The Role of Organosulfur Compounds as Nrf2 Activators and Their Antioxidant Effects
Abstract
:1. Introduction
1.1. Role of Nrf2 in Human (Patho)Physiology
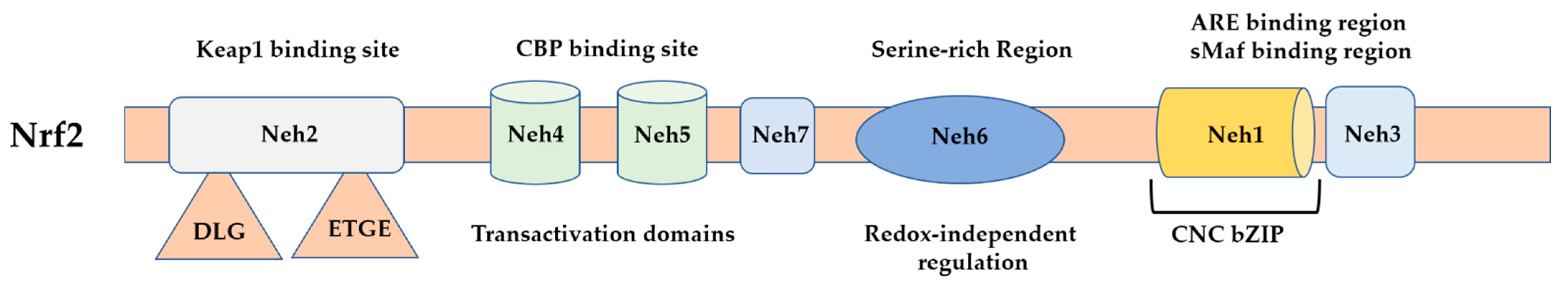
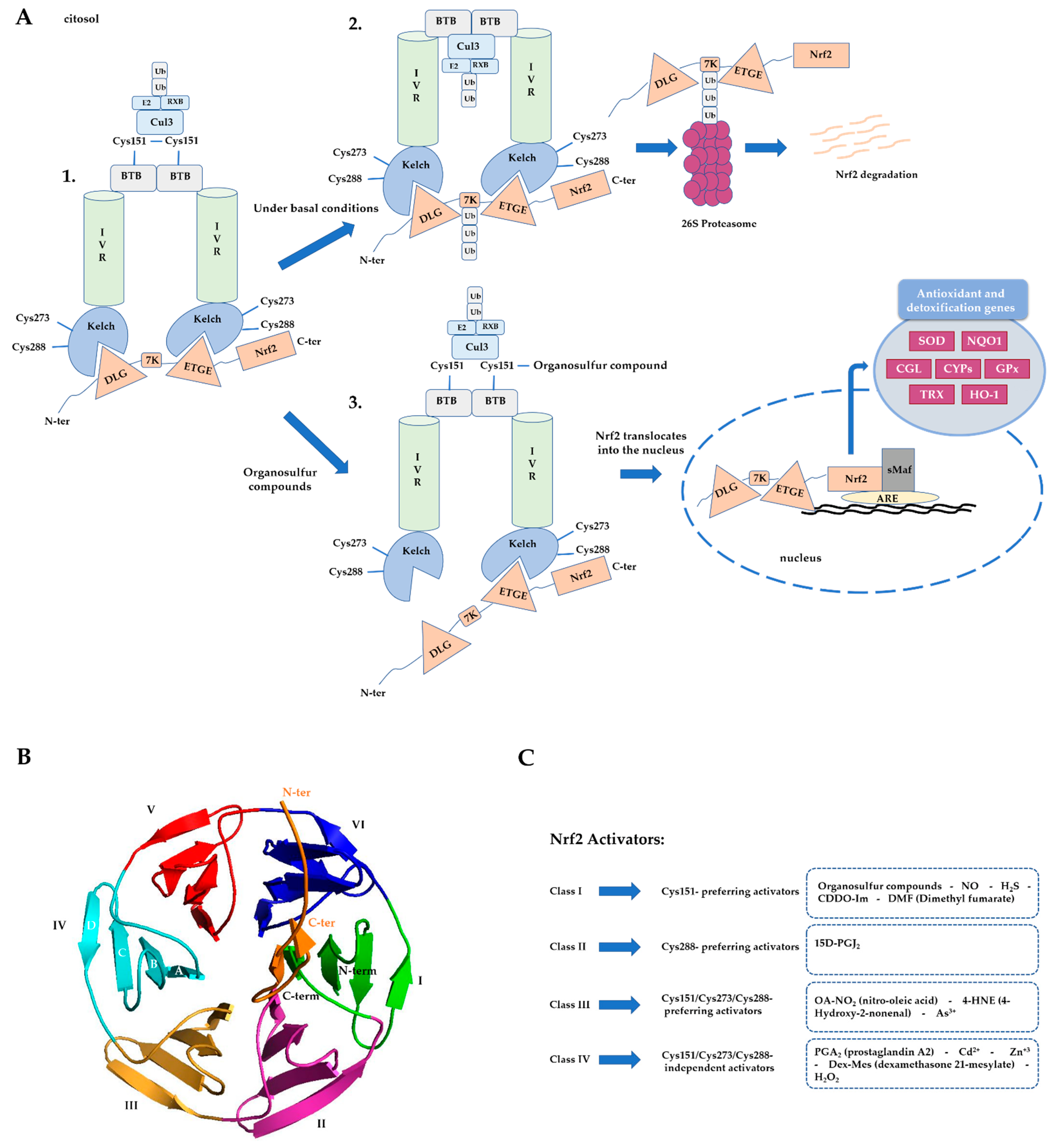
1.2. Nrf2 Regulation
2. Nrf2-Activating Synthetic Organosulfur Compounds
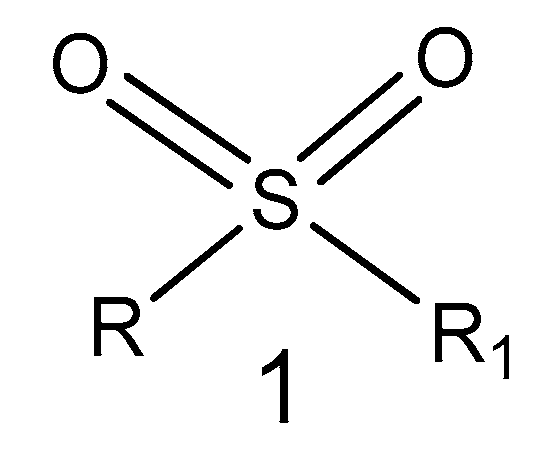
2.1. Sulfonyl Group-Containing Compounds
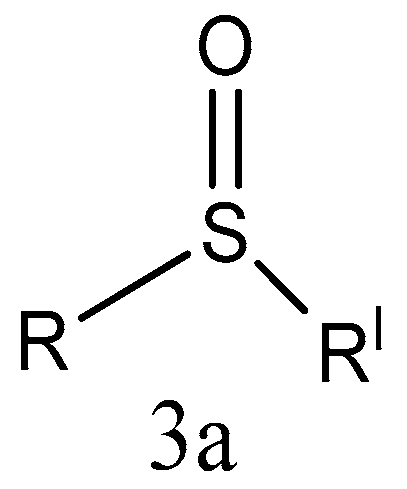
2.1.1. Sulfone Derivatives
2.1.2. Sulfonamide Derivatives
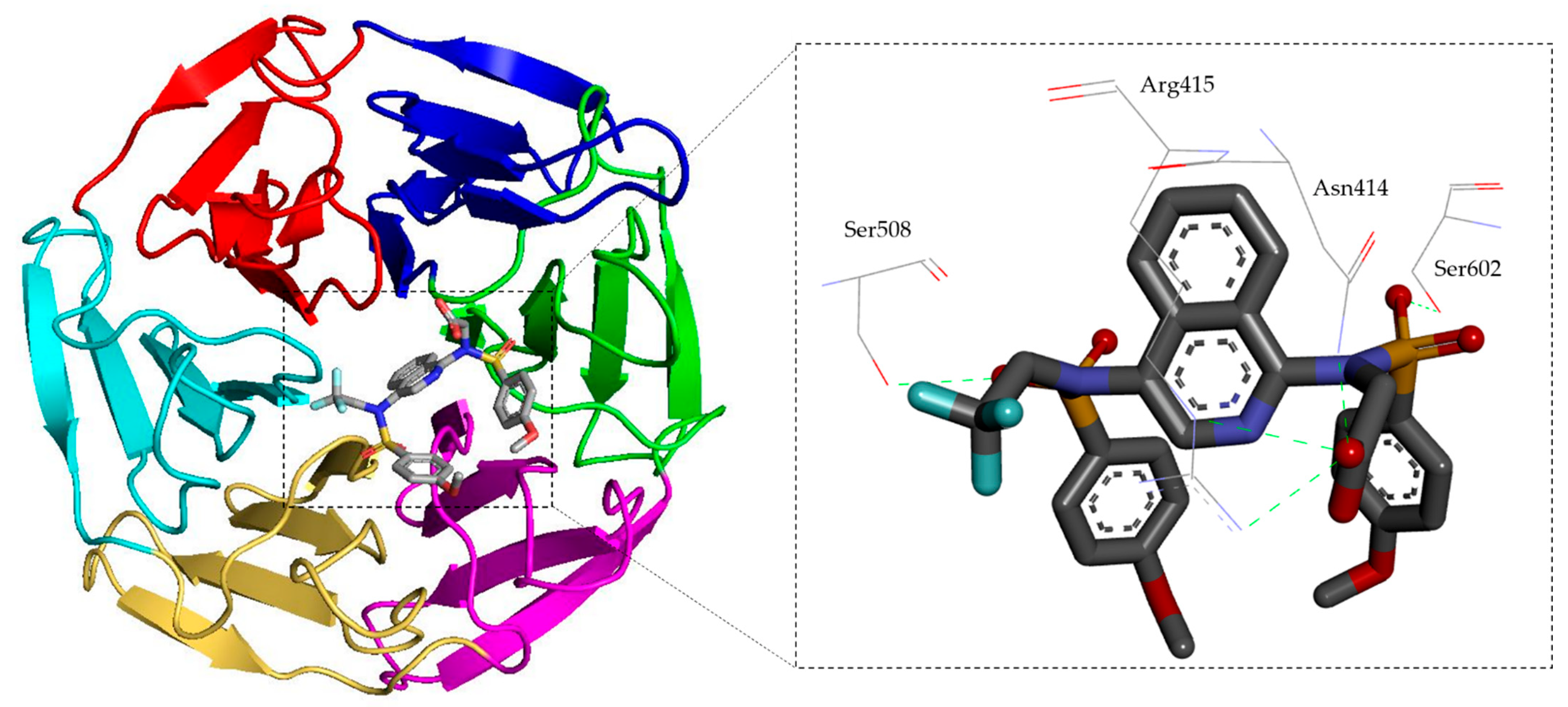
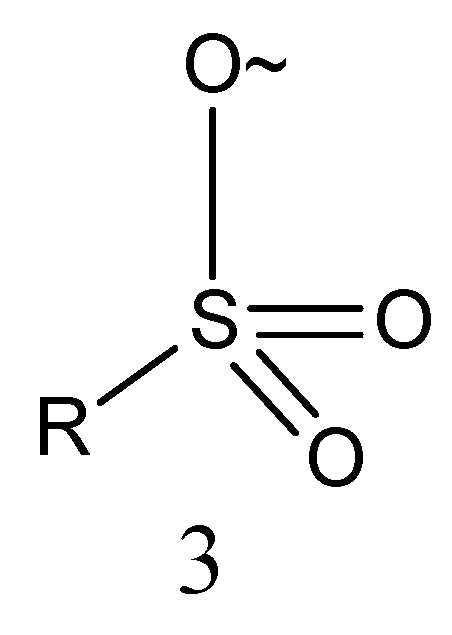
2.1.3. Sulfonate Derivatives
2.2. Sulfinyl Group-Containing Compounds
2.2.1. Sulfoxide Derivatives
2.2.2. Vinyl Sulfoxide Derivatives
2.2.3. Sulfoximine Derivatives
2.3. Oltipraz
| Entry | Compounds | Effective Concentration/Dose | Biological Activity | Study Model | Targeted Diseases | Ref |
|---|---|---|---|---|---|---|
| 4 | SULFONE DERIVATIVES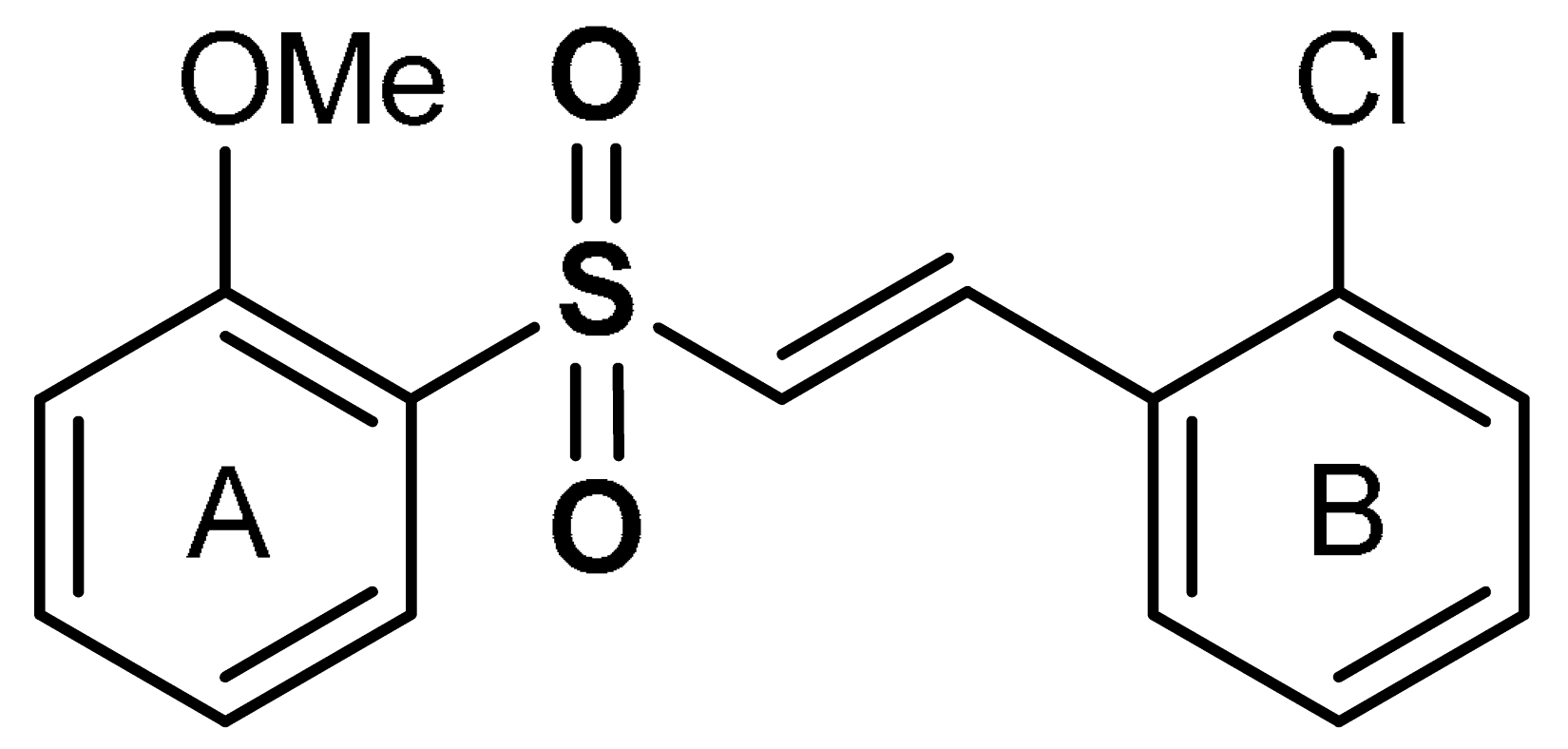 (E)-1-chloro-2-(2-((2-methoxyphenyl)sulfonyl)vinyl)benzene | 1–5 µM | Antioxidant, anti-inflammatory | Microglia, Parkinson’s disease animal model | Parkinson’s disease | [98] |
| 1–10 µM | Antioxidant, neuroprotection | MPTP-induced Parkinson’s disease mouse model | Parkinson’s disease | [99] | ||
| 10 µM | Neuroprotection | HEK293 cells | Traumatic brain injury | [100] | ||
| 5 | 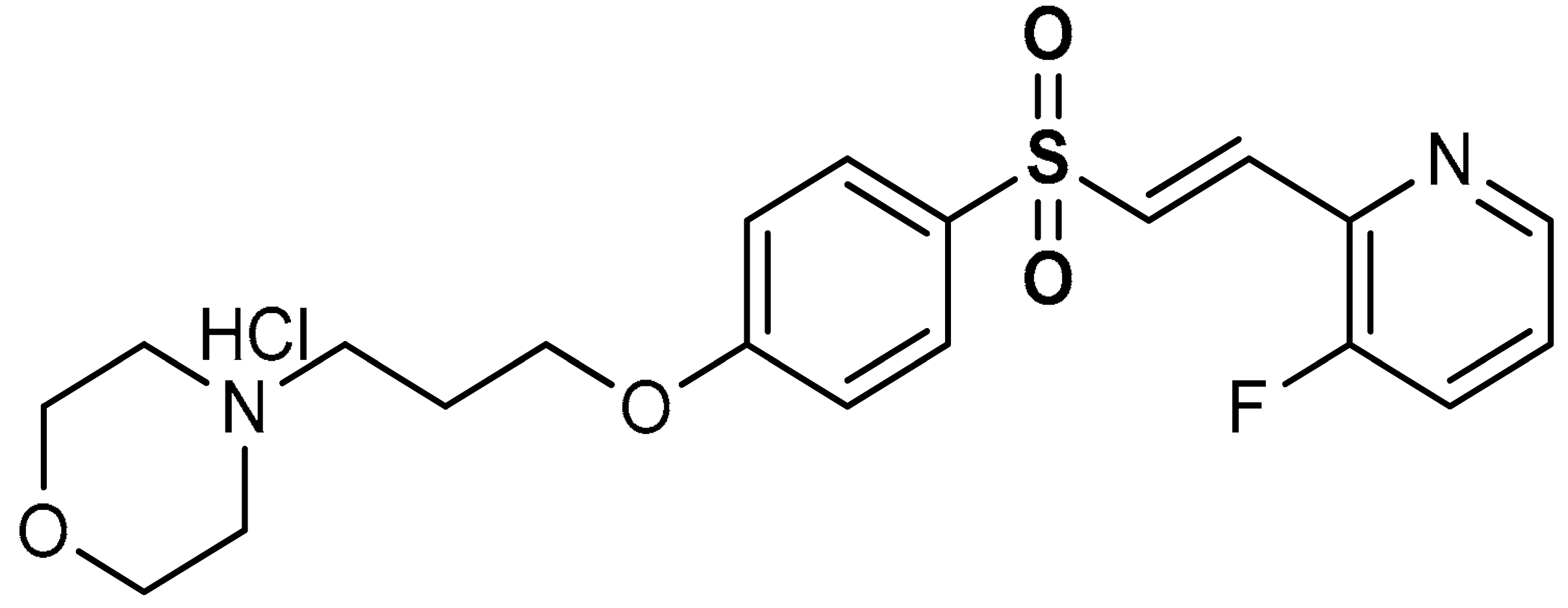 (E)-4-(3-(4-((2-(3-fluoropyridin-2-yl)vinyl)sulfonyl)phenoxy)propyl)morpholine hydrochloride | 10 µM | Antioxidant, neuroprotection | MPTP-induced Parkinson’s disease mouse model | Parkinson’s disease | [102] |
| 6 |  (E)-1-chloro-2-(2-((2-chlorophenyl)sulfonyl)vinyl)benzene | 0.5–1 µM | Antioxidant, neuroprotection | PC12 Cells | Oxidative stress | [32] |
| 7 | 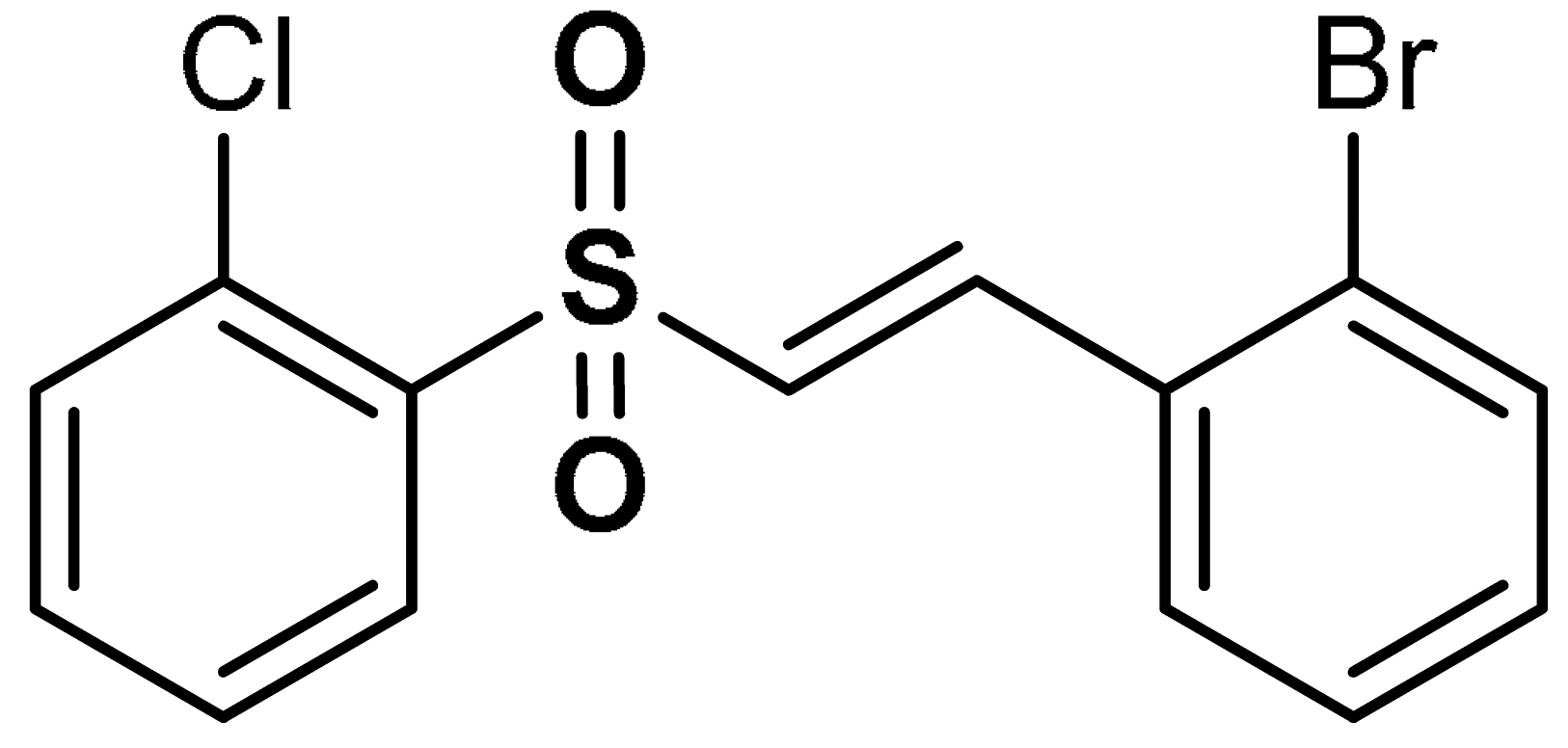 (E)-1-bromo-2-(2-((2-chlorophenyl)sulfonyl)vinyl)benzene | 0.5–1 µM | Antioxidant, neuroprotection | PC12 Cells | Oxidative stress | [32] |
| 8 | SULFONAMIDE DERIVATIVES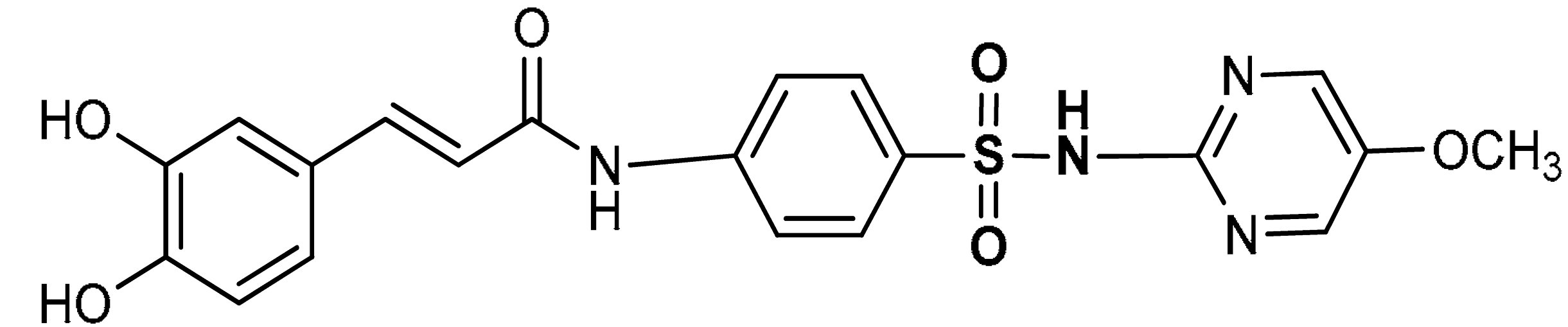 (E)-3-(3,4-dihydroxyphenyl)-N-(4-(N-(5-methoxypyridin-2-yl)sulfamoylphenyl)acrylamide | 50–200 µM | Antioxidant | A549 Cells | Oxidative stress | [111] |
| 9 | 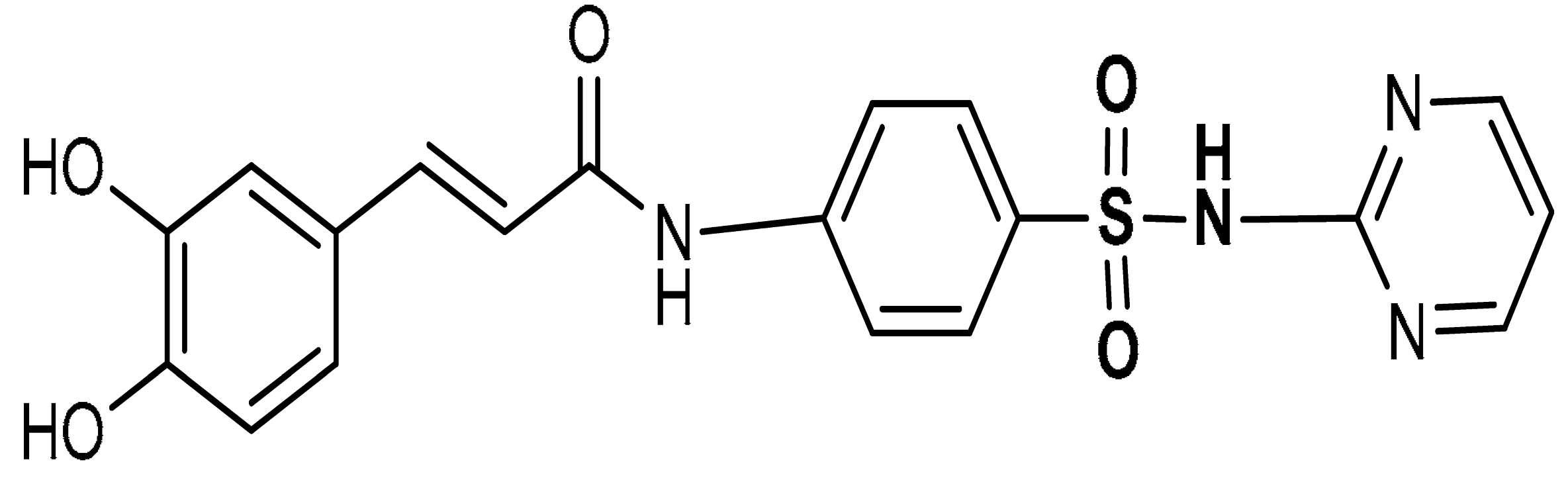 (E)-3-(3,4-dihydroxyphenyl)-N-(4-(N-pyrimidin-2-yl)sulfamoylphenyl)acrylamide | 50–200 µM | Antioxidant | A549 Cells | Oxidative stress | [111] |
| 10 | 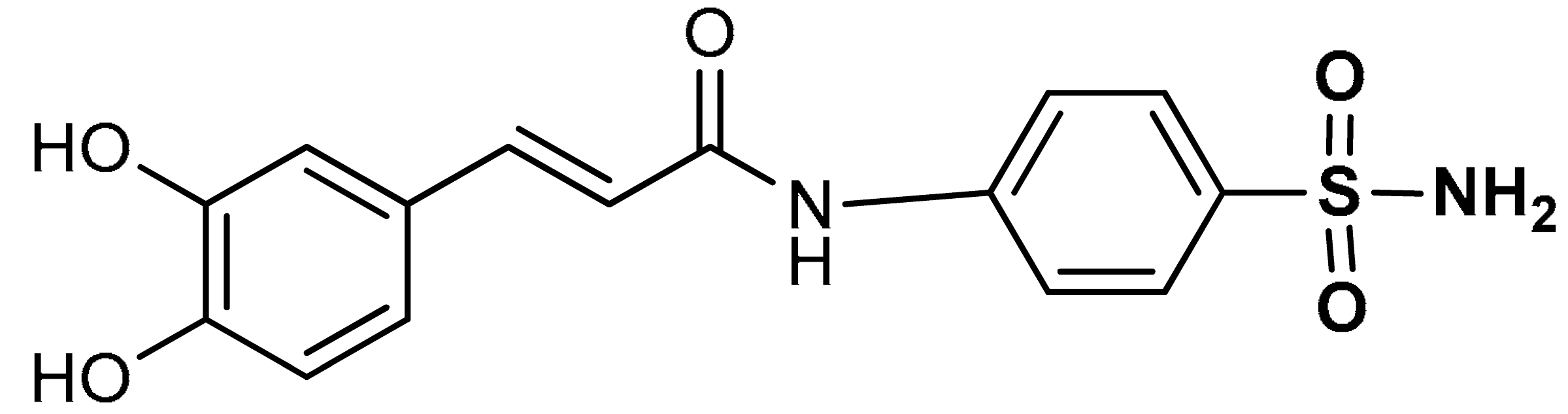 (E)-3-(3,4-dihydroxyphenyl)-N-(4-sulfamoylphenyl)acrylamide | 50–200 µM | Antioxidant | A549 Cells | Oxidative stress | [111] |
| 11 | 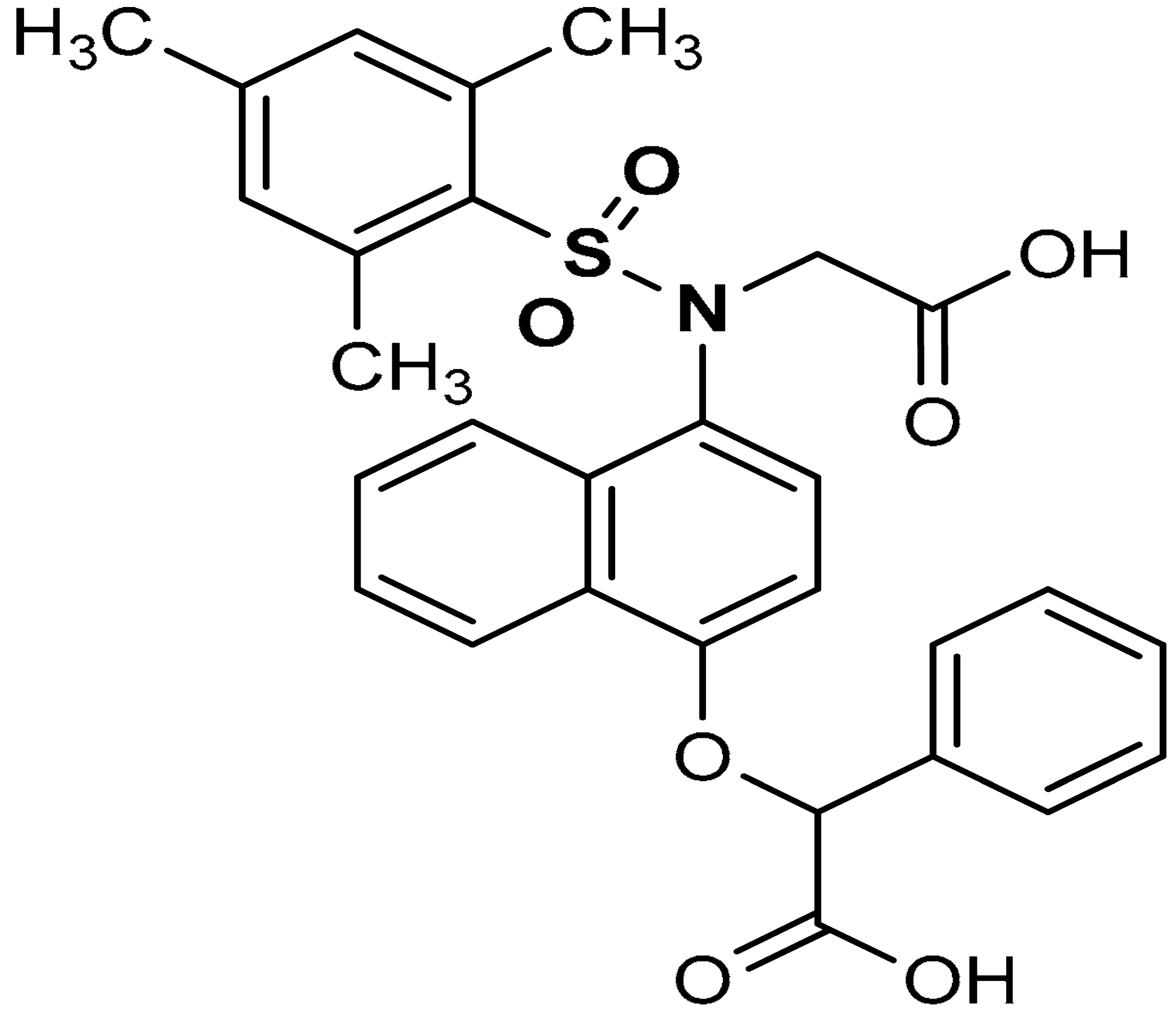 2-((4-(N-(carboxymethyl)-2,4,6-trimethylphenylsulfonamido)naphthalen-1-yl)oxy)-2-phenylacetic acid | 0.58 µM | Antioxidant, Keap1–Nrf2 PPI inhibition | RAW264.7 Cells | Inflammation | [94] |
