PBS) solution. The PBS solution was then removed, and the cell lysates were prepared by scraping each well into 500 μL of RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with Halt™ Protease & Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, MA, USA). The lysates were incubated on ice for 5 min and then clarified by centrifugation at 14,000× g for 5 min. Total protein concentration was determined using a NanoDrop 1000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) at 280 nm. Reading at 260 nm was done to assess the purity of extracted proteins. ACE2 activity was measured using Angiotensin II Converting Enzyme (ACE2) Inhibitor Screening Kit (cat. No. MAK378, Sigma-Aldrich, St. Louis, MO, USA). The assay was performed in triplicate, using an aliquot of 10 μL of each sample and following the manufacturer’s protocol. Fluorescence was read using a Microplate Reader: Infinite® M1000 PRO (Ex: 320 nm, Em: 420 nm, TECAN, Männedorf, Switzerland).
2.5. Target Fishing and Docking Analysis
The Dt molecule interaction with the ACE2 protein was retrieved through an introductory target fishing approach by using the ACID online server [37]. The 3D coordinates of the crystal structure of ACE2 were retrieved from the Protein Data Bank (PDB; PDB id: 1R42). The protein structure was optimized by using UCSF Chimera 1.16 software [38] for removal of all heteroatoms and water molecules, while further polar hydrogen atoms were added to protein to make the receptor molecules suitable for docking. Diatoxanthin (CID 6440986) was prepared as a ligand for docking, following the same procedure. Gasteiger charges were added and the maximum six numbers of active torsions were given to the lead compounds. ACE2 protein was finally docked using the molecular docking program AutoDock Vina 4.2.6 (Scripps Research, San Diego, CA, USA) [39].
2.6. Interleukins IL-6 and IL-10 Concentrations through Enzyme-Linked Immunosorbent Assay (ELISA)
The A549-hACE2 cells (2 × 106 cells wells−1, 2 mL well−1) were seeded in 6-well plates (TPP Techno Plastic Products AG, Trasadingen, Switzerland) and kept overnight for attachment. Cells were incubated with Dt in DMSO at the concentration of 0.1, 1, 10, and 100 ng mL−1, with or without recombinant human coronavirus SARS-CoV-2 spike glycoprotein RBD (His tag) (cat. No. ab275986, Abcam, Darmstadt, Germany). After the 24 h-lasting incubation, cell media were collected and the evaluation of the interleukins IL-6 and IL-10 concentrations was carried out through sandwich ELISA. Human IL-6 Standard ABTS ELISA Development Kit (cat. No. 900-K16, PeproTech, London, UK) and Human IL-10 Standard ABTS ELISA Development Kit (cat. No. 900-K21, PeproTech, London, UK) were used according to the manufacturer’s protocol. The absorbance was measured at 405 nm (with wavelength correction set at 650 nm) using the Microplate Reader: Infinite® M1000 PRO (TECAN, Männedorf, Switzerland).
2.7. RNA Extraction and PCR Array Gene Expression Analysis
The A549-hACE2 cells (2 × 106 cells wells−1, 2 mL well−1) were seeded in 6-well plates (TPP Techno Plastic Products AG, Trasadingen, Switzerland) and kept overnight for attachment. Cells were treated for 2 h with Dt in DMSO at a concentration of 0.1 ng mL−1, while the control condition consisted of A549-hACE2 cells incubated with DMSO without Dt. At the end of the incubation, the medium was removed, and the attached cells were rapidly washed with cold phosphate buffered saline (PBS) solution and lysed directly in plates by adding 500 μL of TRIsure™ reagent (cat. No. BIO-3803, Meridian Bioscience Inc., Cincinnati, OH, USA). RNA was then isolated according to manufacturer’s protocol. RNA concentration and purity were assessed using the NanoDrop 1000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The reverse transcription reaction was carried out using RT2 First Strand Kit (cat. No. 330404, Qiagen, Hilden, Germany). Real-time quantitative polymerase chain reaction (RT-qPCR) was performed using RT2 Profiler PCR Array Human Antiviral Response (384-well format, cat. No. 330231, PAHS-122ZA, Qiagen, Hilden, Germany). Plates were run on a ViiA 7 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). Standard fast PCR cycling protocol was run with 10 µL reaction volumes. Cycling conditions were set up in three stages: the first stage at 50 °C for 2 min and 95 °C for 10 min; the second stage consisted of 40 cycles at 95 °C for 15 s and 60 °C for 1 min; the last stage (melt curve) at 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s. qPCR data (Ct-values) were analyzed with PCR Array Data Analysis Online Software (Qiagen, Hilden, Germany). Relative gene expression values greater or lower than 2.0-expression ratio indicated significant gene expression variations with respect to the control. Genes used as control were actin-beta (ACTB), beta-2-microglobulin (B2M), glyceraldehyde-3 phosphate dehydrogenase (GAPDH), hypoxanthine phosphoribosyl transferase 1 (HPRT1), and ribosomal protein large P0 (RPLP0).