| 12 | 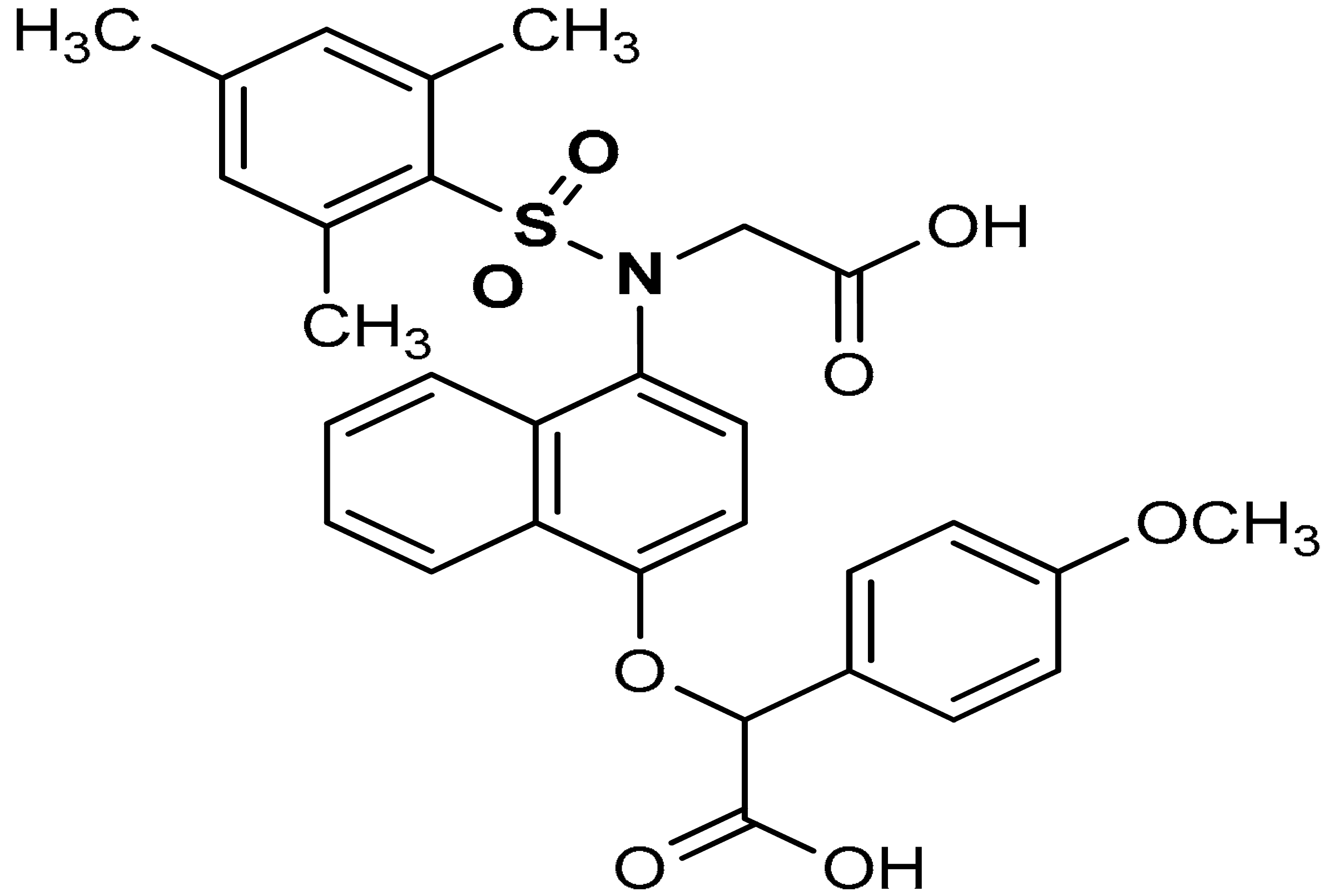 2-((4-(N-(carboxymethyl)-2,4,6-trimethylphenylsulfonamido)naphthalen-1-yl)oxy)-2-(4-methoxyphenyl)acetic acid | ≥500 µL | Antioxidant, Keap1–Nrf2 PPI inhibition | RAW264.7 Cells. HepG2 Cells, mice, rat | Inflammation | [94] |
| 13 | 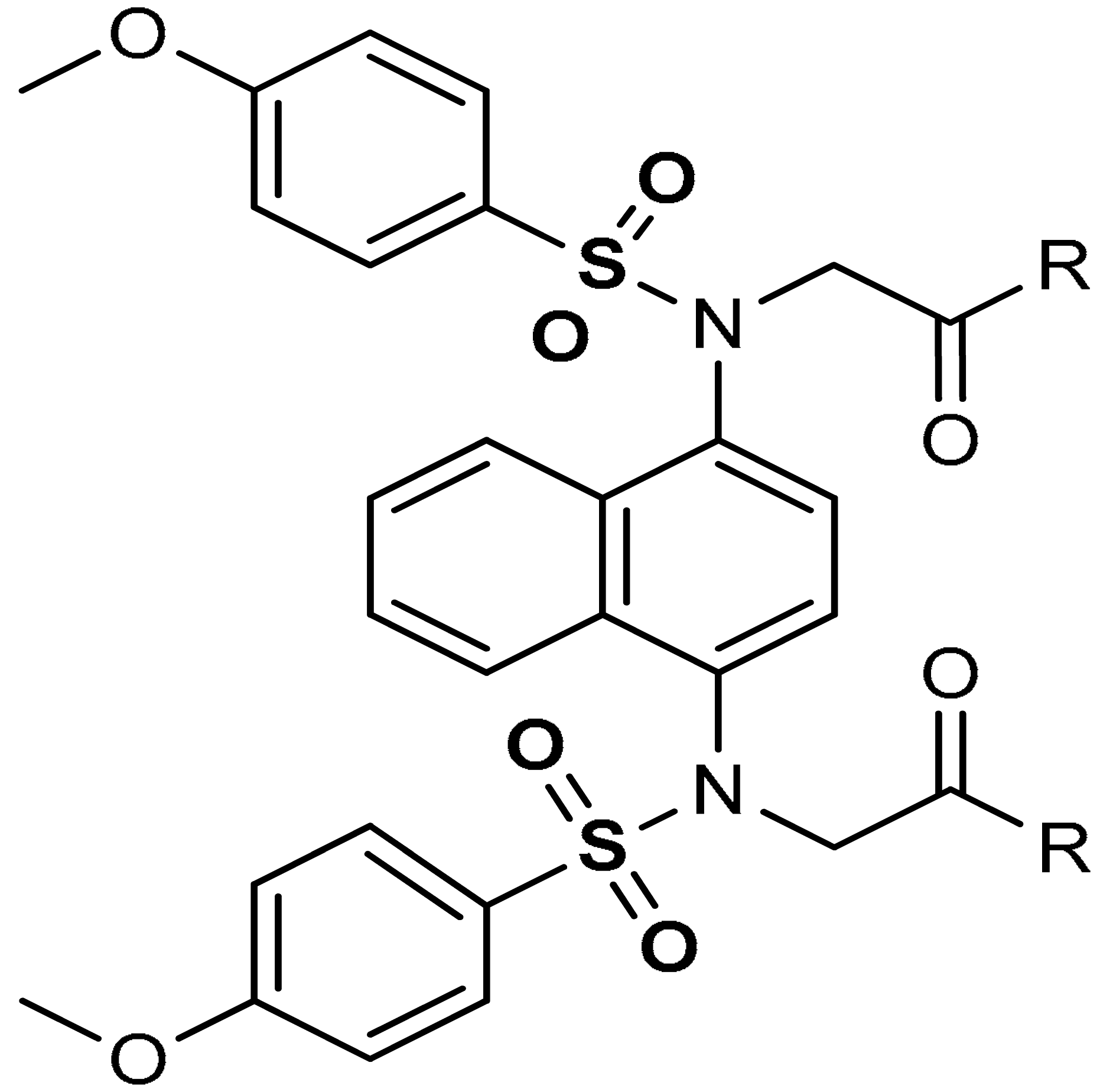 2,2′-(naphthalene-1,4-diylbis(((4-methoxyphenyl)sulfonyl)azanediyl))diacetic acid (R=OH) | ≥500 µL | Antioxidant, Keap1–Nrf2 PPI inhibition | RAW264.7 Cells. HepG2 Cells, mice, rat | Inflammation | [94] |
| 14 |  N-(4-((4-methoxy-N-(2,2,2-trifluoroethyl)phenyl)sulfonamido)isoquinolin-1-yl)-N-((4-methoxyphenyl)sulfonyl)glycine | ≥500 µL | Antioxidant, Keap1–Nrf2 PPI inhibition | RAW264.7 Cells. HepG2 Cells, mice, rat | Inflammation, | [94] |
| 15 | 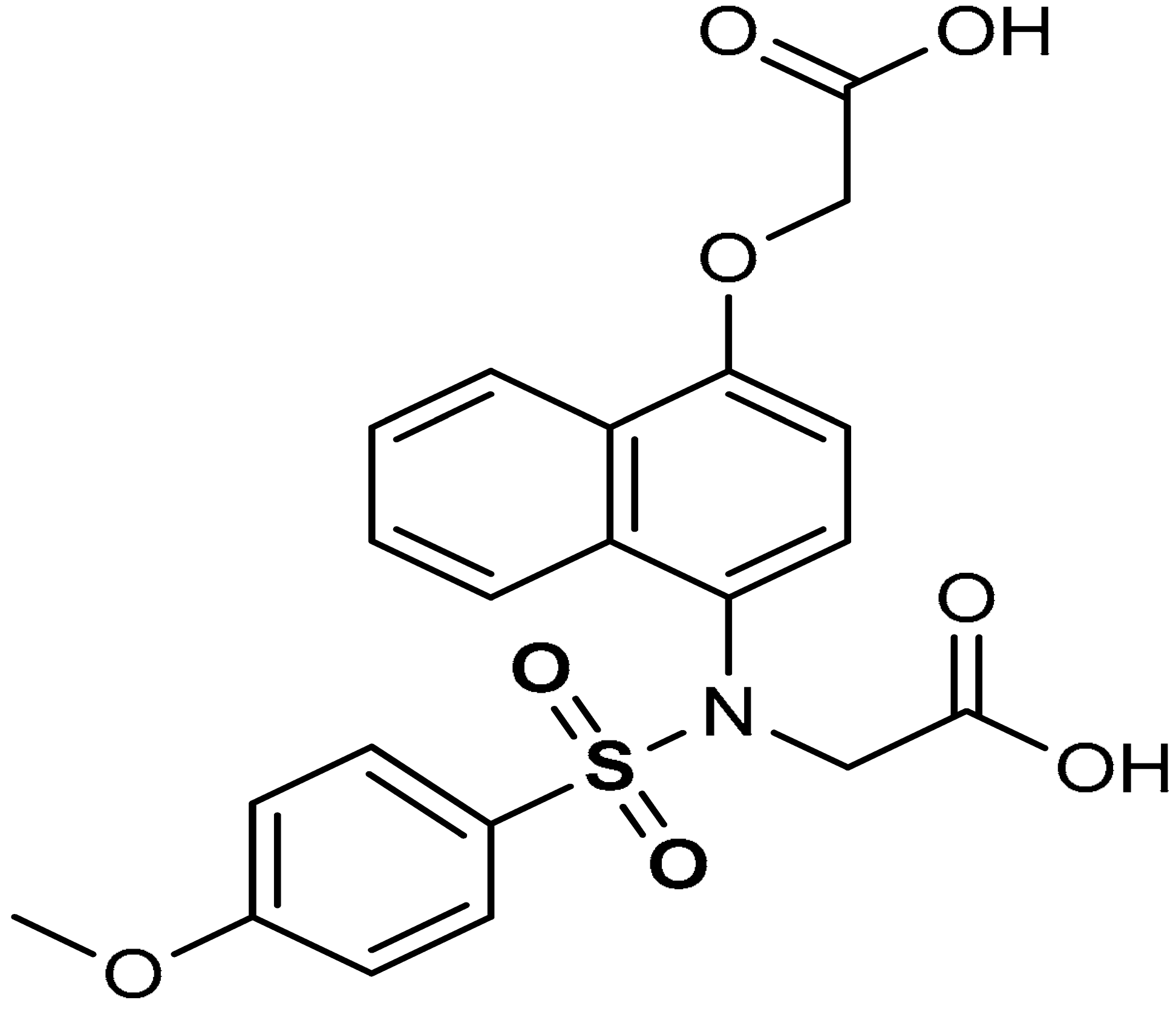 2-(N-(4-(carboxymethoxy)naphthalen-1-yl)-4-methoxyphenylsulfonamido)acetic acid | 8.19 µM | Antioxidant, Keap1–Nrf2 PPI inhibition | RAW264.7 Cells | Inflammation | [94] |
| 16 | 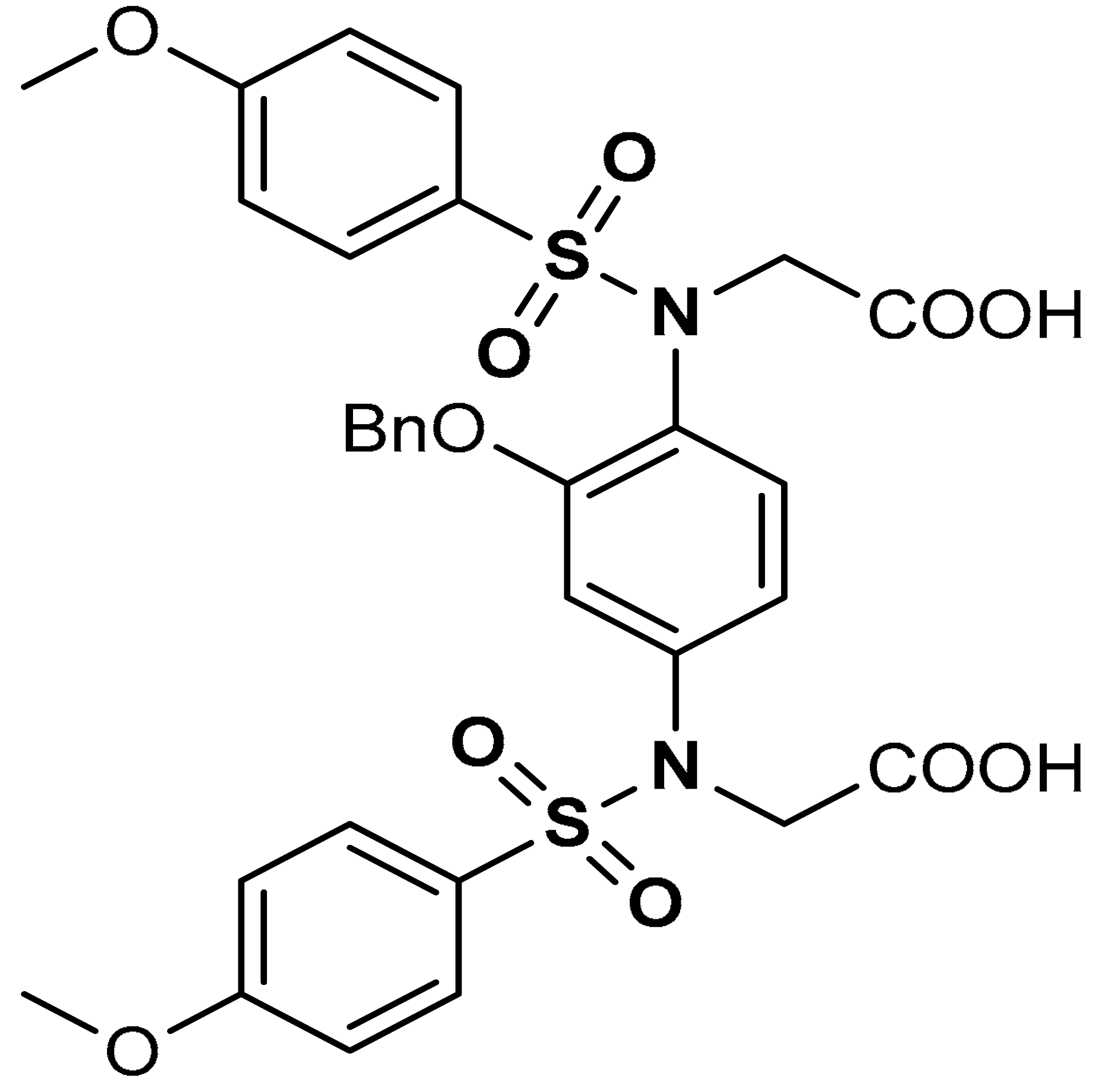 2,2′-((2-(Benzyloxy)-1,4-phenylene)bis(((4- methoxyphenyl)sulfonyl)azanediyl))diacetic acid | 10–100 µM | Keap1–Nrf2 PPI inhibition, Antioxidant, | Hepa1c1c7 mouse hepatic cells | Cytotoxicity | [114] |
| 17 | 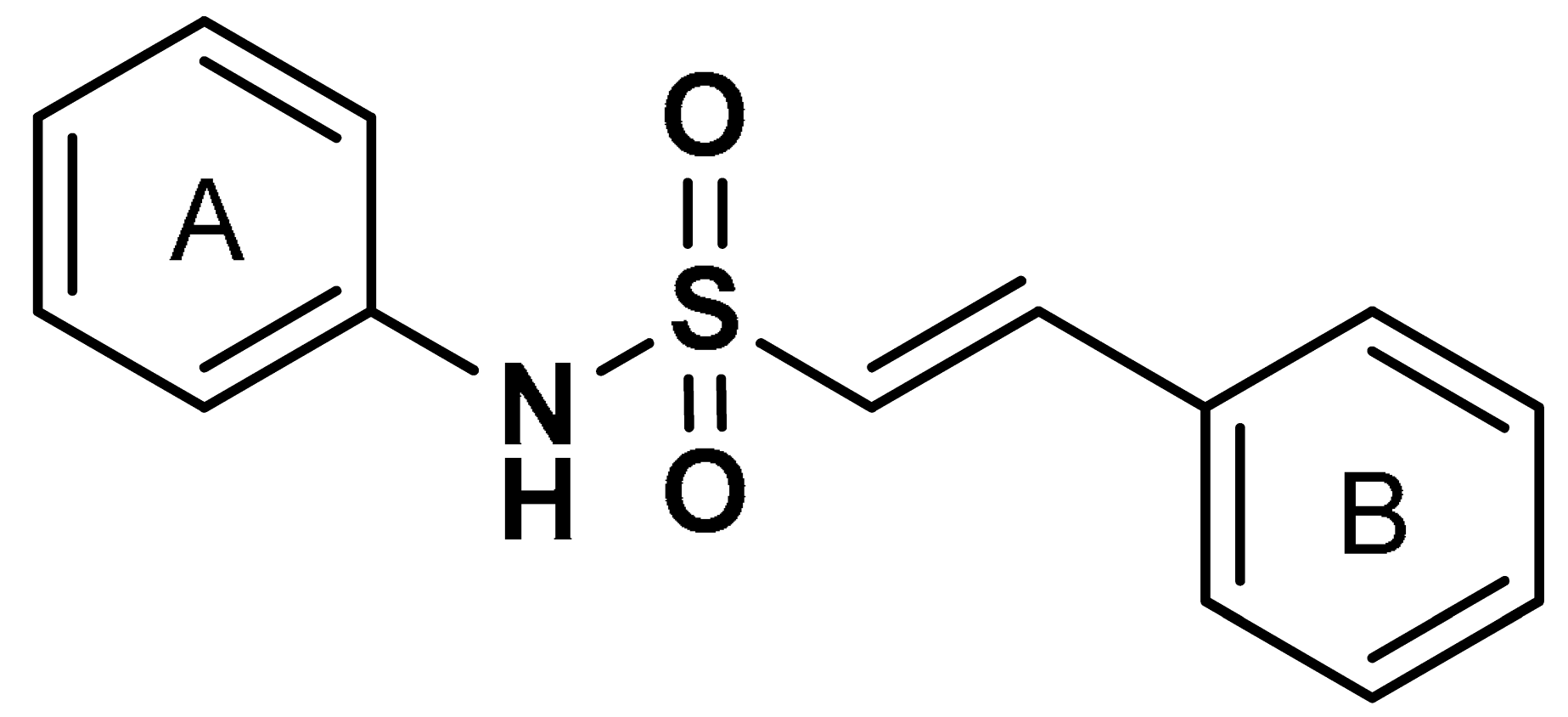 (E)-N,2-diphenylethenesulfonamide | >10 µM | Antioxidant, anti-inflammatory, neuroprotection | MPTP-induced PD mouse model | Parkinson’s disease | [115] |
| 18 | 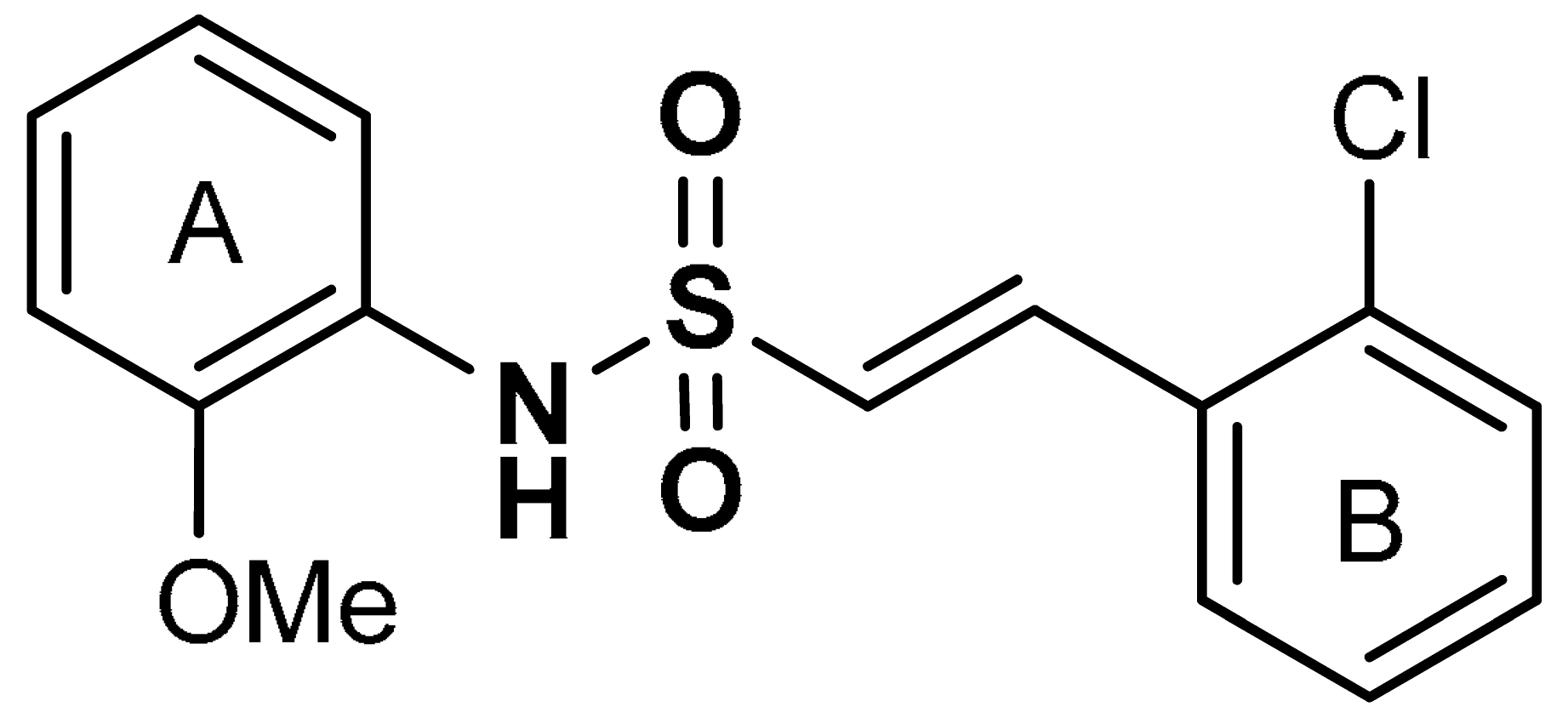 (E)-2-(2-chlorophenyl)-N-(2-methoxyphenyl)ethenesulfonamide | 6.35 µM | Antioxidant, anti-inflammatory, neuroprotection | MPTP-induced PD mouse model | Parkinson’s disease | [115] |
| 19 | SULFONATE DERIVATIVES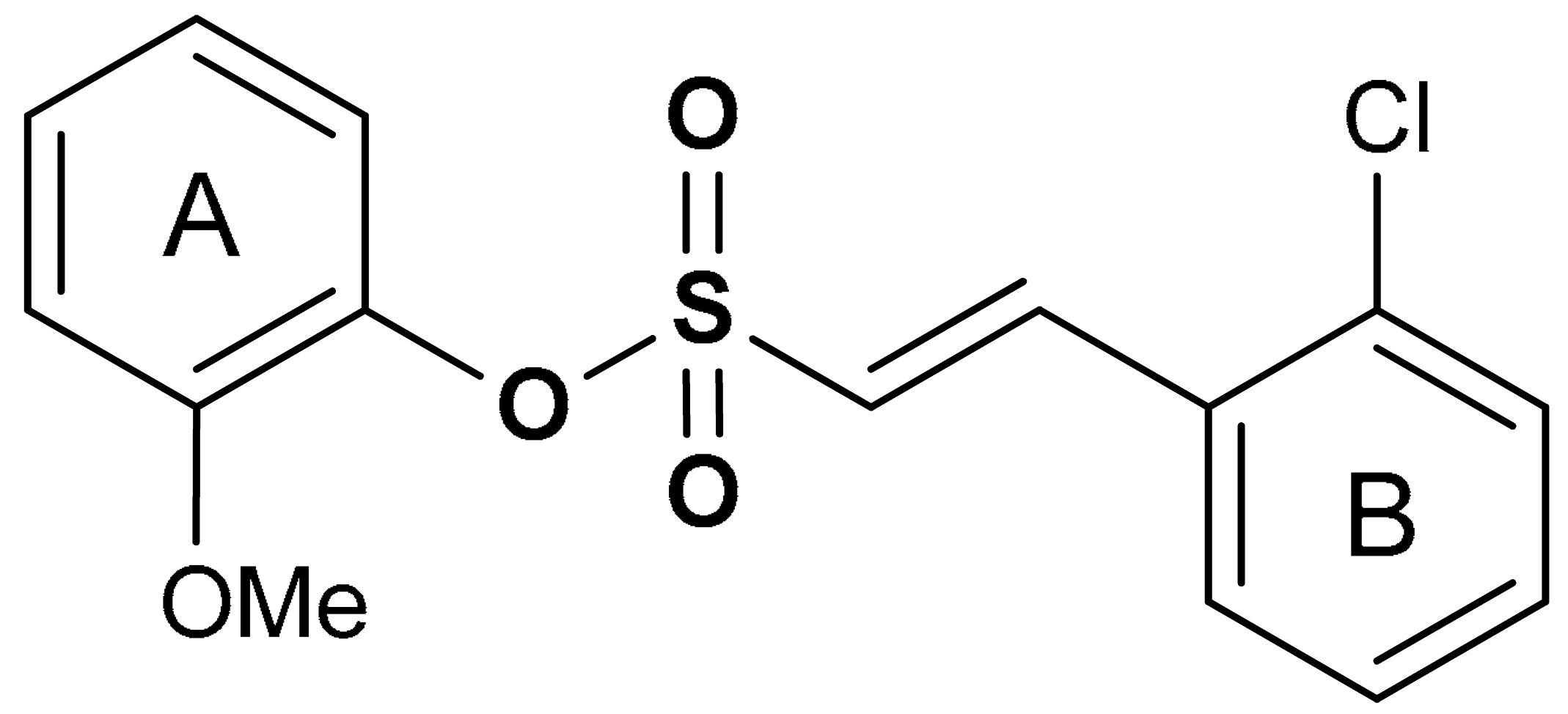 (E)-2-methoxyphenyl 2-(2-chlorophenyl)ethenesulfonate | 0.076 µM | Antioxidant, anti-inflammatory, neuroprotection | MPTP-induced PD mouse model | Parkinson’s disease | [115] |
| 20 |  (E)-3-methoxyphenyl 2-(2-chlorophenyl)ethenesulfonate | 0.165 µM | Antioxidant, anti-inflammatory, neuroprotection | MPTP-induced PD mouse model | Parkinson’s disease | [115] |
| 21 |  (E)-4-methoxyphenyl 2-(2-chlorophenyl)ethenesulfonate | 0.237 µM | Antioxidant, anti-inflammatory, neuroprotection | MPTP-induced PD mouse model | Parkinson’s disease | [115] |
| 22 |  sodium 1,6,6-trimethyl-10,11-dioxo-6,7,8,9,10,11-hexahydrophenanthro [1,2-b]furan-2-sulfonate | 10 µg/mL | Antioxidant | RAW 264.7 Cells, MRC-5 Cells | Silicosis | [121] |
| 23 | SULFOXIDE DERIVATIVES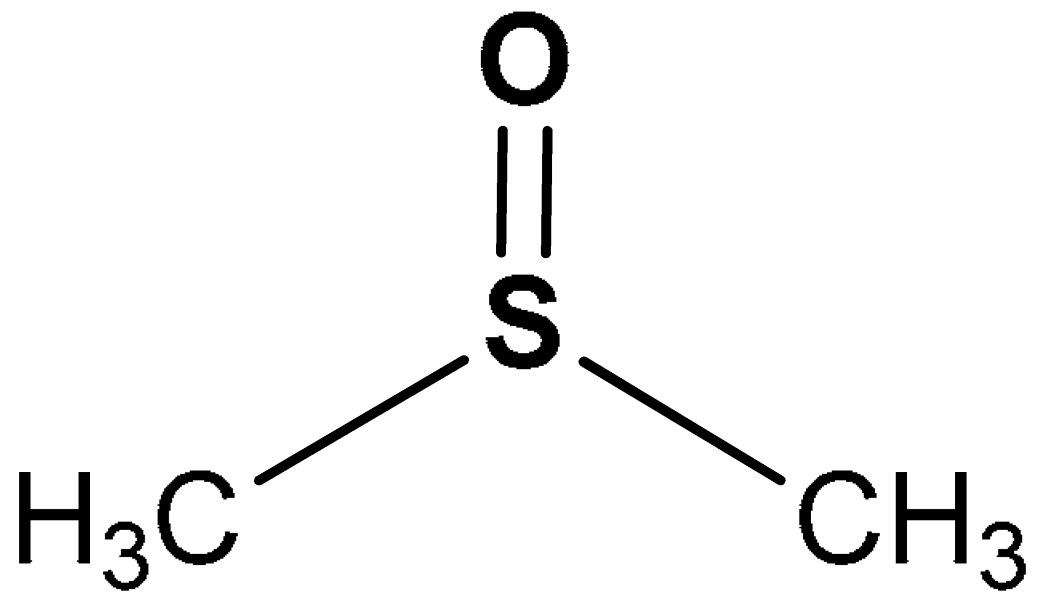 (methylsulfinyl)methane | 0.1–0.8% | Antioxidant, anti-inflammatory | Human umbilical vein endothelial cells (HUVECs) | Oxidative stress | [125] |
| 24 | 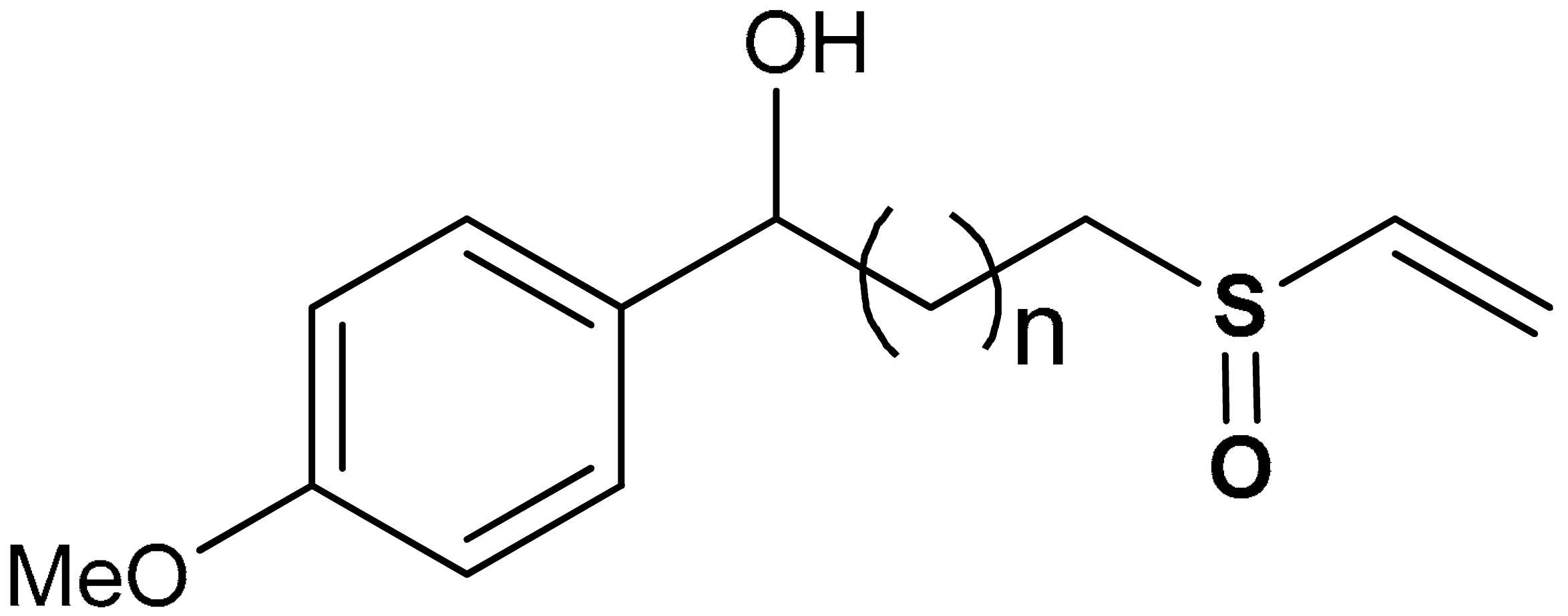 1-(4-methoxyphenyl)-3-(vinylsulfinyl)propan-1-ol | 20 µM | Antioxidant, Nrf2 activation, HO-1 induction | BV-2 Cells | Parkinson’s disease | [135] |
| 25 |  1-(4-fluorophenyl)-3-(vinylsulfinyl)propan-1-ol | 20 µM | Antioxidant, Nrf2 activation, HO-1 induction | BV-2 Cells | Parkinson’s disease | [135] |