2.8. Tumor Necrosis Factor Receptor 1 (TNFR1) and Receptor Interacting Protein 2 (RIP2) Concentration
The production of tumor necrosis factor receptor 1 (TNFR1) and receptor interacting protein 2 (RIP2) was assessed through Western blot analysis. The A549-hACE2 cells (2 × 106 cells wells−1, 1 mL well−1) were seeded in 12-well plates (TPP Techno Plastic Products AG, Trasadingen, Switzerland) and kept overnight for attachment. A group of A549-hACE2 cells was treated with interleukin 1β (IL-1β) at a concentration of 40 pg mL−1 for 24 h. After this time, Dt was added at a concentration of 0.1 ng mL−1 in half of the plates and incubated for 6 h, whereas in the other half of the plates, DMSO was added. Another group of A549-hACE2 cells was incubated for 24 h without IL-1β and then treated with Dt in DMSO at a concentration of 0.1 ng mL−1 for 6 h, while another with DMSO without Dt for 6 h. The medium was then removed, and the cell lysates were prepared by scraping each well into 500 μL of RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific, Waltham, MA, USA), supplemented with Halt™ Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, MA, USA). The lysates were incubated on ice for 5 min and then clarified by centrifugation at 14,000× g for 5 min. Total protein concentration was determined using a NanoDrop 1000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) at 280 nm. Reading at 260 nm was done to assess the purity of extracted proteins. Protein samples were incubated at 95 °C for 5 min before the separation of total polypeptides by SDS-PAGE electrophoresis, using 4–15% Criterion™ TGX Stain-Free™ Protein Gel (cat. No. 5678085, Bio-Rad, Hercules, CA, USA). The SDS-PAGEs were then blotted onto Trans-Blot Turbo Midi 0.2 μm Nitrocellulose membrane (cat. No. 170-4159, Bio-Rad, Hercules, CA, USA) using a Trans-Blot Turbo Transfer System (cat. No. 170-4150, Bio-Rad, Hercules, CA, USA). Membranes were incubated for 1 h in blocking reagent (1X Tris Buffered Saline-TBS), 0.1% Tween-20 with 5% w/v nonfat dry milk and incubated overnight at 4 °C with the primary antibodies diluted in 1X TBS, 0.1% Tween-20 with 5% BSA. TNFR1 antibody (1:1000 diluted, cat. No. 3736, Cell Signaling Technology Inc., Danvers, MA, USA) and RIP2 antibody (1:1000 diluted, cat. No. 4142, Cell Signaling Technology Inc., Danvers, MA, USA) were added and incubated overnight at 4 °C. After incubation, membranes were washed three times for 10 min each with 15 mL of TBS/Tween and then incubated with goat anti-rabbit IgG F(ab’)2 Secondary Antibody, HRP (1:500 dilution, cat. No. 31461, Invitrogen, Waltham, MA, USA) with gentle agitation for 2 h at room temperature. After incubation, membranes were washed three times for 10 min each with 15 mL of TBS/Tween. Blotted membranes were detected by using Clarity Western ECL Substrate (cat. No. 1705061, Bio-Rad, Hercules, CA, USA) and ChemiDoc™ MP Imaging System (cat. No. 120-3154, Bio-Rad, Hercules, CA, USA). Densitometric analysis of five immune-positive bands was performed using ImageLab software (Bio-Rad, Hercules, CA, USA).
2.9. Cytokine Release from A549-hACE2 Cells
The release of 120 cytokines in the media from the A549-hACE2 cells was analyzed through an antibody array using RayBiotech® C-Series Human Cytokine Array C1000 (cat. No. AAH-CYT-1000, RayBiotech, Peachtree Corners, GA, USA). The experimental procedure was the same as previously described for the Western blot analysis. The A549-hACE2 cells (2 × 106 cells wells−1, 1 mL well−1) were seeded in 12-well plates (TPP Techno Plastic Products AG, Trasadingen, Switzerland) and kept overnight for attachment. A group of A549-hACE2 cells was treated with interleukin 1β (IL-1β) at a concentration of 40 pg mL−1 for 24 h in order to simulate the virus intracellular entry effect, when patients have high fever and headache as initial symptoms. After this time, Dt was added at a concentration of 0.1 ng mL−1 in half of the plates and incubated for 6 h, whereas in the other half of the plates, DMSO was added. Another group of A549-hACE2 cells was incubated for 24 h without IL-1β and then treated with Dt in DMSO at a concentration of 0.1 ng mL−1 for 6 h, while another with DMSO without Dt for 6 h. After incubation, cell medium was collected and protein concentration and purity were assessed at 280 and 260 nm, respectively, using a NanoDrop 1000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). For each condition tested, 1 mL of sample was used to perform the antibody array, according to the manufacturer’s protocol. Blots were analyzed using ImageLab software (Bio-Rad, Hercules, CA, USA). Results are represented as the ratio between the expression of cytokines in the medium of A549-hACE2 cells incubated with IL-1β and recovered with Dt vs. cells only incubated with IL-1β.
3. Results
3.1. Dt Effect on the A549-hACE2 Cell Line Viability and ACE2 Activity
Dt did not exert any cytotoxicity on A549-hACE2 cells (Figure 1a). Conversely, it did significantly enhance the metabolic activity of the A549-hACE2 cells, already at the lowest concentration (0.1 ng mL−1) compared to the control (at least p ≤ 0.01; Figure 1a). From this first result, we selected the lowest concentration of Dt, 0.1 ng mL−1, to further evaluate the role and effects of Dt on counteracting inflammation processes in A549-hACE2 cells.
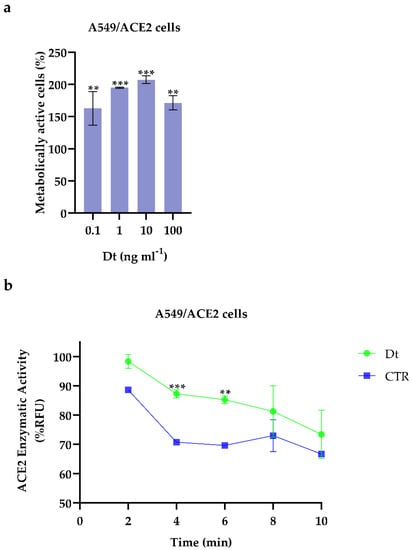
Figure 1.
(a) Cell viability assays on A549-hACE2 cells treated for 48 h at four Dt concentrations. Values are expressed as mean ± SD of metabolically active cells compared to control (no Dt, 100% of metabolically active cells). All concentrations tested showed significant differences compared to the control (no Dt, p-value ≤ 0.0001; Dunnet’s test) (b) ACE2 enzymatic activity screening assay on A549-hACE2 cells treated for 24 h with 0.1 ng mL−1 of Dt. Values are expressed as mean ± SD of %RFU (Relative Fluorescence Units). Asterisks indicate the statistically significant difference compared to the respective control *** p ≤ 0.001, ** p ≤ 0.01; Sidak’s test).
3.2. Angiotensin Converting Enzyme 2 (ACE2) Activity and Molecular Docking between Dt and ACE2
In the presence of Dt, the angiotensin converting enzyme 2 (ACE2) enzymatic activity in A549-hACE2 cells significantly increased (p ≤ 0.01) compared to the control (Figure 1b). Docking analysis revealed that diatoxanthin binds with the ACE2 (Figure 2), a chemical interaction that confirmed the effects of Dt on ACE2. This is due to hydrophobic interaction (ΔE = −7.9 kcal mol−1) established within the alpha-helix in the N domain involving the cleavage site of the enzyme. Docked conformation over-imposes standard ligand conformation (RMSD < 1).
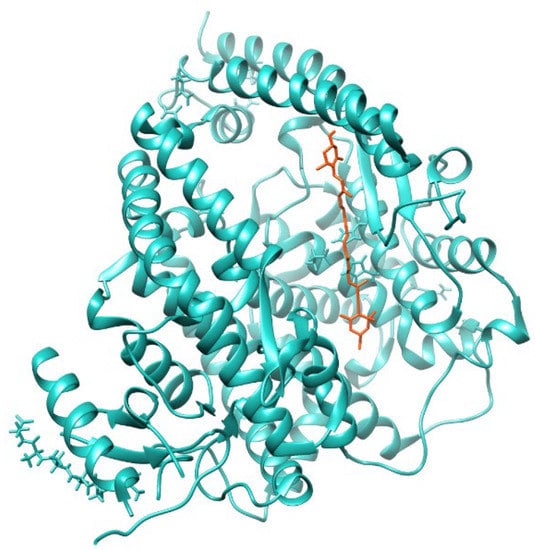
Figure 2.
Molecular docking diagrams of diatoxanthin binding to ACE2.
3.3. Interleukin (IL-6 and IL-10) Production
The Dt mediating effect on both IL-6 and IL-10 release was targeted in the A549-hACE2 cells in the absence or presence of SARS-CoV-2 spike glycoprotein. IL-6 and IL-10 have a role on the cytokine storm occurring in the inflamed cells infected by SARS-CoV-2 [6], and as an anti-inflammatory response affected by SARS-CoV-2 [5,40], respectively. In the absence of SARS-CoV-2 spike glycoprotein, Dt did not significantly affect IL-6 release compared to the control (p > 0.05; Figure 3a), whereas it did significantly increase the IL-10 production when applied at the highest concentration (100 ng mL−1, p < 0.001; Figure 3a).
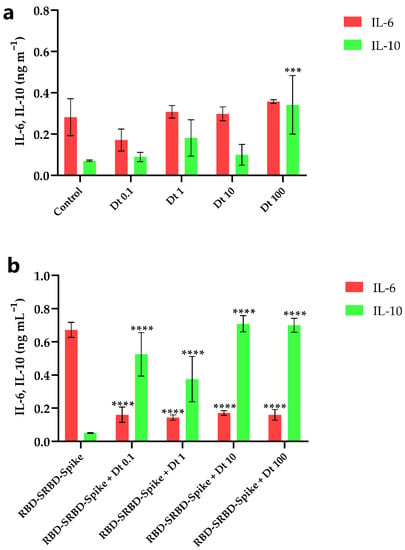
Figure 3.
Extracellular IL-6 and IL-10 concentration in A549-hACE2 cell cultivation treated with four concentrations of Dt, in the absence (a) and presence (b) of recombinant human coronavirus SARS-CoV-2 spike glycoprotein RBD (RBD-SRBD-Spike). Values are expressed as mean ± SD of IL-6 or IL-10 (ng mL−1). Asterisks indicate the statistically significant difference compared to the respective control, namely (a) no Dt in the absence of RBD-SRBD-Spike, and (b) no Dt in the presence of RBD-SRBD-Spike (**** p ≤ 0.0001, *** p ≤ 0.001; Dunnet’s test).
The contact between SARS-CoV-2 spike glycoprotein and A549-hACE2 cells strongly enhanced the extracellular IL-6 release and significantly lowered the IL-10 release (p < 0.001; Figure 3a,b). In this inflammatory context, Dt decreased the IL-6 production (p < 0.0001; Figure 3b) and induced an increase of the IL-10 production (at least p < 0.05; Figure 3b).
3.4. Dt-Modulated Gene Expression in A549-hACE2 Cells
Over the selected 84 genes involved in antiviral innate immunity cell response (Table S1), the A549-hACE2 cells treated with Dt at 0.1 ng mL−1 significantly modulated 15 genes related to the inhibition of the inflammatory response by decreasing all trigger factors in antiviral innate immunity (Figure 4). The A549-hACE2 cells exhibited an upregulation of the genes encoding for the CD80 molecule (CD80), FBJ murine osteosarcoma viral oncogene homolog (FOS), interferon alpha 1 and 2 (IFNA1 and IFNA2) and tumor necrosis factor (TNF) (2.83-, 3.07-, 2.91-, 8.98- and 3.57-fold regulation, respectively). In addition, the downregulation of interferon beta 1 (IFNB1, −7.52-fold regulation) and interleukin 12 alpha (IL12A, −17.2-fold regulation) was recorded. However, Dt treatment induced a down-regulation of interferon regulatory factor 7 (IRF7), mitogen-activated protein kinase 1 (MAP3K1), myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) (MX1), proline-serine-threonine phosphatase interacting protein 1 (PSTP1P1) and Toll-like receptors 7 and 9 (TLR7 and TLR9), (−2.2-, −2.16-, −5.44-, −7.37-, −13.66- and −2.74-fold regulation, respectively). Finally, the treatment with Dt led to a downregulation of Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (APOBEC3G) and chemokine (C-X-C motif) ligand 9 (CXCL9).
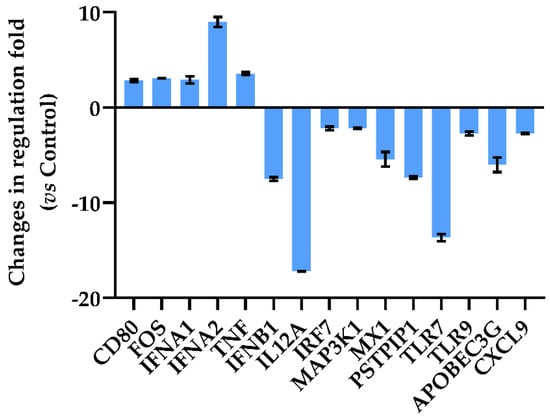
Figure 4.
Quantitative RT-PCR of oxidative stress-related genes in A549-hACE2 cells after 2 h of treatment with 0.1 ng mL−1 of Dt. Values are expressed as mean ± SD of changes in fold-regulation compared to the control (no Dt). All data from gene expression analysis are included in Table S1.
3.5. Anti-Inflammatory Response and Protein Expression
The expression levels of tumor necrosis factor receptor 1 (TNFR1) and receptor-interacting-serine/threonine-protein kinase 2 (RIP2) proteins were investigated in A549-hACE2 cells, treated or not with the pro-inflammatory interleukin 1β (IL-1β) [41] and with or without Dt addition. A549-hACE2 cells treated with Dt increased the TNFR1 protein expression, while no effect on RIP2 protein expression was observed (p < 0.0001 and p > 0.05, respectively; Figure 5). The exposure of A549-hACE2 cells to IL-1β determined a huge increase in TNFR1 protein expression and the decrease of RIP2 protein expression compared to the control (p < 0.0001 and p < 0.001, respectively; Figure 5). In this context, Dt decreased the TNFR1 protein expression to the control level (Figure 5; p < 0.001), and significantly enhanced RIP2 expression (Figure 5; p < 0.0001).