| 26 | 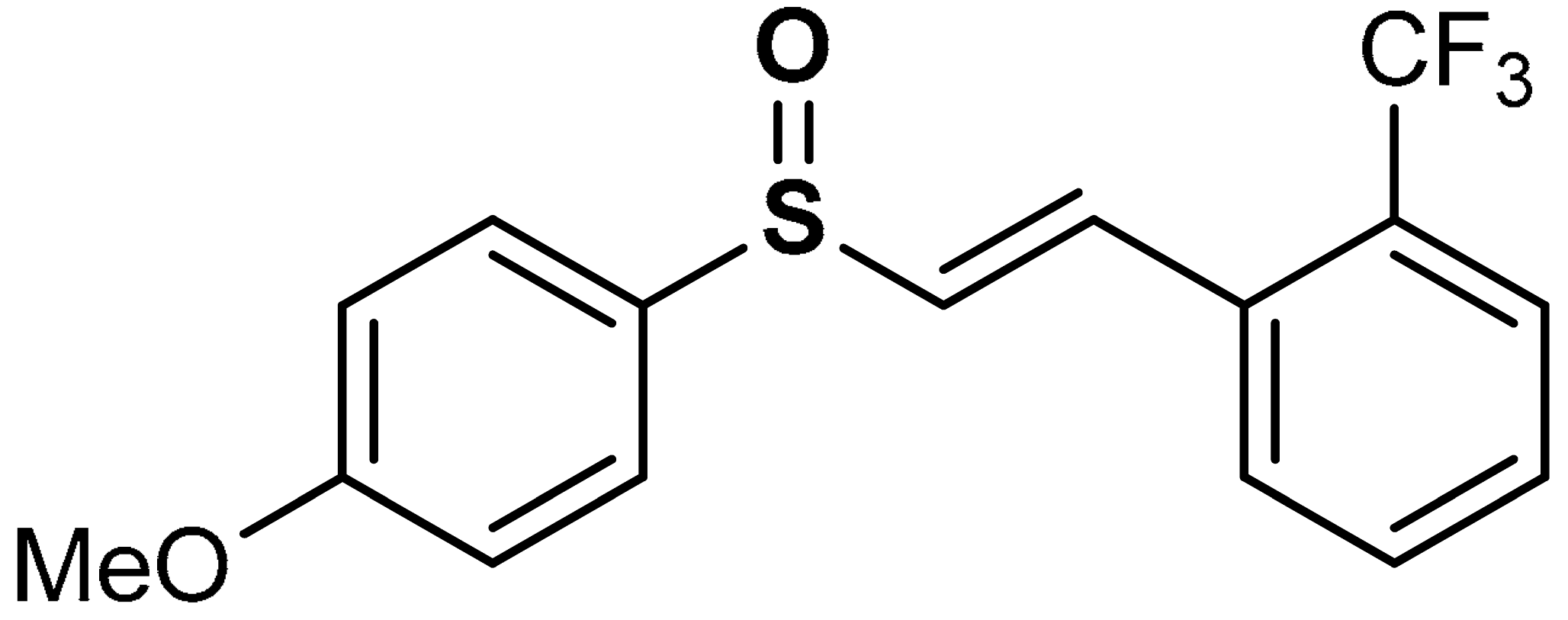 (E)-1-(2-((4-methoxyphenyl)sulfinyl)vinyl)-2-(trifluoromethyl)benzene | 20 µM | Neuroprotection, Antioxidant | BV-2 Cells | Parkinson’s disease | [99] |
| 27 | SULFOXIMINE DERIVATIVES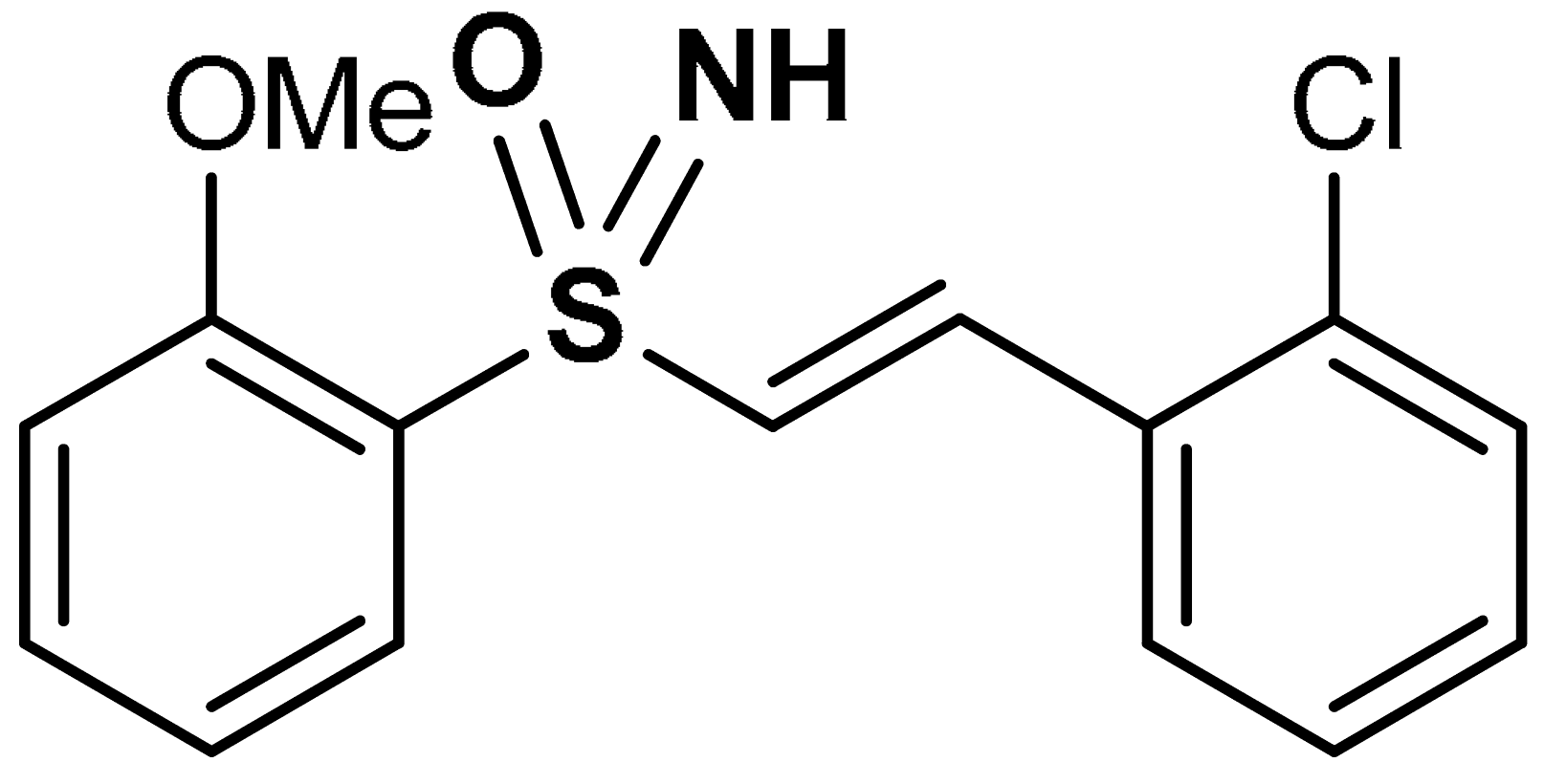 (E)-1-chloro-2-(2-(2-methoxyphenylsulfonimidoyl)vinyl)benzene | 10 µM | Nrf2 activation, Antioxidant | HEK293 Cells | Traumatic brain injury (TBI) | [100] |
| 28 | 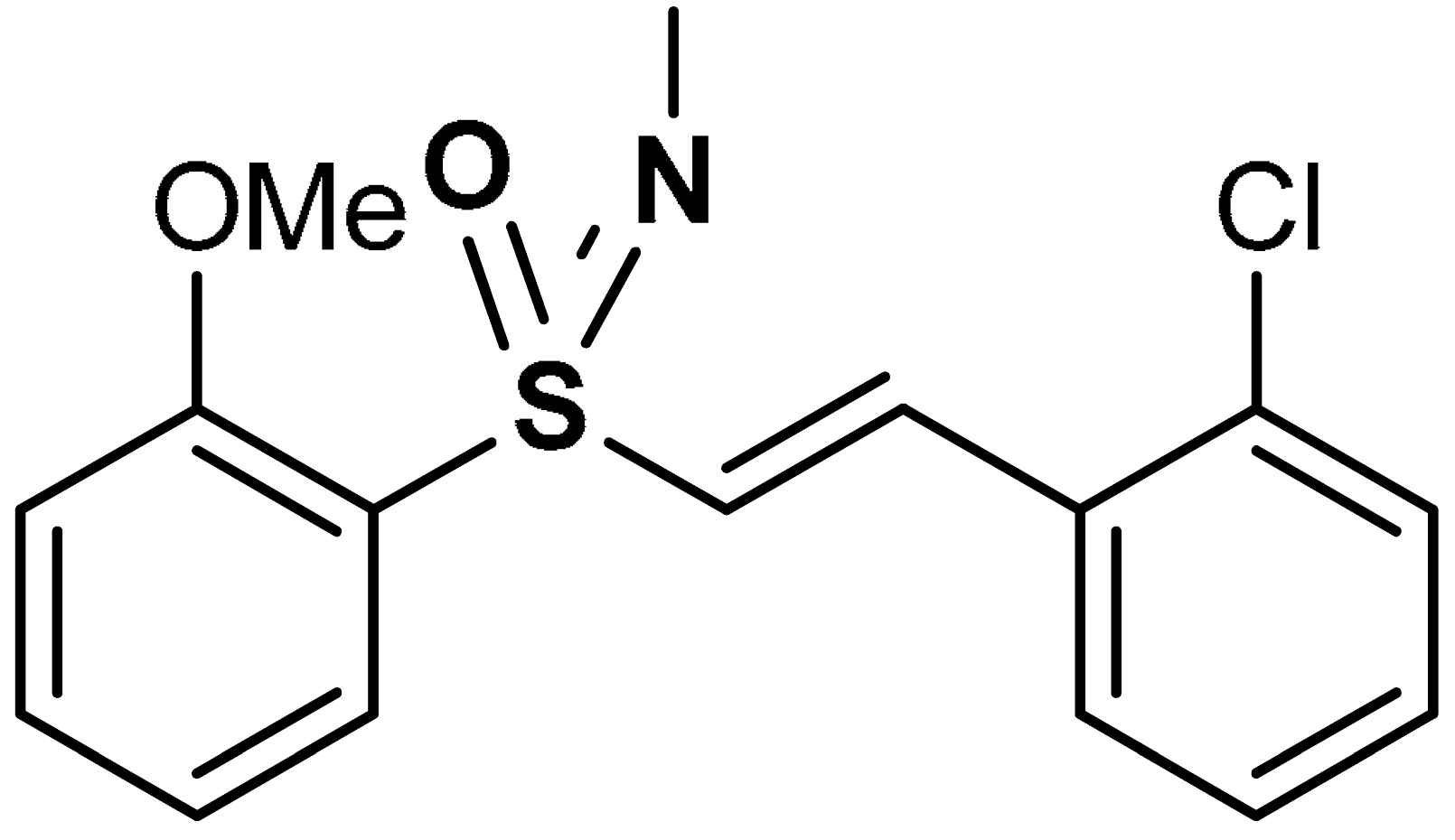 1-chloro-2-{(E)-2-[S-(2-methoxyphenyl)-N-methylsulfonimidoyl]ethenyl}benzene | 10 µM | Nrf2 activation, Antioxidant | HEK293 Cells | Traumatic brain injury (TBI) | [100] |
| 29 | OLTIPRAZ | 150 mg/kg | Antioxidant, Nrf2 activation | HepG2 cells and C57BL/6 mouse liver | oxidative stress | [143] |
| 0.75 g/kg | Antioxidant | C57BL/6J mice | Obesity, insulin resistance, Oxidative stress | [144] | ||
| 20 µM | Antioxidant | RSC96 cells | Oxidative stress, Apoptosis | [145] | ||
| 12 µM | Antioxidant. Nrf2 activation | HEK293 cells | Kidney cell injury | [146] | ||
| 500 mg/kg | Antioxidant, Chemoprevention | Mice | Cancer | [147] | ||
| 250 mg/kg | Anticancer, Antioxidant | Mouse urothelial cells | Urinary bladder carcinogenesis | [148] |
3. Nrf2-Activating Natural Organosulfur Compounds
3.1. Isothiocyanates
3.1.1. Benzyl Isothiocyanate
3.1.2. Allyl Isothiocyanate
3.1.3. Phenylethyl Isothiocyanate
3.1.4. Sulforaphane
3.2. Sulfur-Containing Amino Acids and Derivatives
3.2.1. Methionine
3.2.2. Taurine
3.2.3. Homocysteine
3.2.4. N-Acetylcysteine
3.2.5. Carbocysteine
3.2.6. Ergothioneine
3.3. Allicin
| Entry | Compounds | Effective Concentration/Dose | Biological Activity | Study Model | Targeted Diseases | Ref |
|---|---|---|---|---|---|---|
| 30 | ISOTHIOCYANATES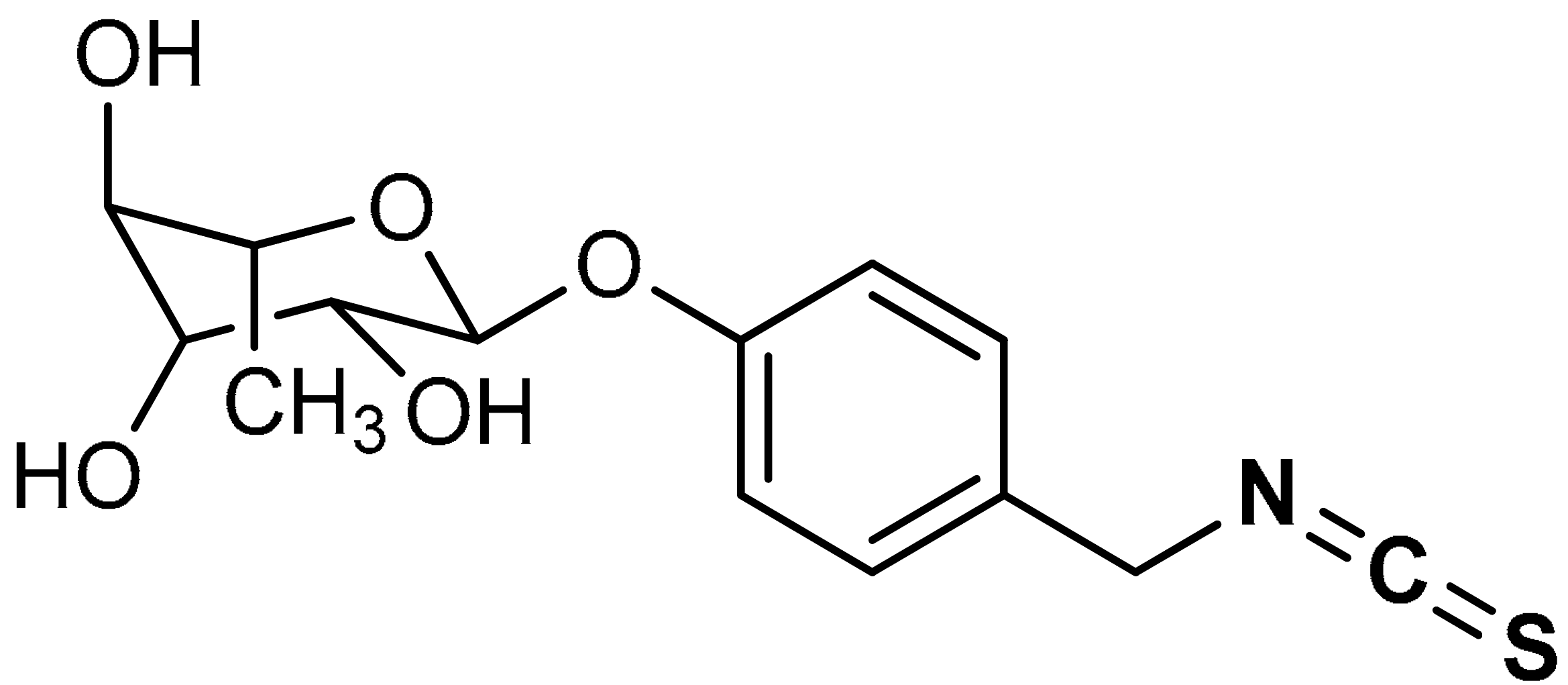 (2S,3R,4R,5R,6S)-2-(4-(isothiocyanatomethyl)phenoxy)-6-methyltetrahydro-2H-pyran-3,4,5-triol | 0.4–100 µM | Antioxidant, anti-inflammatory | HepG2-C8 cells | Diabetic nephropathy, oxidative stress | [151] |
| 31 | 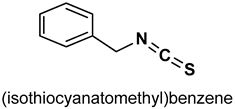 | 5 µM | Antioxidant | Mice | Hyperglycemia, oxidative stress | [152] |
| 0.7 and 1.5 mg/kg | Antioxidant, anti-inflammatory | Rat | Gastric injury | [153] | ||
| 32 | 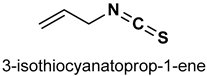 | 10 mg/kg | Antioxidant, anti-inflammatory | C57BL/6J mice | Traumatic brain injury | [156] |
| 40 µM | Antioxidant | 16HBE14o-cells | COPD | [157] | ||
| 25 or 50 mg/kg | Hepatotoxicity, antioxidant | HepG2 and AML12 cells | Acetaminophen-induced liver injury | [158] | ||
| 33 | 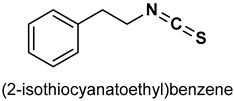 | 1–100 µM | Antioxiodant, cytoprotection | NIH3T3 cells | cytotoxicity | [160] |
| 5 and 10 µM | Antioxidant, anti-inflammatory | C57BL/6J mouse strain | Inflammation | [161] | ||
| 20 and 50 µM | Chemoprevention, antioxidant | HeLa cells | cancer | [162] | ||