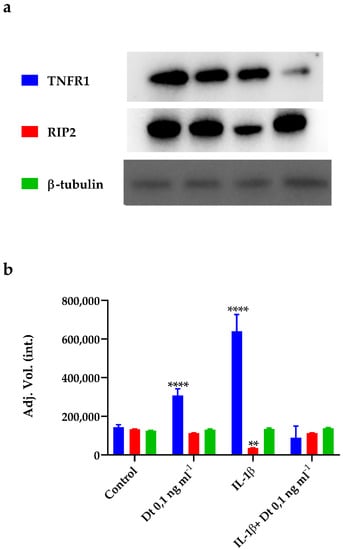
Figure 5.
Expression of TNFR1, RIP2, and β-tubulin proteins in A549-hACE2 cells in different conditions, namely no treatment (Control), treatment for 48 h with Dt at 0.1 ng mL−1, 24 h stimulation with IL-1β (40 pg mL−1), and treatment for 48 h with Dt after a 24 h pre-stimulation with IL-1β (40 pg mL−1). Bands (a) obtained for the proteins were analysed by densitometric analysis to determine the absolute intensity (b). Values are expressed as mean ± SD. Asterisks indicate the statistically significant difference compared to the respective control (**** p ≤ 0.0001, ** p ≤ 0.01; Tukey’s test).
3.6. Cytokine Storm Inhibition
Dt was able to act against the cytokine storm through the inhibition of the expression of the macrophage inflammatory proteins (MIP-1-α, Figure 6), as well of the cytokines interleukin 6 and 8 (IL-6 and IL-8), involved in the onset of the inflammatory storming that characterizes the innate immunity activation. In addition, a significant decrease of the relative expression of monocyte chemoattractant protein-1 (MCP-1, Figure 6) indicated a reduction in proinflammatory response involving immune cell recruitment. In parallel, the Dt-induced lowering of vascular endothelial growth factor (VEGF) production confirmed that inflammation and vasodilation were significantly inhibited (Figure 6).
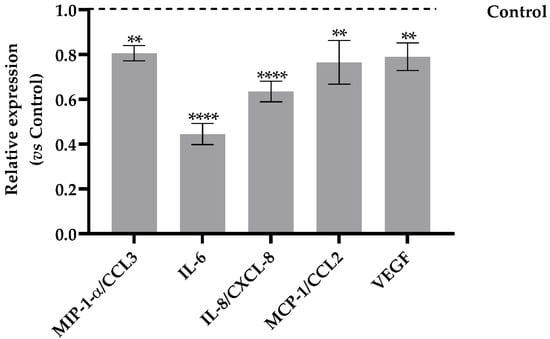
Figure 6.
Human cytokines array analysis of the conditional medium from A549-hACE2 cells treated for 24 h with Dt at 0.1 ng mL−1, after a 24 h pre-stimulation with IL-1β (40 pg mL−1). Values are expressed as relative expression mean levels ± SD, obtained comparing the expression values to those of IL-1β control (**** p ≤ 0.0001, ** p ≤ 0.01; Sidak’s test).
4. Discussion
Coronavirus disease (COVID-19) is the etiologic factor for the pandemic novel severe acute respiratory syndrome-related coronavirus (SARS-CoV-2). The COVID-19 pandemic was similar to a “butterfly effect”, starting with the virus-mediated intracellular cytokine storm in lung cells and generating global economic and social disorders, together with a high mortality rate. This pandemic tells us that modern times’ biological infections might induce unthinkable chaos on Earth, thus critically needing the generation of healthy behaviors and products to enhance prevention against viral attack. The development and administration of vaccines by companies such as AstraZeneca/University of Oxford (UK), Institute Pasteur/Merck/Themis (France/USA/Austria), University of Hong Kong (China), CureVac (Germany), Moderna (USA), Inovio (USA), Clover Biopharmaceuticals (China), Novavax (USA), University of Queensland/CSL (Australia), and Sechenov University (Russia, Sputnik V) reached different phases, but no standard and specific treatment protocol has yet been developed. Vaccine efficacy depends on virus evolution and its mutation [42]. Prevention is worthwhile, both in the vision of National Health Systems towards pandemic shortages and extraordinary expenses, and in economical sustainability for patients. Therefore, it is crucial to invest in preventing actions to inhibit viral entry in the organisms. Targeting ACE2 is one of the promising approaches for preventing COVID-19 disease [43], since small compounds that are able to interact with the ACE2 receptor are likely to inhibit the attachment of SARS-CoV-2 onto the targeted human cells [17,44,45], or block the early interactions of SARS-CoV-2 with ACE2, as demonstrated for SSAA09E2 {N-[[4-(4-methyl-piperazin-1-yl)phenyl]methyl]-1,2-oxazole-5-carboxamide} or NAAE (N-(2-aminoethyl)-l-aziridine-ethanamine) [46]. In addition, chloroquine—a medication used for treating malaria—inhibits SARS-CoV-2 infection and the exacerbation of pneumonia, whereas side effects might be somehow important [47]. Seeking natural and eco-sustainable compounds able to significantly bind ACE2 is therefore essential. In this context, molecular docking to predict the capacity of molecules for binding ACE2, as well as in vitro tests using human cell lines and the SARS-CoV-2 spike protein, are tools allowing the bioprospecting of bioactive molecules potentially useful for health prevention [48,49,50,51,52].