| 34 | 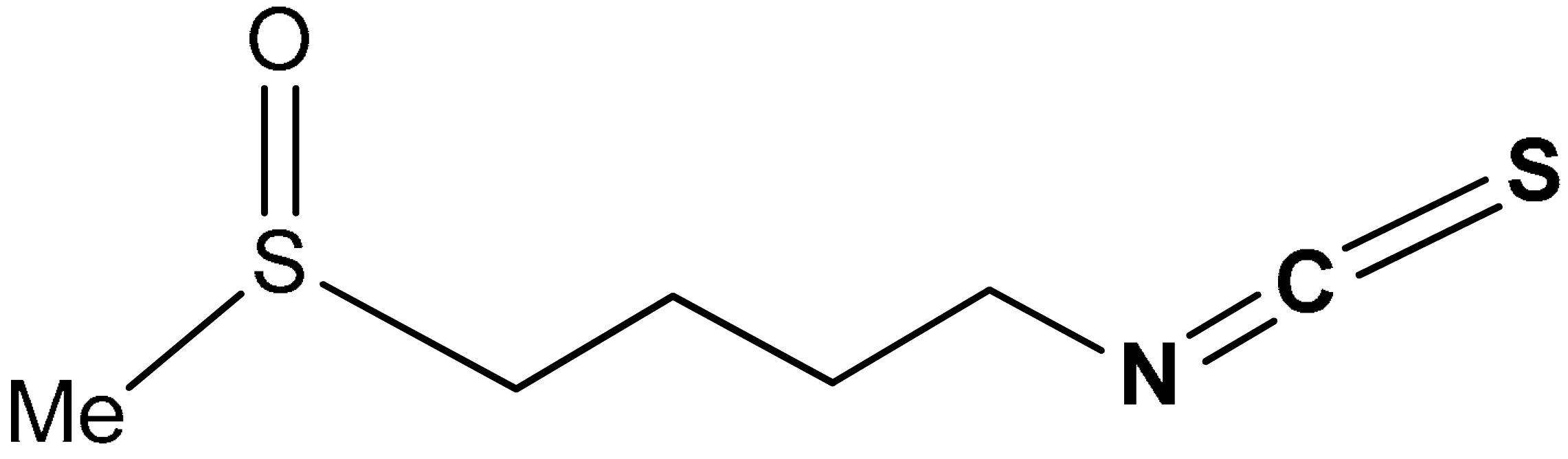 1-isothiocyanato-4-(methylsulfinyl)butane | 3–8 µM | Antioxidant | Human or rat epithelial cells | Oxidative stress | [168] |
| 110–440 µmol/kg | Anticancer, chemoprevention | Wild-type mice | Cancer | [169] | ||
| 9 µmol/day | Anticancer, chemoprevention | Mice | Cancer | [170] | ||
| 1–10 µM | Antioxidant, neuroprotection | PC12 Cells | Oxidative stress, apoptosis | [172] | ||
| 35 | 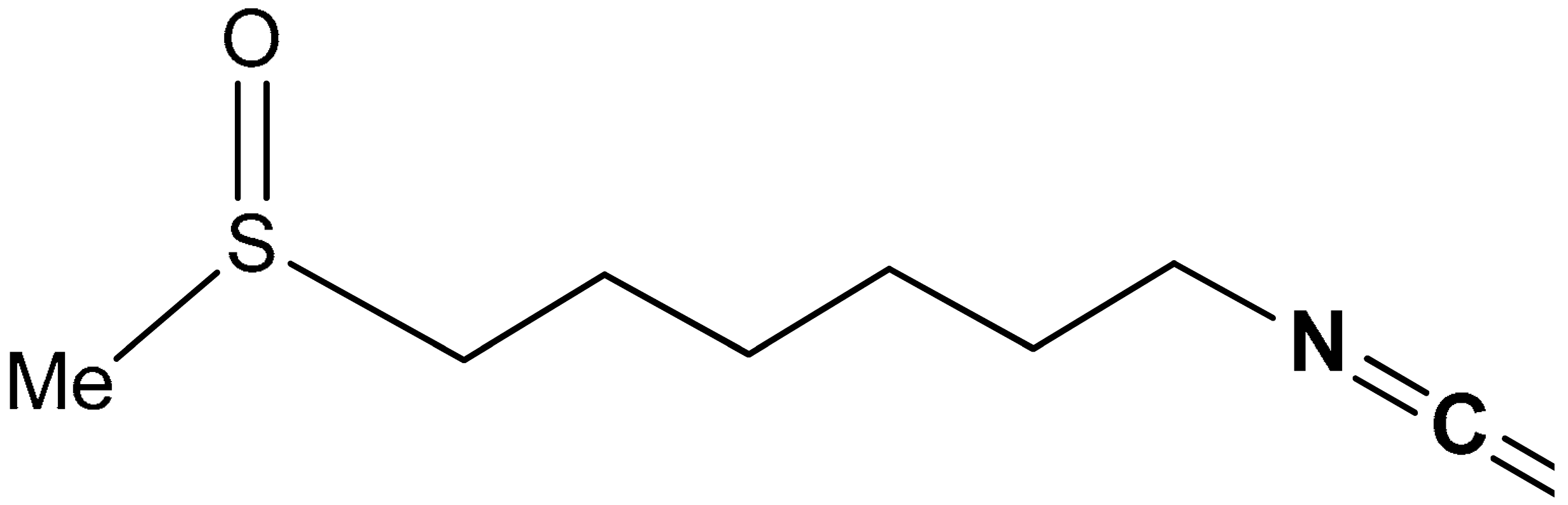 6-Methylsulfinylhexyl isothiocyanate | 5 µM | Anticancer, chemoprevention | RL34 Cells | Cancer | [173] |
| 36 | SULFUR-CONTAINING AMINO ACIDS AND DERIVATIVES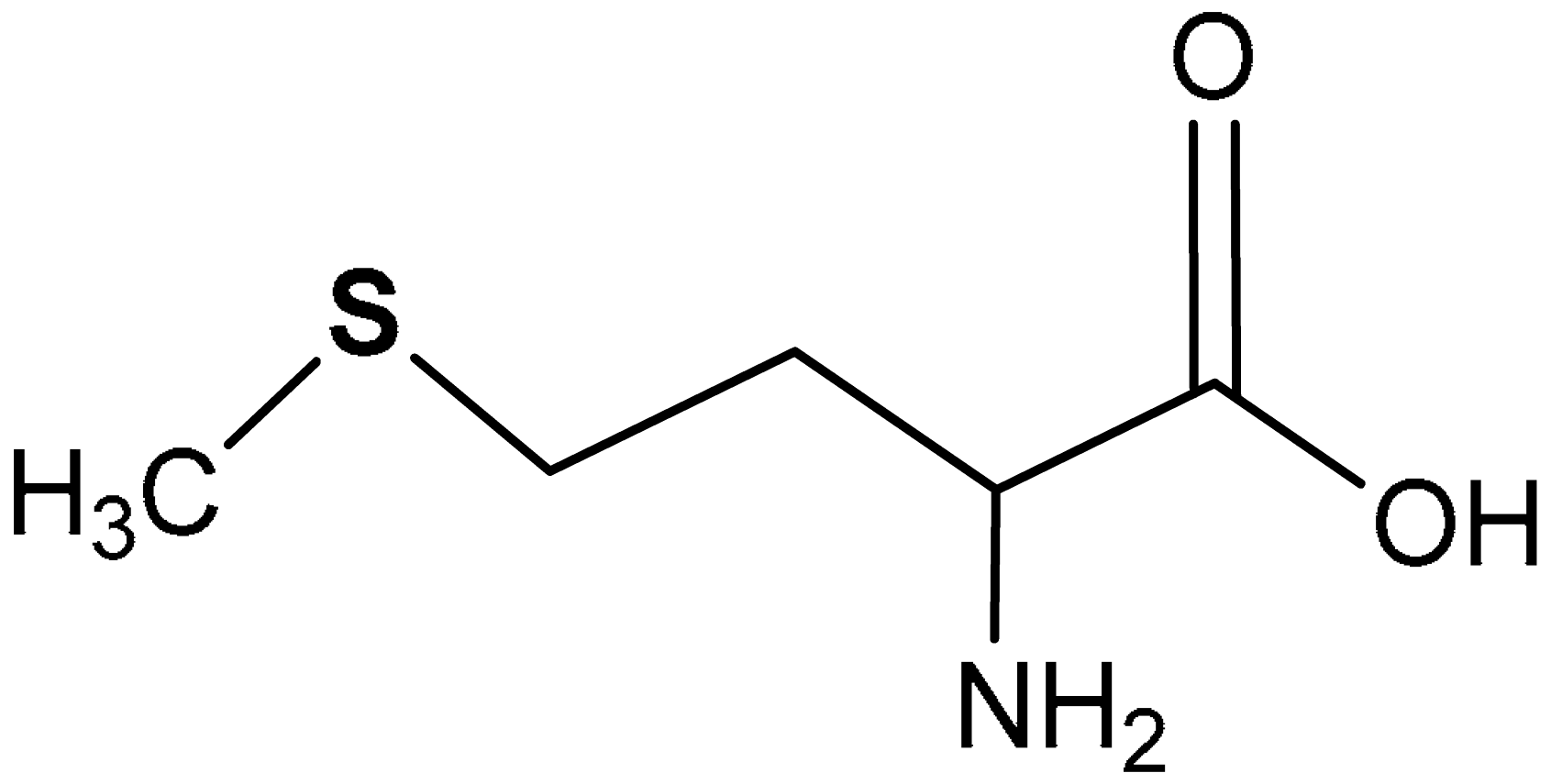 2-amino-4-(methylthio)butanoic acid | 21.50–43.00 mg, 100 g−1 body wt | Antioxidant | Growing rats | Oxidative stress | [183] |
| 21.2 mg/g | Antioxidant | Growing and adult rats | Oxidative stress | [184] | ||
| 0.4–0.91% | Hepatic antioxidant | Lambs | Oxidative stress | [185] | ||
| 37 | 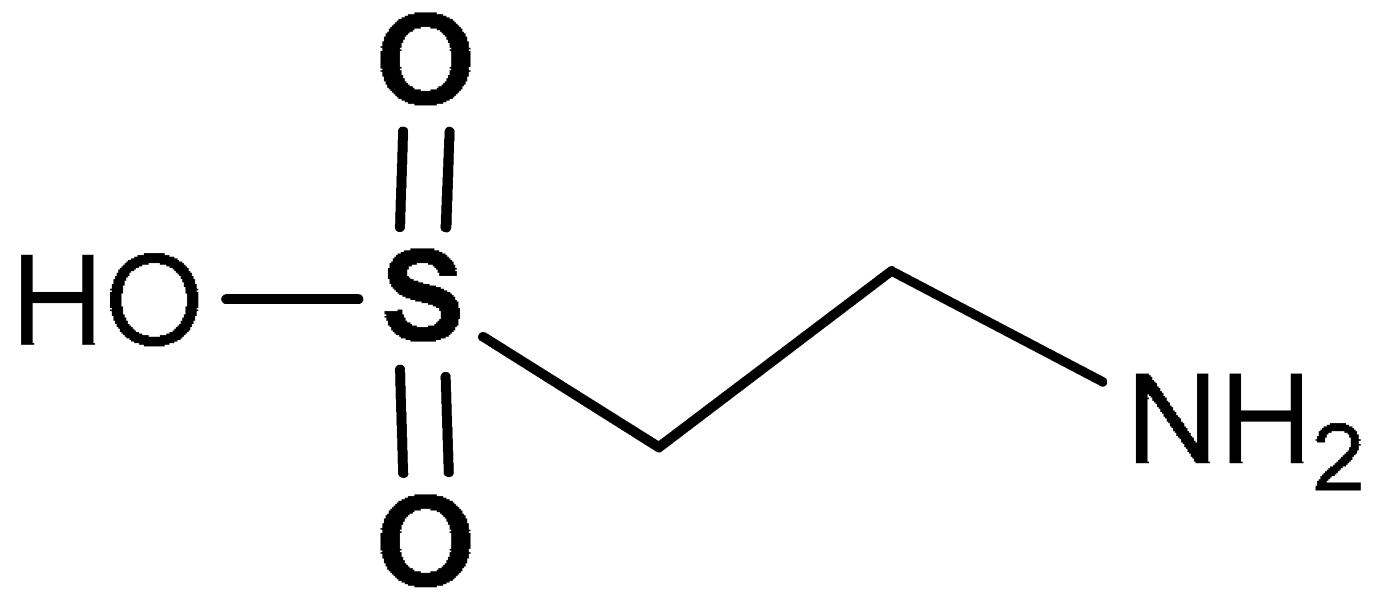 2-aminoethanesulfonic acid | 10–80 mM | Antioxidant | Mouse spermatocytes (GC-2 Cells) | Oxidative stress | [193] |
| 2% w/v | Antioxidant | Diabetic rats | Diabetic neuropathy | [194] | ||
| 38 | 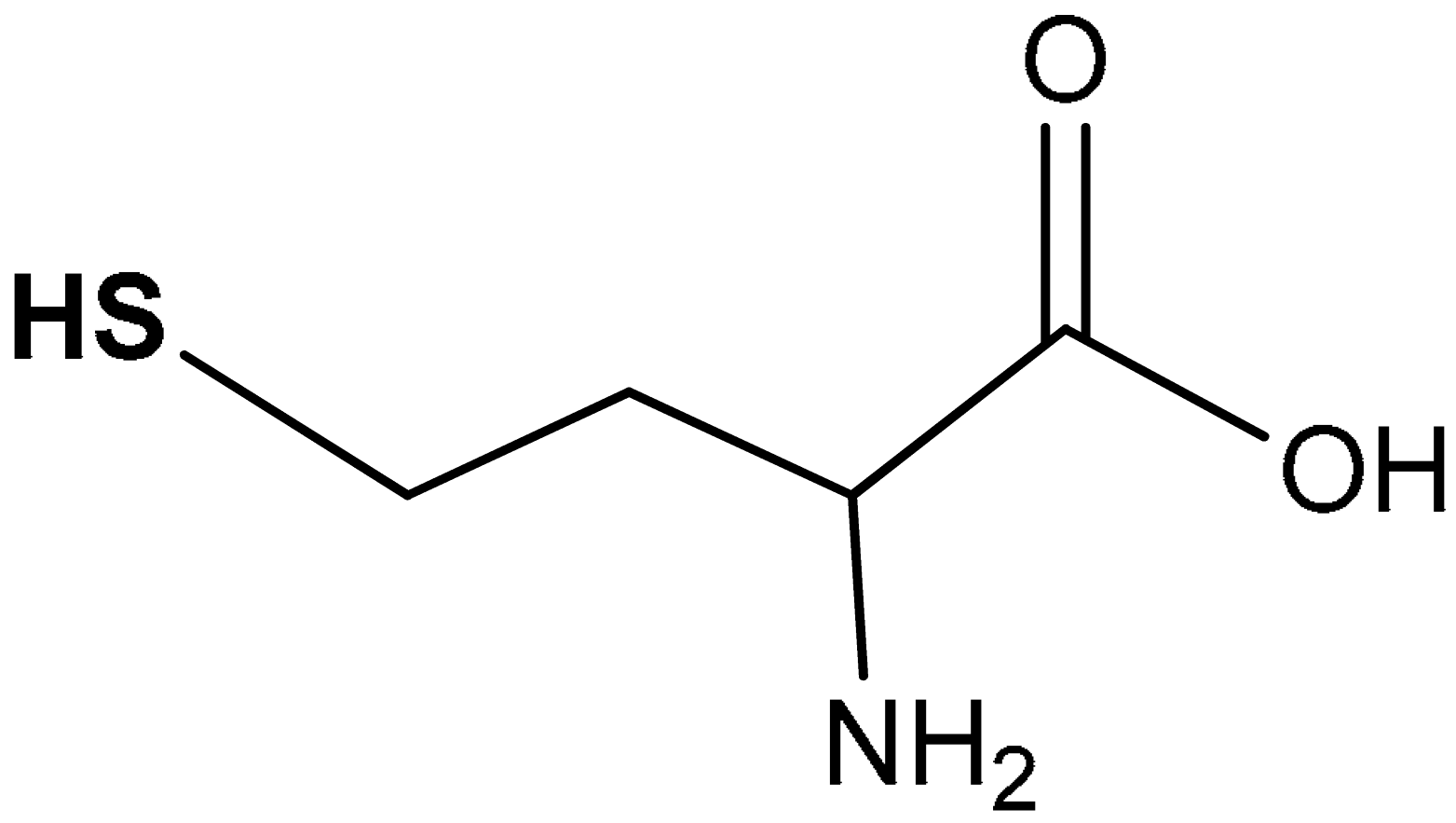 2-amino-4-mercaptobutanoic acid | 50 µMD/L | Antioxidant | Hepatoma cell line (HepG2 Cells) | Oxidative stress | [206] |
| 50 µM | Antioxidant | Hepatoma cell line (HepG2 Cells) | Oxidative stress | [207] | ||
| 0–100 µM | Antioxidant | Hepatoma cell line (HepG2 Cells) | Oxidative stress | [208] | ||
| 50 µM–1 mM | Antioxidant | Muller glial cells | Oxidative stress | [209] | ||
| 39 | 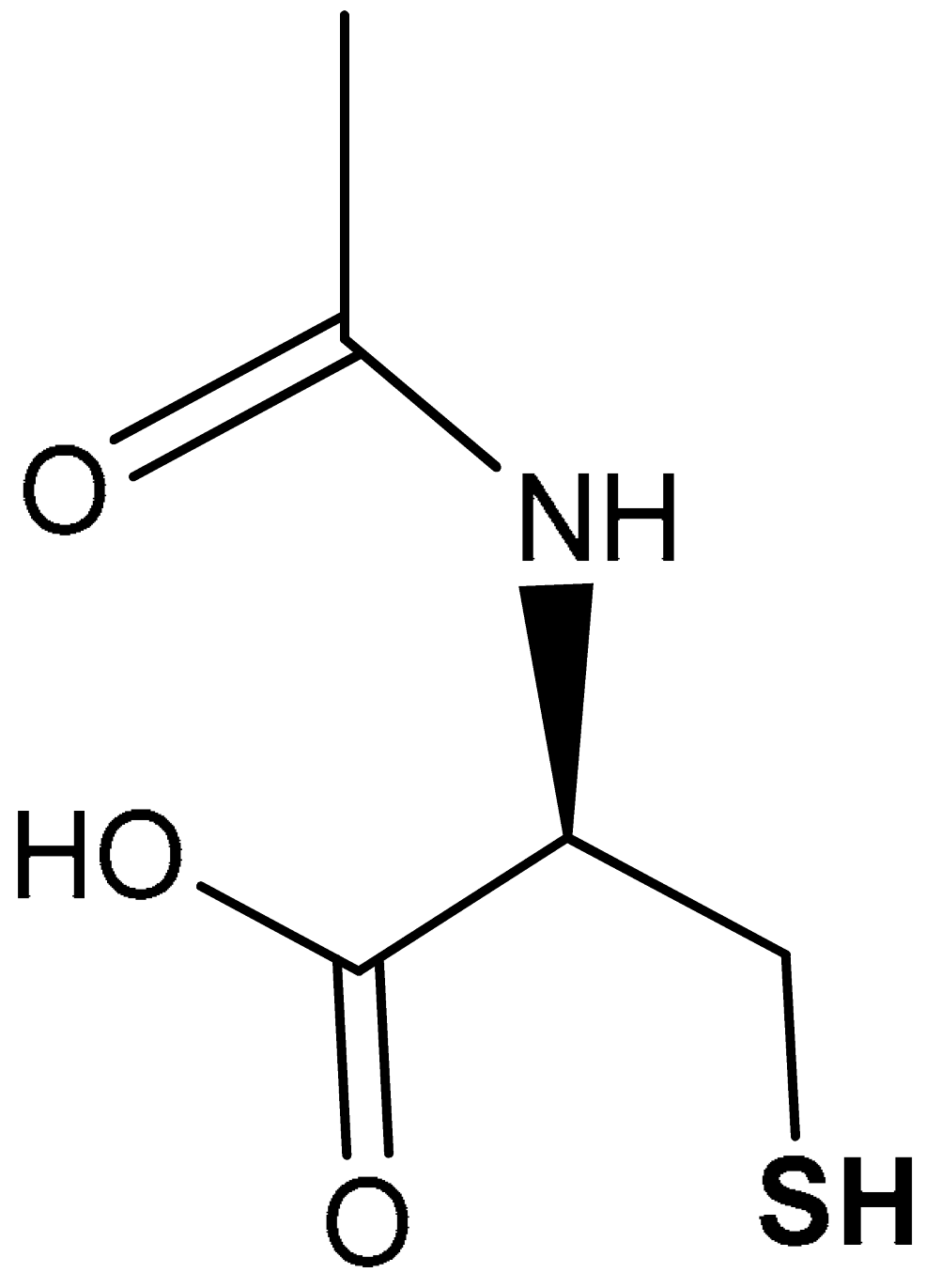 (R)-2-acetamido-3-mercaptopropanoic acid | 600 mg | Antioxidant | Infertile men with asthenoteratozoospermia | Oxidative stress | [213] |
| 40 | 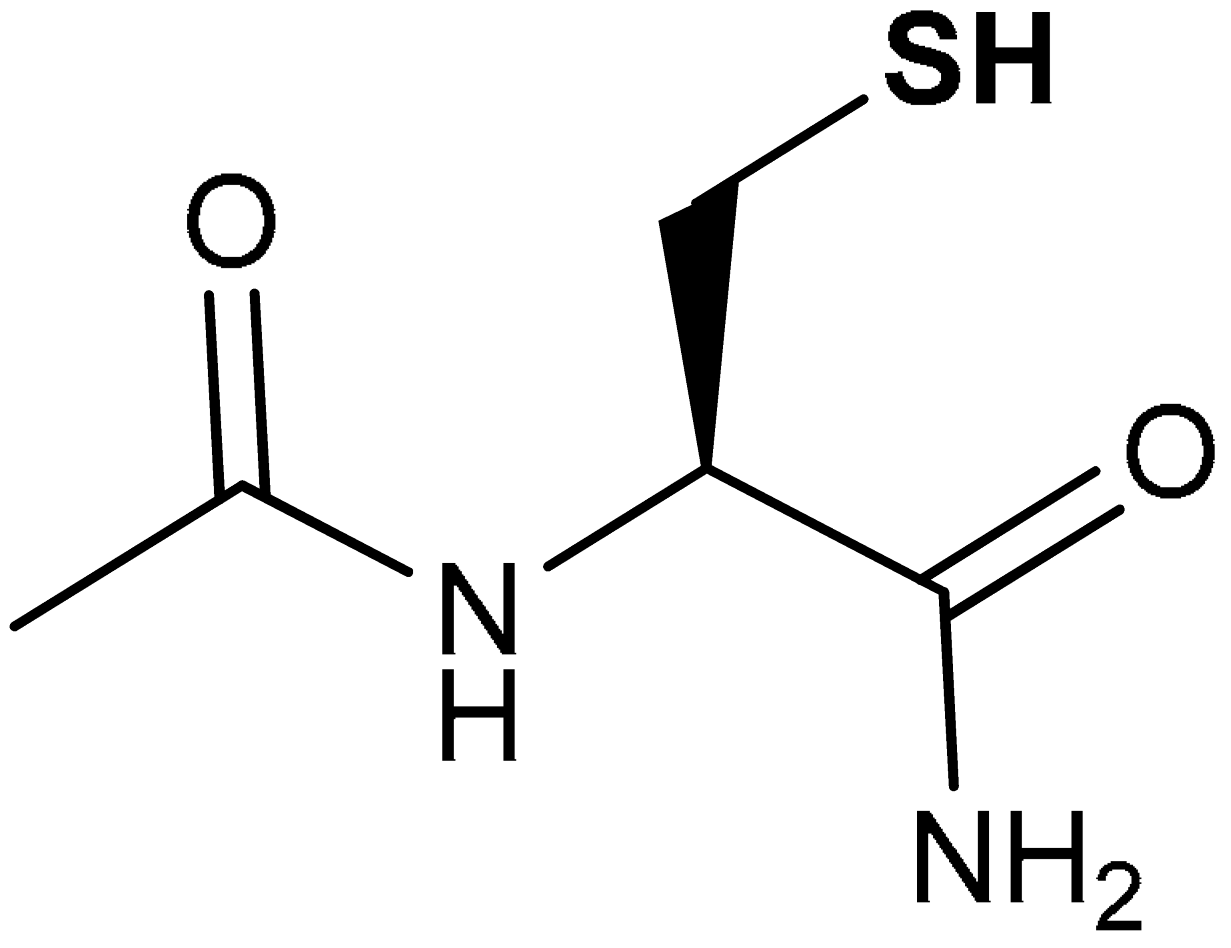 (R)-2-acetamido-3-mercaptopropanamide | 100 mg/kg | Antioxidant, neuroprotection | Mouse model of TBI | Oxidative stress, TBI | [214] |
| 41 | 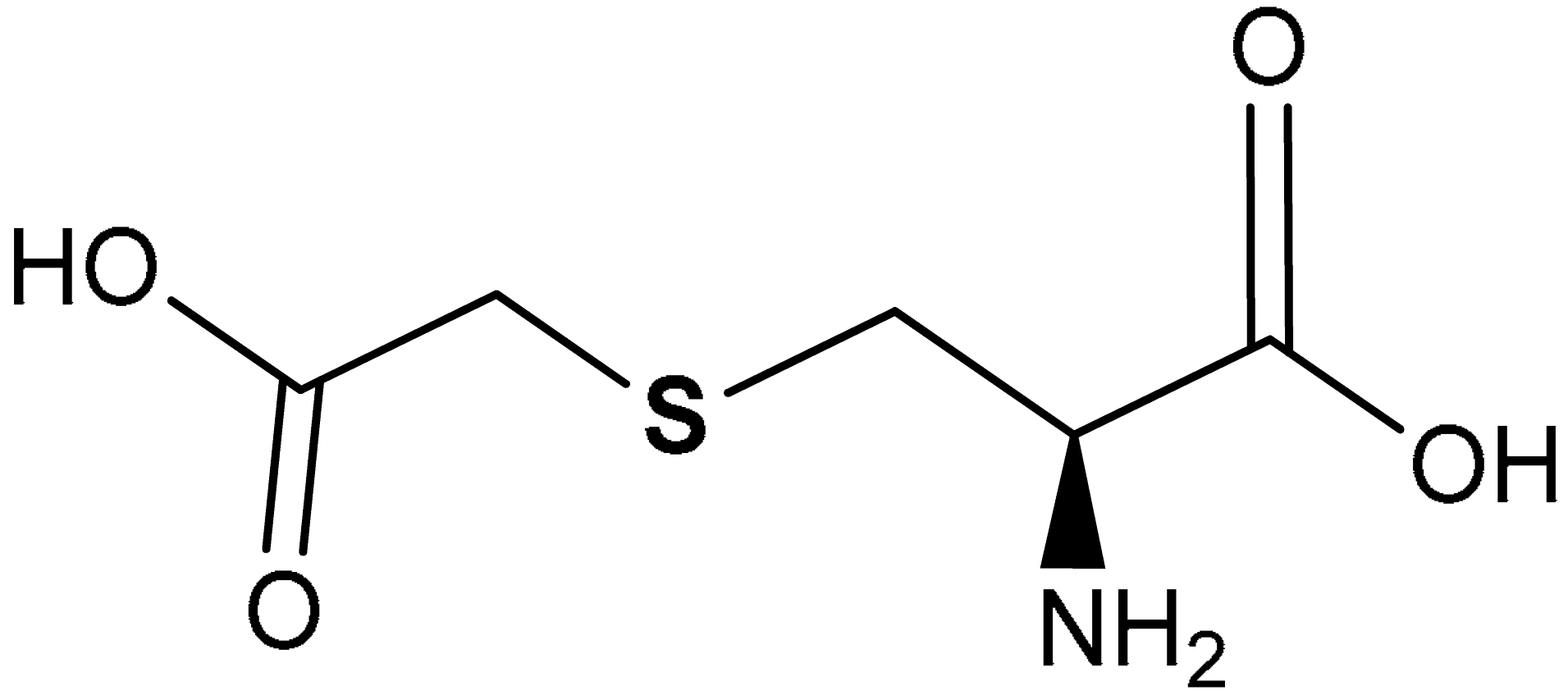 (R)-2-amino-3-((carboxymethyl)thio)propanoic acid | 10−4 M | Antioxidant, Cytoprotection | Bronchial epithelial cells (16-HBE) | Chronic obstructive pulmonary disease (COPD) | [218] |
| 42 | 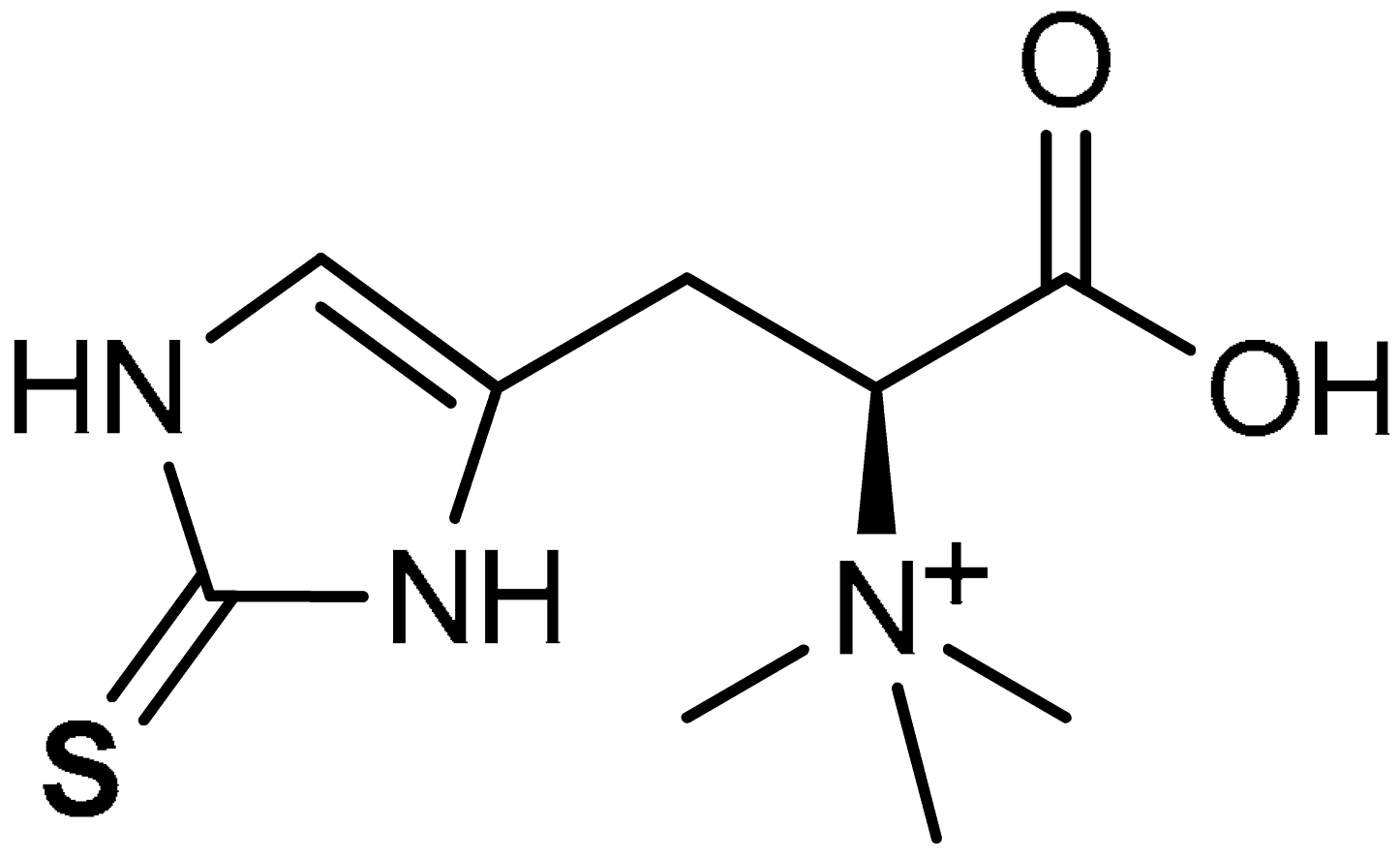 (2S)-3-(2-Sulfanylidene-2,3-dihydro-1H-imidazol-4-yl)-2-(trimethylazaniumyl)propanoate | 70 mg/kg | Antioxidant, Anti-inflammatory | Rat | Nephrotoxicity | [221] |
| 125–500b nM | Antioxidant, Dermato-protection | Human keratinocytes | Skin damage, Oxidative stress | [222] | ||
| 0.1–10 mM | Antioxidant, Anti-inflammatory | Human keratinocytes | Skin damage, Oxidative stress, Inflammation | [223] | ||
| 43 | ALLICIN | 10 µg/mL | Apoptosis | Colon cancer cells (HCT-116) | Colon cancer | [228] |
| 40 µg/mL | Antioxidant, Anti-inflammatory | HUVECs | Oxidative stress, Inflammation | [229] |
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| αGSTA1 | Glutathione S-transferase A1 |