Diatoxanthin is an exclusively aquatic microalgal xanthophyll with a (photo-)protective role [27,28,53]. Recent findings highlighted its outstanding bioactivity, with marked antioxidant, anti-inflammatory, and chemopreventive properties ([27,33] unpublished data). Interestingly, Dt can induce ferroptosis in cancer cell lines [33]. These results, together with the fact that Dt content in microalgae responds to external stresses (e.g., light, nutrients, iron, or viral attack) [31,32,50,54,55] lay the foundation to explore the potential of Dt as an inhibitor of the SARS-CoV-2-induced cytokine storm in lung cells. Results do strongly confirm this hypothesis, with a marked role of Dt in targeting the ACE2 receptor of tumor lung cells and inhibiting the cytokine storm following the contact between cells and the SARS-CoV-2 spike glycoprotein. Dt is eligible as a preventive/therapeutic agent against SARS-CoV-2 infection, cytokine storm consequences at the multiorgan level, and the long-term effects of infection due to the high levels of inflammatory markers in the blood [56]. A recent in vitro study highlighted that Dt displays antioxidant activity, and exerts an anti-inflammatory role [27]. Here, it is shown that Dt inhibits IL-6 release in lung cells exposed to the spike glycoprotein, and increases the IL-10 expression, providing an immunomodulatory signaling role. This result is strongly relevant since IL-6 increase is one of the biological responses responsible for severe respiratory injuries in patients affected by SARS-CoV-2 with dramatic involvement of coagulopathy and lung thrombosis after bilateral pneumonia [2]. The immunostimulant role of Dt involves the up-regulation of interferon alpha 1 and 2 (IFNA1 and IFNA2) and tumor necrosis factor (TNF), belonging to the intracellular innate system for the release of inflammatory mediators that induce specific differentiation in naïve monocytes [50]. The overexpression of interferon suggests an effective protection against ARDS, whereas recent studies demonstrated that patients with IFNAR1 deficiency were prone to develop severe forms of COVID-19 [57].
It is also noteworthy that Dt achieves its bioactive role in contrasting in vitro the first potential effects of SARS-CoV-2 at low concentration (0.1 ng mL−1), which is relevant and promising for potential therapy. A few cubic millimeters of diatom culture are enough to reach such a Dt content [31,32].
Moreover, it is important to consider that (Dt-containing) microalgae offer great opportunities as relevant eco-sustainable resources for the biotechnological panorama [58], being promising cell factories displaying fast growth capacity and being genetically and metabolically engineerable. Indeed, to be attractive—and not only regarding direct health benefit efficacy—the product needs to be sustainable, i.e., with a low productive cost, limited impact on the environment, and using renewable resources. In this framework, terrestrial plants would be limited/avoided as a resource for new compounds due to the environmental costs of terrestrial cultivation and the long time needed for growth (see the review of [53,59]). Microalgae, instead, sharing photosynthetic and microbial properties, are a promising eco-sustainable alternative [60,61,62,63,64,65]. Moreover, the microalgae content and diversity in bioactive compounds—for example, vitamins, carotenoids, or polyphenols [23,27,66,67,68]—make them highly attractive bio-factories [60,67,68,69].
The next step regarding industrial development involves the lowering of the productive cost through technological improvements, and/or the enhancement of the bioactive interests/quality of the biomass [70,71,72,73].
The results from this study, as well as the Dt-producing microalgae opportunities, pave the way for the further investigation, i.e., the in vivo assessment of the antiviral role of Dt, especially against the SARS-CoV-2 virus.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antiox11081515/s1, Table S1: Fold regulation values of genes obtained from PCR-array gene expression analysis. Figure S1: Chemical structure of diatoxanthin (C40H54O2, CAS No. 31063-73-7.
Author Contributions
Conceptualization, C.S., L.C., D.M.N., A.A. and C.B.; data curation, L.P. and C.B.; formal analysis, L.P. and A.D.M.; funding acquisition, A.F. and C.B.; investigation, C.S., L.P., L.C. and A.D.M.; methodology, L.P., L.C. and A.D.M.; project administration, C.S., A.F., A.A. and C.B.; resources, A.A.; supervision, C.S., A.F., D.M.N., A.A. and C.B.; validation, L.P. and C.B.; writing—original draft, C.S. and L.P.; writing—review and editing, C.S., L.P., L.C., A.D.M., A.F., D.M.N., A.A. and C.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Stazione Zoologica Anton Dohrn and by the “Antitumor Drugs and Vaccines from the Sea (ADViSE)” project (PG/2018/0494374). Luigi Pistelli is supported by a PhD fellowship co-funded by the “Antitumor Drugs and Vaccines from the Sea (ADViSE)” project (PG/2018/0494374) and by the Stazione Zoologica Anton Dohrn (PhD Program XXI cycle, Open University, Milton Keynes, UK). Angelo del Mondo is supported by a post-doctoral fellowship funded by the “Antitumor Drugs and Vaccines from the Sea (ADViSE)” project (PG/2018/0494374). This research was partially supported by a grant from the Ministero della Salute COVID-2020-12371849 to D.M.N. Studies are also partially funded by the Italian Ministry of Health Ricerca Corrente—IRCCS MultiMedica.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All of the data is contained within the article and the Supplementary Materials.