| ADME/tox | Absorption, Distribution, Metabolism, and Elimination/Toxicity |
| ARE | Antioxidant response element |
| BAX | BCL2 Associated X, Apoptosis Regulator |
| BBB | Blood-brain barrier |
| Bcl-2 | B-cell lymphoma 2 |
| BTB domain | Broad complex, tramtrack and bric-à-brac domain |
| β-TrCP | β-transducin repeat-containing protein |
| CAT | Catalase |
| CBP | CREB-binding protein |
| CBS | Cystathionine β-synthase |
| CDDO-Im | 1-[2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oyl] imidazole |
| CH3 | Cysteine/histidine-rich domain 3 |
| JNKs | c-Jun N-terminal kinases |
| COX- | 2-Cyclooxygenase-2 |
| CREB | cAMP Responsive Element Binding protein |
| CSE | Cystathionine γ-lyase |
| Cul3 | Cullin3 ligase |
| CYP | Cytochrome P450 |
| Cyt c | Cytochrome c |
| 15d- | PGJ2-15-deoxy-Δ12,14-prostaglandin J2 |
| DA | Dopaminergic |
| DMF | Dimethyl fumarate |
| DMSO | Dimethyl sulfoxide |
| EC50 | Half maximal effective concentration |
| GCL | Glutamate-cysteine ligase |
| GCLM | Glutamate-cysteine ligase regulatory subunit |
| GCS | Gamma-glutamylcysteine synthetase |
| GSH | Glutathione |
| GSSH | Glutathione persulfide |
| GST | Glutathione S-transferase |
| GSTA4 | Glutathione S-transferase Alpha 4 |
| πGSTP1 | Glutathione S-transferase π |
| HDAC-2 | Histone Deacetylase 2 |
| HEK293 | Human embryonic kidney 293 cells |
| 6-HITC | 6-methylsulfinylhexyl isothiocyanate |
| HIF1 | Hypoxia-inducible factor 1 |
| H2O2 | Hydrogen peroxide |
| H2S | Hydrogen sulfide |
| HO-1 | Heme oxygenase-1 |
| HUVEC | Human umbilical vein endothelial cells |
| iNOS | Inducible nitric oxide synthase |
| IVR | Intervening region |
| KD | Dissociation constant |
| Keap1 | Kelch-like ECH associated protein 1 |
| KIX | Kinase-inducible domain interacting |
| LC-MS/MS | High performance liquid chromatography-mass spectrometry |
| LPS | Lipopolysaccharides |
| LXRα | Liver X receptor |
| MAD | Malondialdehyde |
| 3-MP | 3-mercaptopyruvate |
| 3MST | 3-mercaptopyruvate sulfur transferase |
| Maf | Musculoaponeurotic fibrosarcoma |
| METH | Methamphetamine |
| MPTP | 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine |
| MW | Molecular weight |
| NAC | N-acetylcysteine |
| NACA | N-acetylcysteine amide |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NaHS | Sodium hydrosulfide |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NO | Nitric oxide |
| NQOI | NADPH dehydrogenase 1 |
| Nrf2 | Nuclear factor erythroid 2-related factor 2 |
| PD | Parkinson’s disease |
| PPI | Protein–protein interaction |
| Prdx6 | Peroxiredoxin 6 |
| pTRAF | Plasmid for transcription factor reporter activation based upon fluorescence |
| ROS | Reactive oxygen species |
| RP | Rice protein |
| SFN | Sulforaphane |
| SH | Sulfhydryl group |
| siRNA | Small interfering RNA |
| SO | Sulfinyl group |
| SOD | Superoxide dismutase |
| Trx | Thioredoxin |
| TrxR | Thioredoxin reductase |