Acknowledgments
The authors would like to acknowledge the reviewers for their criticisms on the previous version of the manuscript.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Fuentes-Prior, P. Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection. J. Biol. Chem. 2021, 296, 100135. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Yin, Z.; Hu, Y.; Mei, H. Controlling Cytokine Storm Is Vital in COVID-19. Front. Immunol. 2020, 11, 570993. [Google Scholar] [CrossRef] [PubMed]
- Hsu, R.J.; Yu, W.C.; Peng, G.R.; Ye, C.H.; Hu, S.; Chong, P.C.T.; Yap, K.Y.; Lee, J.Y.C.; Lin, W.C.; Yu, S.H. The Role of Cytokines and Chemokines in Severe Acute Respiratory Syndrome Coronavirus 2 Infections. Front. Immunol. 2022, 13, 832394. [Google Scholar] [CrossRef] [PubMed]
- Ragab, D.; Salah Eldin, H.; Taeimah, M.; Khattab, R.; Salem, R. The COVID-19 Cytokine Storm; What We Know So Far. Front. Immunol. 2020, 11, 1446. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Ma, Q.; Li, C.; Liu, R.; Zhao, L.; Wang, W.; Zhang, P.; Liu, X.; Gao, G.; Liu, F.; et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg. Microbes Infect. 2020, 9, 1123–1130. [Google Scholar] [CrossRef]
- Majidpoor, J.; Mortezaee, K. Interleukin-6 in SARS-CoV-2 induced disease: Interactions and therapeutic applications. Biomed. Pharmacother. 2022, 145, 112419. [Google Scholar] [CrossRef]
- Ali, A.; Kamjani, M.H.; Kesselman, M.M. The Role of Tocilizumab in Cytokine Storm and Improving Outcomes in COVID-19. Recent Pat. Antiinfect. Drug Discov. 2020, 15, 104–112. [Google Scholar] [CrossRef]
- Sujan, K.D.; Vishnupriyan, K.; Sharat, D.; Shashi, G.; Manjula, D. IL-6 and IL-10 as predictors of disease severity in COVID-19 patients: Results from meta-analysis and regression. Heliyon 2021, 7, e06155. [Google Scholar] [CrossRef]
- Chen, J.S.; Alfajaro, M.M.; Chow, R.D.; Wei, J.; Filler, R.B.; Eisenbarth, S.C.; Wilen, C.B. Non-steroidal anti-inflammatory drugs dampen the cytokine and antibody response to SARS-CoV-2 infection. J. Virol. 2021, 95, e00014-21. [Google Scholar] [CrossRef]
- Stan, D.; Enciu, A.-M.; Mateescu, A.L.; Ion, A.C.; Brezeanu, A.C.; Stan, D.; Tanase, C. Natural Compounds with Antimicrobial and Antiviral Effect and Nanocarriers Used for Their Transportation. Front. Pharmacol. 2021, 12, 723233. [Google Scholar] [CrossRef]
- Azab, A.; Nassar, A.; Azab, A.N. Anti-Inflammatory Activity of Natural Products. Molecules 2016, 21, 1321. [Google Scholar] [CrossRef] [PubMed]
- Joshi, T.; Joshi, T.; Sharma, P.; Mathpal, S.; Pundir, H.; Bhatt, V.; Chandra, S. In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4529–4536. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Zhang, J.; Bian, Z.; Zhou, H.; Zhang, Z.; Lin, Z.; Xu, H. Bioactive natural compounds against human coronaviruses: A review and perspective. Acta Pharm. Sin. B 2020, 10, 1163–1174. [Google Scholar] [CrossRef]
- Kim, C.-H. Anti–SARS-CoV-2 Natural Products as Potentially Therapeutic Agents. Front. Pharmacol. 2021, 12, 590509. [Google Scholar] [CrossRef] [PubMed]
- Chakravarti, R.; Singh, R.; Ghosh, A.; Deya, D.; Sharmaa, P.; Velayuthama, R.; Roy, S.; Ghosh, D. A review on potential of natural products in the management of COVID-19. RSC Adv. 2021, 11, 16711. [Google Scholar] [CrossRef]
- Chhetri, B.K.; Tedbury, P.R.; Sweeney-Jones, A.M.; Mani, L.; Soapi, K.; Manfredi, C.; Sorscher, E.; Sarafianos, S.G.; Kubanek, J. Marine Natural Products as Leads against SARS-CoV-2 Infection. J. Nat. Prod. 2022, 85, 657–666. [Google Scholar] [CrossRef]
- Isidoro, C.; Chang, A.C.-F.; Sheen, L.-Y. Natural products as a source of novel drugs for treating SARS-CoV2 infection. J. Tradit. Complement. Med. 2022, 12, 1–5. [Google Scholar] [CrossRef]
- Sen, T.; Samanta, S.K. Medicinal plants, human health and biodiversity: A broad review. Adv. Biochem. Eng. Biotechnol. 2015, 147, 59–110. [Google Scholar] [CrossRef]
- Machado Sierra, E.; Serrano, M.C.; Manares, A.; Guerra, A.; Aranguren Díaz, Y. Microalgae: Potential for Bioeconomy in Food Systems. Appl. Sci. 2021, 11, 11316. [Google Scholar] [CrossRef]
- Koyande, A.K.; Chew, K.W.; Rambabu, K.; Tao, Y.; Chu, D.-T.; Show, P.-L. Microalgae: A potential alternative to health supplementation for humans. Food Sci. Hum. Wellness 2019, 8, 16–24. [Google Scholar] [CrossRef]
- Sansone, C.; Brunet, C.; Noonan, D.M.; Albini, A. Marine Algal Antioxidants as Potential Vectors for Controlling Viral Diseases. Antioxidants 2020, 9, 392. [Google Scholar] [CrossRef] [PubMed]
- Panggabean, J.A.; Adiguna, S.P.; Rahmawati, S.I.; Ahmadi, P.; Zainuddin, E.N.; Bayu, A.; Putra, M.Y. Antiviral Activities of Algal-Based Sulfated Polysaccharides. Molecules 2022, 27, 1178. [Google Scholar] [CrossRef] [PubMed]
- Del Mondo, A.; Smerilli, A.; Ambrosino, L.; Albini, A.; Noonan, D.M.; Sansone, C.; Brunet, C. Insights into phenolic compounds from microalgae: Structural variety and complex beneficial activities from health to nutraceutics. Crit. Rev. Biotechnol. 2021, 41, 155–171. [Google Scholar] [CrossRef]
- Talukdar, J.; Bhadra, B.; Dattaroy, T.; Nagle, V.; Dasgupta, S. Potential of natural astaxanthin in alleviating the risk of cytokine storm in COVID-19. Biomed. Pharmacother. 2020, 132, 110886. [Google Scholar] [CrossRef]
- Yim, S.K.; Kim, I.; Warren, B.; Kim, J.; Jung, K.; Ku, B. Antiviral Activity of Two Marine Carotenoids against SARS-CoV-2 Virus Entry In Silico and In Vitro. Int. J. Mol. Sci. 2021, 22, 6481. [Google Scholar] [CrossRef]
- Karpiński, T.M.; Kwaśniewski, M.; Ożarowski, M.; Alam, R. In silico studies of selected xanthophylls as potential candidates against SARS-CoV-2 targeting main protease (Mpro) and papain-like protease (PLpro). Herba Pol. 2021, 67, 1–8. [Google Scholar] [CrossRef]
- Pistelli, L.; Sansone, C.; Smerilli, A.; Festa, M.; Noonan, D.M.; Albini, A.; Brunet, C. MMP-9 and IL-1β as targets for diatoxanthin and related microalgal pigments: Potential chemopreventive and photoprotective agents. Mar. Drugs 2021, 19, 354. [Google Scholar] [CrossRef]
- Brunet, C.; Brylinski, J.M.; Lemoine, Y. In situ variations of the xanthophylls diadinoxanthin and diatoxanthin: Photoadaptation and relationships with an hydrodynamical system of the Eastern English Channel. Mar. Ecol. Prog. Ser. 1993, 95, 69–77. [Google Scholar] [CrossRef]
- Brunet, C.; Casotti, R.; Aronne, B.; Vantrepotte, V. Measured photophysiological parameters used as tools to estimate vertical water movements in the coastal Mediterranean. J. Plankton Res. 2003, 25, 1413–1425. [Google Scholar] [CrossRef] [Green Version]
- Orefice, I.; Chandrasekaran, R.; Smerilli, A.; Corato, F.; Caruso, T.; Casillo, A.; Corsaro, M.M.; Dal Piaz, F.; Ruban, A.V.; Brunet, C. Light-induced changes in the photosynthetic physiology and biochemistry in the diatom Skeletonema marinoi. Algal Res. 2016, 17, 1–13. [Google Scholar] [CrossRef]
- Smerilli, A.; Orefice, I.; Corato, F.; Olea, A.G.; Ruban, A.V.; Brunet, C. Photoprotective and antioxidant responses to light spectrum and intensity variations on a coastal diatom Skeletonema marinoi. Environ. Microbiol. 2017, 19, 611–627. [Google Scholar] [CrossRef] [PubMed]
- Smerilli, A.; Balzano, S.; Maselli, M.; Blasio, M.; Orefice, I.; Galasso, C.; Sansone, C.; Brunet, C. Antioxidant and photoprotection networking in the coastal diatom Skeletonema marinoi. Antioxidants 2019, 8, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, D.; Kepp, O.; Kroemer, G. Ferroptosis becomes immunogenic: Implications for anticancer treatments. Oncoimmunology 2020, 10, 1862949. [Google Scholar] [CrossRef]
- Pawlica, P.; Yario, T.A.; White, S.; Wang, J.; Moss, W.N.; Hui, P.; Vinetz, J.M.; Steitz, J.A. SARS-CoV-2 expresses a microRNA-like small RNA able to selectively repress host genes. Proc. Natl. Acad. Sci. USA 2021, 118, e2116668118. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gilmore, K.; Ramirez, S.; Settels, E.; Gammeltoft, K.A.; Pham, L.V.; Fahnøe, U.; Feng, S.; Offersgaard, A.; Trimpert, J.; et al. In vitro efficacy of artemisinin-based treatments against SARS-CoV-2. Sci. Rep. 2021, 11, 14571. [Google Scholar] [CrossRef]
- Pistelli, L.; Del Mondo, A.; Smerilli, A.; Corato, F.; Piscitelli, C.; Pellone, P.; Carbone, D.A.; Sansone, C.; Brunet, C. Microalgal Co-Cultivation Prospecting to Modulate Vitamin and Bioactive Compounds Production. Antioxidants 2021, 10, 1360. [Google Scholar] [CrossRef]
- Wang, F.; Wu, F.X.; Li, C.Z.; Jia, C.-Y.; Su, S.-W.; Hao, G.-F.; Yang, G.-F. ACID: A free tool for drug repurposing using consensus inverse docking strategy. J. Cheminform. 2019, 11, 73. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Lindner, H.A.; Velásquez, S.Y.; Thiel, M.; Kirschning, T. Lung Protection vs. Infection Resolution: Interleukin 10 Suspected of Double-Dealing in COVID-19. Front. Immunol. 2021, 12, 602130. [Google Scholar] [CrossRef]
- Mantovani, A.; Dinarello, C.A.; Molgora, M.; Garlanda, C. Interleukin-1 and Related Cytokines in the Regulation of Inflammation and Immunity. Immunity 2019, 50, 778–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294. [Google Scholar] [CrossRef]
- Scialo, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Castaldo, G.; Bianco, A. ACE2: The Major Cell Entry Receptor for SARS-CoV-2. Lung 2020, 198, 867–877. [Google Scholar] [CrossRef]
- Singla, R.K.; He, X.; Chopra, H.; Tsagkaris, C.; Shen, L.; Kamal, M.A.; Shen, B. Natural Products for the Prevention and Control of the COVID-19 Pandemic: Sustainable Bioresources. Front. Pharmacol. 2021, 12, 758159. [Google Scholar] [CrossRef] [PubMed]
- Villa, A.; Brunialti, E.; Dellavedova, J.; Meda, C.; Rebecchi, M.; Conti, M.; Donnici, L.; De Francesco, R.; Reggiani, A.; Lionetti, V.; et al. DNA aptamers masking angiotensin converting enzyme 2 as an innovative way to treat SARS-CoV-2 pandemic. Pharmacol. Res. 2022, 175, 105982. [Google Scholar] [CrossRef]
- Liu, M.; Wang, T.; Zhou, Y.; Zhao, Y.; Zhang, Y.; Li, J. Potential role of ACE2 in coronavirus disease 2019 (COVID-19) prevention and management. J. Transl. Int. Med. 2020, 8, 9–19. [Google Scholar] [CrossRef]
- Meyerowitz, E.A.; Vannier, A.G.L.; Friesen, M.G.N.; Schoenfeld, S.; Gelfand, J.A.; Callahan, M.V.; Kim, A.Y.; Reeves, P.M.; Poznansky, M.C. Rethinking the role of hydroxychloroquine in the treatment of COVID-19. FASEB J. 2020, 34, 6027–6037. [Google Scholar] [CrossRef] [PubMed]
- Basu, A.; Sarkar, A.; Maulik, U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci. Rep. 2020, 10, 17699. [Google Scholar] [CrossRef]
- Takayama, K. In Vitro and Animal Models for SARS-CoV-2 research. Trends Pharmacol. Sci. 2020, 41, 513–517. [Google Scholar] [CrossRef]
- Lazniewski, M.; Dermawan, D.; Hidayat, S.; Muchtaridi, M.; Dawson, W.K.; Plewczynski, D. Drug repurposing for identification of potential spike inhibitors for SARS-CoV-2 using molecular docking and molecular dynamics simulations. Methods 2022, 203, 498–510. [Google Scholar] [CrossRef]