| UGT | UDP-glucuronosyltransferase |
| UVB | Ultraviolet B radiation |
References
- Cuadrado, A.; Manda, G.; Hassan, A.; Alcaraz, M.J.; Barbas, C.; Daiber, A.; Ghezzi, P.; Leon., R.; Lopez, M.G.; Oliva, B.; et al. Transcription factor Nrf2 as a therapeutic target for chronic diseases: A system medicine approach. Pharmacol. Rev. 2018, 70, 348–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukwevho, E.; Ferreira, Z.; Ayeleso, A. Potential role of sulfur-containing antioxidant systems in highly oxidative environments. Molecules 2014, 19, 19376–19389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parcell, S.; Cand, N.D. Sulfur in human nutrition and applications in medicine. Altern. Med. Rev. 2002, 7, 22–44. [Google Scholar] [PubMed]
- Cuadrado, A.; Rojo, A.I.; Wells, G.; Hayes, J.D.; Cousin, S.P.; Rumsey, W.L.; Attucks, O.C.; Franklin, K.S.; Levonen, A.; Kensler, T.W.; et al. Therapeutic targeting of the Nrf2 and Keap1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019, 18, 295–317. [Google Scholar] [CrossRef] [Green Version]
- Egbujor, M.C.; Saha, S.; Buttari, B.; Profumo, E.; Saso, L. Activation of Nrf2signaling pathway by natural and synthetic chalcones: A therapeutic road map for oxidative stress. Expert Rev. Clin. Pharmacol. 2021, 14, 465–480. [Google Scholar] [CrossRef]
- Saha, S.; Buttari, B.; Panieri, E.; Profumo, E.; Saso, L. An overview of Nrf2 signaling pathway and its role in flammation. Molecules 2020, 25, 5474. [Google Scholar] [CrossRef]
- He, Y.; Jiang, J.; He, B.; Shi, Z. Chemical activators of the Nrf2 signalling pathway in nonalcoholic fatty liver disease. Nat. Prod. Commun. 2021, 16, 1934578X20987095. [Google Scholar]
- Imai, T.; Matsubara, H.; Hara, H. Potential therapeutic effects of Nrf2 activators on intracranial hemorrhage. J. Cereb. Blood Flow Metab. 2021, 41, 1483–1500. [Google Scholar] [CrossRef]
- Liebman, S.E.; Le, T.H. Eat your Broccoli: Oxidative stress, Nrf2, and sulforaphane in chronic kidney disease. Nutrients 2021, 13, 266. [Google Scholar] [CrossRef]
- Russo, M.; Spagnuolo, C.; Russo, G.L.; Skalicka-Woźniak, K.; Daglia, M.; Sobarzo-Sánchez, E.; Nabavi, S.F.; Nabavi, S.M. Nrf2 targeting by sulforaphane: A potential therapy for cancer treatment. Crit. Rev. Food Sci. Nutr. 2018, 58, 1391–1405. [Google Scholar] [CrossRef]
- Sun, Q.; Shen, X.; Ma, J.; Lou, H.; Zhang, Q. Activation of Nrf2 signaling by oltipraz inhibits death of human macrophages with mycobacterium tuberculosis infection. Biochem. Biophys. Res. Commun. 2020, 531, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Hseu, Y.-C.; Yang, H.-L. Use of ergothioneine for inducing activity of nuclear factor erythroid 2-related factor-2 (Nrf2) in cell: EP2992882A1. In Inventors; Taiwan Assignee; China Medical University: Taiwan, China, 2016; Available online: https://patents.google.com/patent/EP2992882A1/en (accessed on 22 June 2022).
- Abdelhamid, A.M.; Youssef, M.E.; Cavalu, S.; Mostafa-Hedeab, G.; Youssef, A.; Elazab, S.T.; Ibrahim, S.; Allam, S.; Elgharabawy, R.M.; El-Ahwany, E.; et al. Carbocisteine as a Modulator of Nrf2/HO-1 and NFκB Interplay in Rats: New Inspiration for the Revival of an Old Drug for Treating Ulcerative Colitis. Front. Pharmacol. 2022, 13, 887233. [Google Scholar] [CrossRef]
- Mahn, A.; Castillo, A. Potential of sulforaphane as a natural immune system enhancer: A review. Molecules 2021, 26, 752. [Google Scholar] [CrossRef] [PubMed]
- Clifford, T.; Acton, J.P.; Cocksedge, S.P.; Davis, K.A.B.; Balley, S.J. The effect of dietary phytochemicals on nuclear factor erythroid 2-related factor 2 (Nrf2) activation: A systemic review of human intervention trials. Mol. Biol. Rep. 2021, 48, 1745–1761. [Google Scholar] [CrossRef] [PubMed]
- Motohashi, H.; O’Connor, T.; Katsuoka, F.; Engel, J.D.; Yamamoto, M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 2002, 294, 1–12. [Google Scholar] [CrossRef]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef] [Green Version]
- Katoh, Y.; Itoh, K.; Yoshida, E.; Miyagishi, M.; Fukamizu, A.; Yamamoto, M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 2001, 6, 857–868. [Google Scholar] [CrossRef]
- Jaramillo, M.C.; Zhang, D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013, 27, 2179–2191. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Liu, K.; Geng, M.; Gao, P.; Wu, X.; Hai, Y.; Li, Y.; Li, Y.; Luo, L.; Hayes, J.D.; et al. RXRα inhibits the NRF2-ARE signaling pathway through a direct interaction with the Neh7 domain of NRF2. Cancer Res. 2013, 73, 3097–3108. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, M.; Kuga, A.; Suzuki, M.; Panda, P.; Kitamura, H.; Motohashi, H.; Yamamoto, M. Microenvironmental Activation of Nrf2 Restricts the Progression of Nrf2-Activated Malignant TumorsNrf2-High Microenvironment Restricts Nrf2-Activated Tumors. Cancer Res. 2020, 80, 3331–3344. [Google Scholar] [CrossRef]
- Singh, S.; Nagalakshmi, D.; Sharma, K.K.; Ravichandiran, V. Natural autioxidants for neuroinflammatory disorders and possible involvement of Nrf2 pathway: A review. Heliyon 2021, 7, e06216. [Google Scholar] [CrossRef]
- Yao, W.; Lin, S.; Su, J.; Cao, Q.; Chen, Y.; Chen, J.; Zhang, Z.; Hashimoto, K.; Qi, Q.; Zhang, J. Activation of BDNF by transcription factor Nrf2 contributes to antidepressant –Like actions in rodent. Transl. Psychiatry 2021, 11, 140. [Google Scholar] [CrossRef] [PubMed]
- Dhaliwal, N.; Dhaliwal, J.; Singh, A.; Chopra, K. Dimethyl fumarate attenuates 2-vo-induced vascular dementia via activating the Nrf2 signaling pathway in rates. Inflammopharmacology 2021, 29, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Shao, J.P.; Zhao, F.-Q.; Liu, H. Nrf2-ARE signalling partially attenuates lipopolysaccharide-induced mammary lesions via regulation of oxidative and organelle stresses but not inflammatory response in mice. Oxid. Med. Cell. Longev. 2021, 2021, 8821833. [Google Scholar]
- Zhang, W.-B.; Lai, X.; Guo, X.-F. Activation of Nrf2 by miR-152 inhibits doxorubicin-induced cardiotoxicity via attenvation of oxidative stress, inflammation and apoptosis. Oxid. Med. Cell. Longev. 2021, 2021, 8860883. [Google Scholar] [PubMed]
- KO, E.; Kim, D.; Min, D.W.; Kwon, S.H.; Lee, J.-Y. Nrf2 regulates cell motility through RhoA-Rock1 signaling in non-small-cell lung cancer cells. Sci. Rep. 2021, 11, 1247. [Google Scholar] [CrossRef] [PubMed]
- Moi, P.; Chan, K.; Aunis, I.; Cao, A.; Kan, Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2 –like basic leucine zipper transcriptional activator that bionds to the tandem NF-E2/API repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930. [Google Scholar] [CrossRef] [Green Version]
- Chan, K.; Li, R.; Chang, J.C.; Kan, Y.W. Nrf2 a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. USA 1996, 93, 13943–13948. [Google Scholar] [CrossRef] [Green Version]
- Kerins, M.J.; Ooi, A. The Roles of NRF2 in Modulating Cellular Iron Homeostasis. Antioxid. Redox Signal. 2018, 29, 1756–1773. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Zhu, J.l.; In, H.; Gu, K.; Feng, F. Recent progress in the development of small molecule Nrf2 modulators: A patent review (2012–2016). Expert Opin. Ther. Pat. 2017, 27, 763–785. [Google Scholar] [CrossRef]
- Song, Z.-L.; Hou, Y.; Bai, F.; Fang, J. Generation of potent Nrf2 activators via tuning the electrophilicity and steric hinderance of vinyl sulfones for neuroprotection. Bioorg. Chem. 2020, 107, 104520. [Google Scholar] [CrossRef] [PubMed]
- Jaganjac, M.; Milkovic, L.; Sunjic, S.B.; Zarkovic, N. The NRF2, thioredoxin, and glutathione system in tumorigenesis and anticancer therapies. Antioxidants 2020, 9, 1151. [Google Scholar] [CrossRef] [PubMed]
- Harvey, C.J.; Thimmulappa, R.K.; Singh, A.; Blake, D.J.; Ling, G.; Wakabyashi, N.; Fujii, J.; Myers, A.; Biswal, S. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic. Biol. Med. 2009, 46, 443–453. [Google Scholar] [CrossRef] [Green Version]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef] [PubMed]
- Barid, L.; Dinkova-Kostova, A.T. The cytoprotective role of the Keapl-Nrf2 pathway. Arch. Toxicol. 2011, 85, 241–272. [Google Scholar] [CrossRef]
- Magesh, S.; Chen, Y.; Hu, L. Small molecule modulators of Keapl-Nrf2 ARE pathway as potential preventive and therapeutic agents. Med. Res. Rev. 2012, 32, 687–726. [Google Scholar] [CrossRef] [Green Version]
- Done, A.J.; Traustadottir, T. Nrf2 mediates redox adaptations to exercise. Redox Biol. 2016, 10, 191–199. [Google Scholar] [CrossRef] [Green Version]
- Bauer, M.; Bauer, I. Heme oxygenase-1: Redox regulation and role in the hepatic response to oxidative stress. Antioxid. Redox Signal. 2002, 4, 749–758. [Google Scholar] [CrossRef]
- Bellezza, I.; Tucci, A.; Galli, F.; Grottellli, S.; Mieria, A.L.; Piloll, F.; Minelti, A. Inhibition of NF-κB nuclear translocation via HO-1 activation underlies α-tocopheryl succinate toxicity. J. Nutr. Biochem. 2012, 23, 1585. [Google Scholar] [CrossRef]
- Yerra, V.G.; Negi, G.; Sharma, S.S.; Kumar, A. Potential therapeutic effect of the simultaneous targeting of the Nrf2 and NF-KB pathways in dietary neuropathy. Redox Biol. 2013, 1, 394–397. [Google Scholar] [CrossRef] [Green Version]
- Hourihan, J.M.; Kenna, J.G.; Hayes, J.D. The gasotransmitter hydrogen sulfide induces Nrf2-target genes by inactivating the Keap1 ubiquitin ligase substrate adaptor through formation of a disulphide bond between cys-226 and cys-613. Antioxid. Redox Signal. 2013, 19, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.D.; Dinkova-Kostova, A.T. The Nrf2 regulatory network provides and interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014, 39, 199–218. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.C.; Liu, J.J.; Klaassen, C.D. Nrf2 activation prevents cadmium-induced acute liver injury. Toxicol. Appl. Pharmacol. 2012, 263, 14–20. [Google Scholar] [CrossRef]
- Sears, M.E. Chelation: Harnessing and enhancing heavy metal detoxification—A review. Sci. World J. 2013, 2013, 219840. [Google Scholar] [CrossRef] [Green Version]
- Shen, G.; Kong, A.N. Nrf2 plays an important role in coordinated regulation of Phase II drug metabolism enzymes and Phase III drug transporters. Biopharm. Drug Dispos. 2009, 30, 345–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayes, J.D.; Dinkova-Kostova, A.T.; McMahon, M. Cross-talk between Transcription Factors AhR and Nrf2: Lessons for Cancer Chemoprevention from Dioxin. Toxicol. Sci. 2009, 111, 199–201. [Google Scholar] [CrossRef]
- Shaw, P.; Mondal, P.; Bandyopadhyay, A.; Chattopadhyay, A. Environmentally relevant concentration of chromium activates Nrf2 and alters transcription of related XME genes in liver of zebrafish. Chemosphere 2019, 214, 35–46. [Google Scholar] [CrossRef]
- Kawajiri, K.; Fujii-Kuriyama, Y. The aryl hydrocarbon receptor: A multifunctional chemical sensor for host defense and homeostatic maintenance. Exp. Anim. 2017, 66, 75–89. [Google Scholar] [CrossRef] [Green Version]
- Ooi, B.K.; Chan, K.G.; Goh, B.H.; Yap, W.H. The Role of Natural Products in Targeting Cardiovascular Diseases via Nrf2 Pathway: Novel Molecular Mechanisms and Therapeutic Approaches. Front. Pharmacol. 2018, 9, 1308. [Google Scholar] [CrossRef] [Green Version]
- Rojo de la Vega, M.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef]
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 2004, 279, 31556–31567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.K.M.; Chen, N.L.; Chan, A.; Wang, H.J. The many blades of the β-propeller proteins: Conserved but versatile. Trends Biochem. Sci. 2011, 36, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, D.; Hannink, M.; Beamer, L.J. Crystal structure of the Kelch domain of human Keap1. J. Biol. Chem. 2004, 279, 54750–54758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, S.C.; Li, X.; Henzl, M.T.; Beamer, L.J.; Hannink, M. Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 2006, 25, 3605–3617. [Google Scholar] [CrossRef] [Green Version]
- Eggler, A.L.; Liu, G.; Pezzuto, J.M.; Van Breemen, R.B.; Mesecar, A.D. Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. USA 2005, 102, 10070–10075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, K.I.; Katoh, Y.; Kusunoki, H.; Itoh, K.; Tanaka, T.; Yamamoto, M. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: Characterization of the two-site molecular recognition model. Mol. Cell. Biol. 2006, 26, 2887–2900. [Google Scholar] [CrossRef] [Green Version]
- Fukutomi, T.; Takagi, K.; Mizushima, T.; Ohuchi, N.; Yamamoto, M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol. Cell. Biol. 2014, 34, 832–846. [Google Scholar] [CrossRef] [Green Version]
- Horie, Y.; Sazuki, T.; Inoue, J.; Iso, T.; Wells, G.; Moore, T.W.; Mizushuma, T.; Dinkova-Kostova, A.T.; Kasai, T.; Kamei, T.; et al. Molecular basis for the disruption of Keap1–Nrf2 interaction via Hinge & Latch mechanism. Commun. Biol. 2021, 4, 576. [Google Scholar]
- Padmanabhan, B.; Tong, K.I.; Ohta, T.; Nakamura, Y.; Scharlock, M.; Ohtsuji, M.; Kang, M.I.; Kobayashi, A.; Yokoyama, S.; Yamamoto, M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 2006, 21, 689–700. [Google Scholar] [CrossRef]
- Yamamoto, T.; Suzuki, T.; Kobayashi, A.; Wakabayashi, J.; Maher, J.; Motohashi, H.; Yamamoto, M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell. Biol. 2008, 28, 2758–2770. [Google Scholar] [CrossRef] [Green Version]
- Saito, R.; Suzuki, T.; Hiramoto, K.; Asami, S.; Naganuma, E.; Suda, H.; Iso, T.; Yamamoto, H.; Morita, M.; Baird, L.; et al. Characterizations of Three Major Cysteine Sensors of Keap1 in Stress Response. Mol. Cell. Biol. 2015, 36, 271–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robledinos-Antón, N.; Fernández-Ginés, R.; Manda, G.; Cuadrado, A. Activators and Inhibitors of NRF2: A Review of Their Potential for Clinical Development. Oxid. Med. Cell. Longev. 2019, 2019, 9372182. [Google Scholar] [CrossRef] [PubMed]
- Tran, K.T.; Pallesen, J.S.; Solbak, S.M.Ø.; Narayanan, D.; Baig, A.; Zang, J.; Aguayo-Orozco, A.; Carmona, R.M.C.; Garcia, A.D.; Bach, A. A Comparative Assessment Study of Known Small-Molecule Keap1-Nrf2 Protein-Protein Interaction Inhibitors: Chemical Synthesis, Binding Properties, and Cellular Activity. J. Med. Chem. 2019, 62, 8028–8052. [Google Scholar] [CrossRef]
- McMahon, M.; Lamont, D.J.; Beattie, K.A.; Hayes, J.D. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. USA 2010, 107, 18838–18843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplan, P.; Tatarkova, Z.; Sivonova, M.K.; Racay, P.; Lehotsky, J. Homocysteine and mitochondria in cardiovascular and cerebrovascular systems. Int. J. Mol. Sci. 2020, 21, 7698. [Google Scholar] [CrossRef] [PubMed]
- Kwon, K.-B.; Yoo, S.-J.; Ryu, D.-G.; Yang, J.-Y.; Rho, H.-W.; Kim, J.-S.; Park, J.-W.; Kim, H.R.; Park, B.H. Induction of apoptosis by diallyl disulfide through activation of caspase-3 in human leukemia HL-60 cells. Biochem. Pharmacol. 2002, 63, 41–47. [Google Scholar] [CrossRef]
- Filomeni, G.; Aquilano, K.; Rotilio, G.; Ciriolo, M.R. Reactive oxygen species-dependent c-Jun NH2-terminal kinase/c-Jun signaling cascade mediates neuroblastoma cell death induced by diallyl disulfide. Cancer Res. 2003, 63, 5940–5949. [Google Scholar]
- Haskew-Layton, R.E.; Payappilly, J.B.; Smirnova, N.A.; Maa, T.C.; Chana, K.K.; Murphy, T.H.; Guoa, H.; Langley, B.; Sultana, R.; Butterfield, D.A.; et al. Controlled enzymatic production of astrocytic hydrogen peroxide protects neurons from oxidative stress via an Nrf2-independent pathway. Proc. Natl. Acad. Sci. USA 2010, 107, 17385–17390. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Masayuki, Y. Stress-sensing mechanisms and the physiological roles of the Keap1–Nrf2 system during cellular stress. J. Biol. Chem. 2017, 292, 16817–16824. [Google Scholar] [CrossRef] [Green Version]
- Brennan, M.S.; Matos, M.F.; Li, B.; Hronowski, X.; Gao, B.; Juhasz, P.; Rhodes, K.J.; Scannevin, R.H. Dimethyl fumarate and monoethyl fumarate exhibit differential effects on KEAP1, NRF2 activation, and glutathione depletion in vitro. PLoS ONE 2015, 10, e0120254. [Google Scholar] [CrossRef]
- Takaya, K.; Suzuki, T.; Motohashi, H.; Onodera, K.; Satomi, S.; Kensler, T.W.; Yamamoto, M. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2012, 53, 817–827. [Google Scholar] [CrossRef] [Green Version]
- Dayalan, N.S.; Muramatsu, A.; Saito, R.; Asami, S.; Honda, T.; Hosoya, T.; Itoh, K.; Yamamoto, M.; Suzuki, T.; Dinkova-Kostova, A.T. C151 in KEAP1 is the main cysteine sensor for the cyanoenone class of NRF2 activators, irrespective of molecular size or shape. Sci. Rep. 2018, 8, 8037. [Google Scholar] [CrossRef] [PubMed]
- Baird, L.; Yamamoto, M. The molecular mechanisms regulating the KEAP1-NRF2 pathway. Mol. Cell. Biol. 2020, 40, e00099-20. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Sherratt, P.J.; Pickett, C.B. Regulatory mechanisms controlling gene expression mediated by the antioxidant response element. Annu. Rev. Pharmacol. Toxicol. 2003, 43, 233–260. [Google Scholar] [CrossRef]
- Raghunath, A.; Sundarraj, K.; Nagarajan, R.; Arfuso, F.; Bian, J.; Kumar, A.P.; Sethi, G.; Perumal, E. Antioxidant response elements: Discovery, classes, regulation and potential applications. Redox Biol. 2018, 17, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.L.; Dodd, G.; Thomas, S.; Zhang, X.; Wasserman, M.A.; Rovin, B.H.; Kunsch, C. Activation of Nrf2/ARE pathway protects endothelial cells from oxidant injury and inhibits inflammatory gene expression. Am. J. Physiol. Heart Circ. Physiol. 2006, 290, H1862-70. [Google Scholar] [CrossRef] [Green Version]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, T.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hussain, S.; Parveen, S.; Zhang, S.; Yang, Y.; Zhu, C. Sulfonyl group-containing compounds in the design of potential drugs for the treatment of diabetes and its complications. Curr. Med. Chem. 2012, 19, 3578–3604. [Google Scholar] [CrossRef] [PubMed]
- Egbujor, M.C.; Okoro, U.C.; Okafor, S. Design, synthesis, molecular docking, antimicrobial and antioxidant activities of new phenylsulfamoyl carboxylic acids of pharmacological interest. Med. Chem. Res. 2019, 28, 2118–2127. [Google Scholar] [CrossRef]
- Egbujor, M.C.; Okoro, U.C. New methionine-based p-toluenesulphonamoyl carboxamide derivatives as antimicrobial and antioxidant agents: Design, synthesis. J. Pharm. Res. Int. 2019, 28, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Egbujor, M.C.; Okoro, U.C.; Okafor, S.; Nwankwo, N.E. Design, synthesis and molecular docking of novel serine-based sulphonamide bioactive compounds as potential antioxidant and antimicrobial agents. Indo Am. J. Pharm. Sci. 2019, 6, 12232–12240. [Google Scholar]
- Egbujor, M.C.; Nwobodo, D.C.; Egwuatu, P.I.; Abu, I.P.; Ezeagu, C.U. Sulphonamide drugs and Pseudomonas aeruginosa resistance: A review. Int. J. Mod. Pharm. Res. 2020, 4, 78–83. [Google Scholar]
- Egbujor, M.C.; Okoro, U.C.; Okafor, S. Novel alanine-based antimicrobial and antioxidant agents: Synthesis and molecular docking. Indian J. Sci. Technol. 2020, 13, 1003–1014. [Google Scholar] [CrossRef]
- Egbujor, M.C.; Okoro, U.C.; Nwobodo, D.C.; Ezeagu, C.U.; Amadi, U.B.; Okenwa-Ani, C.G.; Ugwu, J.I.; Okoye, I.G.; Abu, I.P.; Egwuatu, P.I. Design, synthesis, antimicrobial and antioxidant activities of novel threonine-based sulfonamide derivatives. J. Pharm. Res. Int. 2020, 32, 51–61. [Google Scholar] [CrossRef]
- Humljan, J.; Kotnik, M.; Constreras-Martel, C.; Blanot, D.; Urleb, U.; Dessen, A.; Olmajer, T.; Gobec, S. Novel naphthalene-N-Sulfonyl-D-glutamic acid derivatives as inhibitors of MurD, a Key peptidoglycan biosynthesis enzyme. J. Med. Chem. 2008, 51, 7486–7494. [Google Scholar] [CrossRef] [PubMed]
- Bachovchin, D.A.; Zuhl, A.M.; Speers, A.E.; Wolfe, M.R.; Weerapana, E.; Brown, S.J.; Rosen, H.; Cravatt, B.F. Discovery and optimisation of sulfonyl acrylonitriles as selective, covalent inhibitors of protein phosphatese-methylesterase-1. J. Med. Chem. 2011, 54, 5229–5236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alqasoumi, S.I.; Al-Taweed, A.M.; Alafeefy, A.M.; Ghorab, M.M.; Noaman, E. Discovering some novel tetrahydroquinoline derivatives bearing the biologically active sulphonamide moiefy as a new class of antitumor agents. Eur. J. Med. Chem. 2010, 45, 1849–1853. [Google Scholar] [CrossRef] [PubMed]
- Silvestri, R. Boom in the development of non-peptidic beta-secretase (BACE1) inhibitors for the treatment of Alzheimer’s disease. Med. Res. Rev. 2009, 29, 295–338. [Google Scholar] [CrossRef] [PubMed]
- Egbujor, M.C.; Okoro, U.C.; Okafor, S.; Nwankwo, N.E. Synthesis, characterization and in silico studies of novel alkanoylated 4-methylphenyl sulphonamoyl carboxylic acids as potential antimicrobial and antioxidant agents. Int. J. Pharm. Phytopharm. Res. 2019, 9, 89–97. [Google Scholar]
- Egbujor, M.C.; Egu, S.A.; Okonkwo, V.I.; Jacob, A.D.; Egwuatu, P.I.; Amasiatu, I.S. Antioxidant drug design: Historical and recent developments. J. Pharm. Res Int. 2021, 32, 36–56. [Google Scholar] [CrossRef]
- Egbujor, M.C.; Okoro, U.C.; Okafor, S.N.; Amasiatu, I.S.; Amadi, U.B.; Egwuatu, P.I. Synthesis, molecular docking and pharmacological evaluation of new 4-methylphenylsulphamoyl carboxylic acids analogs. Int. J. Res. Pharm. Sci. 2020, 11, 5357–5366. [Google Scholar] [CrossRef]
- Egbujor, M.C.; Okoro, U.C.; Egu, S.A.; Egwuatu, P.I.; Eze, F.U.; Amasiatu, I.S. Design, synthesis and biological evaluation of alanine-based sulphonamide derivatives. Int. J. Res. Pharm. Sci. 2020, 11, 6449–6458. [Google Scholar] [CrossRef]