- Biswas, S.; Mahmud, S.; Mita, M.A.; Afrose, S.; Hasan, M.R.; Sultana Shimu, M.S.; Saleh, M.A.; Mostafa-Hedeab, G.; Alqarni, M.; Obaidullah, A.J.; et al. Molecular Docking and Dynamics Studies to Explore Effective Inhibitory Peptides Against the Spike Receptor Binding Domain of SARS-CoV-2. Front. Mol. Biosci. 2022, 8, 791642. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.K.B.; Olson, K.M.; Wotring, J.W.; Sexton, J.Z.; Carlson, H.A.; Traynor, J.R. In silico analysis of SARS-CoV-2 proteins as targets for clinically available drugs. Sci. Rep. 2022, 12, 5320. [Google Scholar] [CrossRef] [PubMed]
- Brunet, C.; Johnsen, G.; Lavaud, J.; Roy, S. Pigments and photoacclimation processes. In Phytoplankton Pigments; Roy, S., Johnsen, G., Llewellyn, C., Skarstad, E., Eds.; Cambridge University: Cambridge, UK, 2011; pp. 445–471. [Google Scholar]
- Llewellyn, C.A.; Evans, C.; Airs, R.L.; Cook, I.; Bale, N.; Wilson, W.H. The response of carotenoids and chlorophylls during virus infection of Emiliania huxleyi (Prymnesiophyceae). J. Exp. Mar. Biol. Ecol. 2007, 344, 101–112. [Google Scholar] [CrossRef]
- Beer, A.; Juhas, M.; Büchel, C. Influence of Different Light Intensities and Different Iron Nutrition on The Photosynthetic Apparatus In The Diatom Cyclotella Meneghiniana (Bacillariophyceae). J. Phycol. 2011, 47, 1266–1273. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Xie, X.; Tu, Z.; Fu, J.; Xu, D.; Zhou, Y. The signal pathways and treatment of cytokine storm in COVID-19. Signal Transduct. Target Ther. 2021, 6, 255. [Google Scholar] [CrossRef]
- Khanmohammadi, S.; Rezaei, N.; Khazaei, M.; Shirkani, A. A Case of Autosomal Recessive Interferon Alpha/Beta Receptor Alpha Chain (IFNAR1) Deficiency with Severe COVID-19. J. Clin. Immunol. 2022, 42, 19–24. [Google Scholar] [CrossRef]
- Lu, Y.; Zhang, X.; Gu, X.; Lin, H.; Melis, A. Engineering microalgae: Transition from empirical design to programmable cells. Crit. Rev. Biotechnol. 2021, 41, 1233–1256. [Google Scholar] [CrossRef]
- Viana, C.M.; Freire, D.; Abrantes, P.; Rocha, J.; Pereira, P. Agricultural land systems importance for supporting food security and sustainable development goals: A systematic review. Sci. Total Environ. 2022, 806, 150718. [Google Scholar] [CrossRef]
- Garcıa, J.L.; de Vicente, M.; Galan, B. Microalgae, old sustainable food and fashion nutraceuticals. Microb. Biotechnol. 2017, 10, 1017–1024. [Google Scholar] [CrossRef] [Green Version]
- Fabris, M.; Abbriano, R.M.; Pernice, M.; Sutherland, D.L.; Commault, A.S.; Hall, C.C.; Labeeuw, L.; McCauley, J.I.; Kuzhiuparambil, U.; Ray, P.; et al. Emerging Technologies in Algal Biotechnology: Toward the Establishment of a Sustainable, Algae-Based Bioeconomy. Front. Plant Sci. 2020, 11, 279. [Google Scholar] [CrossRef] [Green Version]
- Araújo, R.; Vázquez Calderón, F.; Sánchez López, J.; Azevedo, I.C.; Bruhn, A.; Fluch, S.; Garcia Tasende, M.; Ghaderiardakani, F.; Ilmjärv, T.; Laurans, M.; et al. Current Status of the Algae Production Industry in Europe: An Emerging Sector of the Blue Bioeconomy. Front. Mar. Sci. 2021, 7, 626389. [Google Scholar] [CrossRef]
- Rotter, A.; Barbier, M.; Bertoni, F.; Bones, A.M.; Cancela, M.L.; Carlsson, J.; Carvalho, M.F.; Cegłowska, M.; Chirivella-Martorell, J.; Dalay, M.C.; et al. The Essentials of Marine Biotechnology. Front. Mar. Sci. 2021, 8, 629629. [Google Scholar] [CrossRef]
- Levasseur, W.; Perré, P.; Pozzobon, V. A review of high value-added molecules production by microalgae in light of the classification. Biotechnol. Adv. 2020, 41, 107545. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Melchor-Martínez, E.M.; Saxena, A.; Kapoor, N.; Singh, K.J.; Saldarriaga-Hernández, S.; Parra-Saldívar, R.; Iqbal, H.M.N. Therapeutic attributes and applied aspects of biological macromolecules (polypeptides, fucoxanthin, sterols, fatty acids, polysaccharides, and polyphenols) from diatoms—A review. Int. J. Biol. Macromol. 2021, 171, 398–413. [Google Scholar] [CrossRef]
- Del Mondo, A.; Smerilli, A.; Sané, E.; Sansone, C.; Brunet, C. Challenging microalgal vitamins for human health. Microbial. Cell Factories 2020, 19, 201. [Google Scholar] [CrossRef] [PubMed]
- Sañé, E.; Del Mondo, A.; Ambrosino, L.; Smerilli, A.; Sansone, C.; Brunet, C. The Recent Advanced in Microalgal Phytosterols: Bioactive Ingredients Along with Human-Health Driven Potential Applications. Food Rev. Int. 2021, 1–20. [Google Scholar] [CrossRef]
- Del Mondo, A.; Sansone, C.; Brunet, C. Insights into the biosynthesis pathway of phenolic compounds in microalgae. Computat. Struct. Biotechnol. J. 2022, 20, 1901–1913. [Google Scholar] [CrossRef]
- Butler, T.; Kapoore, R.V.; Vaidyanathan, S. Phaeodactylum tricornutum: A Diatom Cell Factory. Trends Biotechnol. 2020, 38, 606–622. [Google Scholar] [CrossRef]
- Williams, P.J.B.; Laurens, L.M.L. Microalgae as biodiesel & biomass feedstocks: Review & analysis of the biochemistry, energetics & economics. Energy Environ. Sci. 2010, 3, 554–590. [Google Scholar] [CrossRef]
- Ruiz, J.; Olivieri, G.; de Vree, J.H.; Bosma, R.; Willems, P.; Reith, J.H.; Barbosa, M.J.; Eppink, M.H.M.; Kleinegris, D.M.M.; Wijffels, R.H. Towards industrial products from microalgae. Energy Environ. Sci. 2016, 9, 3036–3043. [Google Scholar] [CrossRef] [Green Version]
- Kroumov, A.D.; Scheufele, F.B.; Trigueros, D.E.G.; Modenes, A.N.; Zaharieva, M.; Najdenski, H. Chapter 11—Modeling and Technoeconomic Analysis of Algae for Bioenergy and Coproducts. In Algal Green Chemistry; Rastogi, R.P., Madamwar, D., Pandey, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 201–241. [Google Scholar]
- Fernandez, F.G.A.; Reis, A.; Wijffels, R.H.; Barbosa, M.; Verdelho, V.; Llamas, B. The role of microalgae in the bioeconomy. New Biotechnol. 2021, 61, 99–107. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).