- Lu, M.-C.; Shao, H.-L.; Liu, T.; You, Q.-D.; Jian, Z.-Y. Discovery of 2-oxy-2-phenylacetic acid substituted naphthalene sulphonamide derivatives as potent keapl-Nrf2 protein-protein interaction inhibitors for inflammatory conditions. Eur. Med. Chem. 2020, 207, 112734. [Google Scholar] [CrossRef] [PubMed]
- Voutyritsa, E.; Triandafillidi, I.; Kokotos, C.G. Green Organocatalytic Oxidation of Sulfides to Sulfoxides and Sulfones. Synthesis 2017, 49, 917–924. [Google Scholar]
- Kirihara, M.; Itou, A.; Noguchi, T.; Yamamoto, J. Tantalum carbide or niobium carbide catalyzed oxidation of sulphides with hydsrogen peroxide: High efficient and chemo selective syntheses of sulfoxides and sulfones. Synlett 2010, 2010, 57–1561. [Google Scholar] [CrossRef]
- Palmer, J.T.; Rasnick, D.; Klaus, J.L.; Brömme, D. Vinyl sulfones as mechanism-based cysteine protease inhibitors. J. Med. Chem. 1995, 38, 3193–3196. [Google Scholar] [CrossRef]
- Lee, J.A.; Kim, J.H.; Woo, S.Y.; Son, H.J.; Han, S.H.; Jang, B.K.; Choi, J.W.; Kim, D.J.; Park, K.D.; Hwang, O. A novel compound VSC2 has anti-inflammatory and antioxidant properties in microglia and in Parkinson’s disease model. Br. J. Pharmacol. 2015, 172, 1087–1100. [Google Scholar] [CrossRef] [Green Version]
- Woo, S.Y.; Kim, J.H.; Moon, M.K.; Han, S.-H.; Yeon, S.K.; Choi, J.W.; Jang, B.K.; Song, J.; Kang, Y.G.; Kim, J.W.; et al. Discovery of vinyl sulfones as a novel class of neuroprotective agents towards Parkinson’s disease therapy. J. Med. Chem. 2014, 57, 1473–1487. [Google Scholar] [CrossRef]
- Carlstrom, K.E.; Chinthakindi, P.K.; Espinosa, B.; Nimer, F.A.; Arner, E.S.J.; Arvidsson, P.I.; Piehl, F.; Johansson, K. Characterization of more selective central nervous system Nrf2- activating novel vinyl sulfoxime compounds compared to dimethyl fumarate. Neurotherapeutics 2020, 17, 1142–1152. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Rosell, G.; Sanchez, S.; Boixareu, N.; Pors, K.; Pouplana, R.; Campanera, J.M.; Pujol, M.D. Synthesis and biological properties of aryl methyl sulfones. Bioorg. Med. Chem. 2018, 26, 4113–4126. [Google Scholar] [CrossRef]
- Choi, J.W.; Kim, S.; Park, J.-H.; Kim, H.J.; Shin, S.J.; Kim, J.W.; Woo, S.Y.; Lee, C.; Han, S.M.; Lee, J.; et al. Optimization factor erythroid 2-related factor 2 (Nrf2) activators for Parkinson’s disease therapy. J. Med. Chem. 2019, 62, 811–830. [Google Scholar] [CrossRef] [PubMed]
- Begnini, F.; Geschwindner, S.; Johansson, P.; Wissler, L.; Lewis, R.J.; Danelius, E.; Luttens, A.; Matricon, P.; Carlsson, J.; Lenders, S.; et al. Importance of Binding Site Hydration and Flexibility Revealed When Optimizing a Macrocyclic Inhibitor of the Keap1-Nrf2 Protein-Protein Interaction. J. Med. Chem. 2022, 65, 3473–3517. [Google Scholar] [CrossRef]
- Ovung, A.; Bhattacharyya, J. Sulfonamide drugs: Structure, antibacterial property, toxicity, and biophysical interactions. Biophys. Rev. 2021, 13, 259–272. [Google Scholar] [CrossRef] [PubMed]
- De Luca, L.; Giacomelli, G. An easy microwave-assisted synthesis of sulfonamides directly from sulfonic acids. J. Org. Chem. 2008, 73, 3967–3969. [Google Scholar] [CrossRef] [PubMed]
- Eze, F.U.; Okoro, U.C.; Ugwu, D.I.; Okafor, S.N. New carboxamides bearing benzene sulfonamides: Synthesis, molecular docking and pharmacological properties. Bioorg. Chem. 2019, 92, 103265. [Google Scholar] [CrossRef]
- Nadeem, R.A.; Qadir, M.A.; Ahmed, M.; Sajid, I. Cephalosporin conjugated sulphonamides: Synthesis, characterization and anticancer activities. Lett. Drug Des. Discov. 2020, 17, 264–270. [Google Scholar] [CrossRef]
- Abdel-Aziz, A.A.; Angeli, A.; El-Azab, A.S.; Hammouda, M.E.A.; El-sherbeny, M.A.; Supuran, C.T. Synthesis and auti-inflammatory activity of sulphonamides and carboxygenase/carbonic anhydrase inhibitory actions. Bioorg. Chem. 2019, 84, 260–268. [Google Scholar] [CrossRef]
- Dodson, M.; De la Vega, M.R.; Cholanians, A.B.; Schmidlin, C.J.; Chapman, E.; Zhang, D.D. Modulating Nrf2 in disease: Timing is everything. Annul. Rev. Pharmacol. Toxicol. 2019, 59, 555–575. [Google Scholar] [CrossRef]
- Egbujor, M.C.; Garrido, J.; Borges, F.; Saso, L. Sulfonamide a valid scaffold for antioxidant drug development. Mini-Rev. Org. Chem. 2022, 19. [Google Scholar] [CrossRef]
- Peng, X.; Hu, T.; Zhang, Y.; Zhao, A.; Natarajan, B.; Wei, J.; Yan, H.; Chen, H.; Lin, C. Synthesis of caffeic acid sulphonamide derivatives and their protective effect against H2O2 induced oxidative damage in A549 cells. RSC Adv. 2020, 10, 9924–9933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.-Y.; Lu, M.-C.; Xu, L.-L.; Yang, T.-T.; Xi, M.-Y.; Xu, X.-L.; Guo, X.-K.; Zhang, X.-J.; You, Q.-D.; Sun, H.-P. Discovery of potent Keap1-Nrf2 protein-protein interaction inhibitor based on molecular binding determinants analysis. J. Med. Chem. 2014, 57, 2736–2745. [Google Scholar] [CrossRef] [PubMed]
- Naidu, S.D.; Suzuki, T.; Dikovskaya, D.; Knatko, E.V.; Higgins, M.; Sato, M.; Novak, M.; Villegas, J.A.; Moore, T.W.; Yamamoto, M.; et al. The isoquinoline PRL-295 increases the thermostability of Keap1 and disrupts its interaction with Nrf2. iScience 2021, 25, 103703. [Google Scholar] [CrossRef]
- Georgakopoulos, N.; Torgakopoulos, N.; Talapatra, S.; Dikovskaya, D.; Naidu, S.D.; Higgins, M.; Gatliff, J.; Ayhan, A.; Nikoloudaki, R.; Schaap, F.; et al. Phenyl bis-sulfonamide keap1-Nrf2 protein-protein interaction inhibitors with an alternative binding mode. J. Med. Chem. 2022, 65, 7380–7398. [Google Scholar] [CrossRef]
- Choi, J.W.; Shin, S.J.; Kim, H.J.; Park, J.H.; Kim, H.J.; Lee, E.H.; Pae, A.N.; Bahn, Y.S.; Park, K.D. Antioxidant, Anti-inflammatory, and Neuroprotective Effects of Novel Vinyl Sulfonate Compounds as Nrf2 Activator. ACS Med. Chem. Lett. 2019, 10, 1061–1067. [Google Scholar] [CrossRef]
- Strecker, A. Ueber eine neue bildungsweise und die constitution der sulfosauren. Ann. Chem. Pharm. 1868, 148, 90–96. [Google Scholar]
- Sacoman, J.L.; Badish, L.N.; Sharkey, T.D.; Hollingsworth, R.I. The metabolic and biochemical impact of glucose 6-sulfonate (sulfoquinovose), a dietary sugar, on carbohydrate metabolism. Carbohydr. Res. 2012, 362, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Luan, F.; Wei, L.; Zhang, J.; Mi, Y.; Dong, F.; Li, Q.; Guo, Z. Antioxidant activity and antifungal activity of chitosan derivatives with propane sulfonate groups. Polymers 2018, 10, 395. [Google Scholar] [CrossRef] [Green Version]
- Arshia, A.J.; Faheem, A.; Khan, K.M.; Shah, S.; Perveen, S. Benzophenone esters and sulfonates: Synthesis and their potential as anti-inflammatory agents. Med. Chem. 2019, 15, 162–174. [Google Scholar] [CrossRef]
- Jeelani, R.; Jahanbakhsh, S.; Kohan-Ghadr, H.-R.; Thakur, M.; Khan, S.; Aldhaheri, S.R.; Yang, Z.; Andreana, P.; Morris, R.; Abu-soud, H.M. Mesna(2-mercaptoethane sodium sulfonate) functions as a regulator of myeloperoxidase. Free Radic. Biol. Med. 2017, 110, 54–62. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, V.; Liang, D.; Yang, G.; Li, C.; Niu, P.; Tian, L. Sodium Tanshinone IIA sulfonate suppresses pulmonary fibroblas proliferation and activation induced by silica: Role of the Nrf2/Trx pathway. Toxicol. Res. 2016, 5, 116. [Google Scholar] [CrossRef]
- Gilchrist, T.L. Comprehensive Organic Functional Group Transformations, Synthesis: Carbon with Three or Four Attached Heteroatoms; Elsevier Science: Oxford, UK, 2017; Volume 6. [Google Scholar]
- Otocka, S.; Kwiatkowska, M.; Madalinska, L.; Kielbasinski, P. Chiral organosulfur ligands/catalysts with a stereogenic sulfur atom: Applications in asymmetric synthesis. Chem. Rev. 2017, 117, 4147–4181. [Google Scholar] [CrossRef]
- Kielbasinski, P.; Kwiatkowska, M.; Mikolajczyk, M. Organic Heteroatom–Containing compounds. In Future Directions in Biocatalsis, 2nd ed.; Matsuda, T., Ed.; Elsevier: Oxford, UK, 2017; Volume 1, pp. 191–250. [Google Scholar]
- Liang, C.; Xue, Z.; Cang, J.; Wang, H.; Li, P. Dimethyl sulfoxide induces heme oxygenase-1 expression via JNKs and Nrf2 pathways in human umbilical vein endothelial cells. Mol. Cell. Biochem. 2011, 355, 109–115. [Google Scholar] [CrossRef]
- Kensler, T.W.; Egner, P.A.; Agyeman, A.S.; Visvanathan, K.; Groopman, J.D.; Chen, J.-G.; Chen, T.-Y.; Fahey, J.W.; Talalay, P. Keapl-Nrf2 signaling: A target for cancer prevention by sulforaphane. Top. Curr. Chem. 2013, 329, 163–178. [Google Scholar] [PubMed] [Green Version]
- Khiar, N.; Werner, S.; Mallouk, S.; Lieder, F.; Alcudia, A.; Fernandez, I. Enantiopure sulforaphane analogues with various substituents at the sulfinyl sulfur: Asymmetric synthesis and biological activities. J. Org. Chem. 2009, 74, 6002–6009. [Google Scholar] [CrossRef] [PubMed]
- Kielbasinski, P.; Kwiatkowska, M.; Cierpial, T.; Rachwalski, M.; Lesniak, S. The sulfinyl group: Biological activity. Phosphorus Sulfur Silicon Relat. Elem. 2019, 194, 649–653. [Google Scholar] [CrossRef]
- Patai, S.; Rappoport, Z. (Eds.) Syntheses of Sulphones, Sulphoxides and Cyclic Sulphides; John Wile & Sons: Hoboken, NJ, USA, 1995. [Google Scholar]
- Sanmartin-Suarez, C.; Soto-Otero, R.; Sanchez-Sellero, I.; Mendez-Alvarez, E. Antioxidant properties of dimethyl sulfoxide and its viability as a solvent in the evaluation of neuroprotective antioxidants. J. Pharmacol. Toxicol. Methods 2011, 63, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Unnikrishna, M.K.; Rao, M.N.A. Anti-inflammatory activity of methionine, methionine sulfoxide and methionine sulfone. Agents Actions 1990, 31, 110–112. [Google Scholar] [CrossRef]
- Novak, K.M. Drug Facts and Comparisons, 56th ed.; Wolters Klumer Health: St. Louis, MO, USA, 2002; p. 619. [Google Scholar]
- Gavrilin, M.V.; Sen’ Chukova, G.V.; Kompantseva, E.V. Structure of Chemical compounds, methods of analysis and process control. Methods for the synthesis and analysis of dimethyl sulfoxide (a review). Pharm. Chem. J. 2000, 34, 35–38. [Google Scholar]
- Steffel, J.; Hermann, M.; Greutert, H.; Gay, S.; Luscher, T.F.; Ruschitzka, F.; Tanner, F.C. Celecoxib decreases endothelial tissue factor expression through inhibition of c-Jun terminal NH2 Kinase phosphorylation. Circulation 2005, 111, 1685–1689. [Google Scholar] [CrossRef] [Green Version]
- Shim, Y.S.; Hwangm, H.S.; Nam, G.; Choi, K. Synthesis and Nrf2 activating ability of thiourea and vinyl sulfoxide derivatives. Bull. Korean Chem. Soc. 2013, 34, 2317. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Xing, K.; Zhang, J.; Tong, T.; Shi, Y.; Cao, H.; Yu, H.; Zhang, Y.; Liu, D.; Zhao, L. Application of sulfoximines in medicinal chemistry from 2013 to 2020. Eur. J. Med. Chem. 2021, 209, 112885. [Google Scholar] [CrossRef] [PubMed]
- Peters, T. Methionine Sulfoximine: A Novel Anti-Inflammatory Agent. Ph.D. Thesis, Wayne State University, Detroit, MI, USA, 2018; p. 2124. [Google Scholar]
- Tota, A.; Zensola, M.; Chawner, S.J.; John-Acmpbell, S.; Carlucci, C.; Romanazzi, G.; Degennaro, L.; Bull, J.A.; Luisi, R. Synthesis of NH-sulfoximines from sulfides by chemoselective one-pot N-and O-transfers. Chem. Commun. 2017, 53, 348–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdovinos-Flores, C.; Limon-Pacher, J.H.; Leon-Rodriguez, R.; Petrosyan, P.; Garza-Lombo, C.; Gonsebatt, M.E. Systemic L-buthionine-S-R-sulfoximine treatment increases plasma NGF and upregulates L-cys/L-cys2 transporter and γ-glutamylcysteine ligase mRNAs through the NGF/TrKA/AKt/Nrf2 pathway in the striatum. Front. Cell. Neurosci. 2019, 13, 325. [Google Scholar] [CrossRef] [Green Version]
- Ansari, M.I.; Khan, M.M.; Saquib, M.; Khatoon, S.; Husssain, M.K. Dithiolethiones: A privileged pharmacophore for anticancer therapy and chemoprevention. Future Med. Chem. 2018, 10, 1241–1260. [Google Scholar] [CrossRef]
- Tew, K.D. Oltipraz; Reference Module in Biomedical Sciences; Elsevier: Amsterdam, Switzerland, 2016. [Google Scholar]
- Kim, J.W.; Choi, K.D.; Lim, J.W.; Lee, K.H.; Lee, S.H. Method for Preparing Oltipraz. U.S. Patent 7288652B2, 30 October 2007. [Google Scholar]
- Merrelle, M.D.; Jackson, J.P.; Augustine, L.M.; Fisher, C.D.; Slitt, A.L.; Maher, J.M.; Huana, W.; Moore, D.D.; Zhang, Y.; Klaassen, C.D.; et al. the Nrf2 activator oltipraz also activates the constitutive androstance receptor. Drug Metab. Dispos. 2008, 36, 1716–1721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Z.; Shao, Z.; Ling, W.; Fantus, I.G.; Jin, T. Oltipraz upregulates the nuclear factor (erythroid–derived 2)-like 2 (Nrf2) antioxidant system and prevents insulin resistance and obesity induced by a high-fat diet in C57BL/6J mice. Diabetologia 2011, D, 922–934. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.; Bian, M.; Wu, J.; Li, D.; Ding, L.; Zeng, Q. Oltipraz prevents high glucose-induced oxidative stress and apoptosis in RSC96 cells through the Nrf2/NQO1 signaling pathway. BioMed Res. Int. 2020, 2020, 5939815. [Google Scholar] [CrossRef]
- Antilano-Roque, A.; Wen, X.; Alekunes, L.M.; Joy, M.S. Nrf2 activators as potential modulators of injury in human kidney cells. Toxicol. Rep. 2016, 3, 153–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos-Gomez, M.; Dolan, P.M.; Itoh, K.; Yamamoto, M.; Kensler, T.W. Interactive effects of nrf2 genotype and oltipraz on benzo[a]pyrene-DNA adducts and tumor yield in mice. Carcinogenesis 2003, 24, 461–467. [Google Scholar] [CrossRef] [Green Version]
- Iida, K.; Itoh, K.; Kumagai, Y.; Oyasu, R.; Hattori, K.; Kawai, K.; Shimazui, I.; Akaza, H.; Yamamoto, M. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 2004, 64, 6424–6431. [Google Scholar] [CrossRef] [Green Version]
- Faley, J.W.; Zalcmann, A.T.; Talalay, P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 2001, 56, 5–51. [Google Scholar]
- Dinkova-Kostova, A.T. Chemoprotection against cancer by isothiocyanate: A focus on the animal models and protective mechanisms. In Natural Products in Cancer Prevention and Therapy; Springer: Berlin/Heidelberg, Germany, 2012; pp. 179–201. [Google Scholar]
- Cheng, D.; Gao, L.; Su, S.; Sargsyan, D.; Wu, R.; Raskin, H.; Koug, A.-N. Moringa isothiocyanate activates Nrf2: Potential role in diabetic nephropathy. AAPs J. 2019, 21, 31. [Google Scholar] [CrossRef] [PubMed]
- Chuang, W.-T.; Yen, C.-C.; Huang, C.-S.; Chen, H.-W.; Lii, C.-K. Benzyl isothiocyanate ameliorates high-fat diet-induced hyperglycemia by enhancing Mrf2-dependant antioxidant defuse-mediated IRS-1/AKT/TBC1D1 signaling and GLUT4 expression in skeletal muscle. J. Agric. Food Chem. 2020, 68, 15228–15238. [Google Scholar] [CrossRef] [PubMed]
- El Badawy, S.A.; Ogaly, H.A.; Abd-Elsalam, R.M.; Azouz, A.A. Benzyl isothiocyanates modulate inflammation, oxidative stress and apoptosis via Nrf2/ho-1 and NF-KB signaling pathways on indomethacin-induced gastric injury in rats. Food Funct. 2021, 12, 6001–6013. [Google Scholar] [CrossRef]
- Miyoshi, N.; Takabayashi, S.; Osawa, T.; Nakamura, Y. Benzyl isothiocyanate inhibits excessive superoxide generation in inflammatory leukocytes. Implication for prevention against inflammation-related carcinogenesis. Carcinogenesis 2004, 25, 567–575. [Google Scholar] [CrossRef] [Green Version]
- Das, B.N.; Kim, Y.-W.; Keum, Y.-S. Mechanisms of Nrf2/Keap1-Dependent phase II cytoprotective and detoxifying gene expression and potential cellular targets of chemo preventive isothiocyanates. Oxid. Med. Cell. Longev. 2013, 2013, 839409. [Google Scholar]
- Caglayan, B.; Kilic, E.; Dalay, A.; Altunay, S.; Tuzcu, M.; Erten, F.; Orhan, C.; Gunal, M.Y.; Yulug, B.; Juturu, V.; et al. Allyl isothiocyanate attenuates oxidative stress and inflammation by modulating Nrf2/Ho-1 and NF-KB pathways in traumatic brain injury in mice. Mol. Biol. Rep. 2019, 46, 241–250. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, S.; Wang, X.; Xu, X.; Yao, Z.; Fang, W.; Wu, J.; Wu, Q.; Li, Z.; Wang, D. Allyl isothiocyanate increases MRP1 expression in cigarate extract-simulated human bronchial epithelial cells via the JNK/Nrf2 pathway. Exp. Ther. Med. 2021, 4, 409. [Google Scholar] [CrossRef]
- Kim, M.W.; Kang, J.-H.; Jung, H.J.; Park, S.Y.; Lephan, T.H.; Namgung, J.-H.; Seo, S.-Y.; Yoon, Y.S.; Oh, S.H. Allyl isothiocyanate protects acetaminophen-induced liver injury via Nrf2 activation by decreasing spontaneous degradation in hepatocyte. Nutrients 2020, 12, 3585. [Google Scholar] [CrossRef]
- Naidu, S.D.; Suzuki, T.; Yamamoto, M.; Fahey, J.W.; Dinkova-Kostova, A.T. Phenyl isothiocyanate, a dual activator of transcription factors Nrf2 and HSF1. Mol. Nutr. Food Res. 2018, 62, 1700908. [Google Scholar] [CrossRef] [Green Version]
- Ernst, I.M.; Wagner, A.E.; Schuemann, C.J.; Storm, N.; Hoppner, W.; Doring, F.; Stocker, A.; Rimbach, G. Allyl-, butyl-and phenylethyl–isothiocyanate activative Nrf2 in cultured fibroblasts. Pharmacol. Res. 2011, 63, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Boyanapalli, S.S.S.; Paredes-Gonzalez, X.; Fuetes, F.; Zhang, C.; Guo, Y.; Pung, D.; Saw, C.L.L.; Kong, A.N.T. Nrf2 knockout attenuates the anti-inflammatory effects of phenylethyl isothiocyanates and curcumin. Chem. Res. Toxicol. 2014, 27, 2036–2043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keum, Y.-S.; Owuor, E.D.; Kim, B.-R.; Hu, R.; Kong, A.N.T. Involvement of NrF2 and JNK1 in the activation of antioxidant response element (ARE) by chemopreventive agent phenylethyl isothiocyanate (PEITC). Pharm. Res. 2003, 20, 1351–1356. [Google Scholar] [CrossRef] [PubMed]
- Houghton, C.A.; Fassett, R.G.; Combes, J.S. Sulforaphane and other nutrigenomic Nrf2 activators: Can the clinician’s expectation be matched by the reality? Oxid. Med. Cell. Longev. 2016, 2016, 7857186. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Wang, W.; Tang, J.; Bowater, R.P.; Bao, Y. Antioxidant effects of sulforaphane in human HepG2 cells and immortalised hepatocytes. Food Chem. Toxicol. 2019, 128, 129–136. [Google Scholar] [CrossRef] [Green Version]
- Ruhee, R.T.; Suzuki, K. The integrative role of sulforaphane in preventing inflammation, oxidative stress and fatique: A review of a potential protective phytochemical. Antioxidants 2020, 9, 521. [Google Scholar] [CrossRef]
- Hua, D.; Luo, A.; Wu, Z.; Huang, C.; Li, S.; Xu, X.; Xu, J.; Yang, C.; Wang, D.; Liu, C. Sulforaphane improves cognitive dysfunction after surgery and anaesthesia in mice: The role of Keap1-Nrf2 signaling. Brain Res. Bull. 2022, 181, 1–11. [Google Scholar] [CrossRef]
- Li, X.; Ying, H.; Zhang, Z.; Yang, Z.; You, C.; Cai, X.; Lin, Z.; Xiao, Y. Sulforaphane attenuates chronic intermittent hypoxia-induced brain damage in mice via augmenting Nrf2 nuclear translocation and autophagy. Front. Cell. Neurosci. 2022, 16, 827527. [Google Scholar] [CrossRef]
- Kubo, E.; Chhunchha, B.; Singh, P.; Sasaki, H.; Singh, D.P. Sulforaphane reactivates cellular antioxidant defence by inducing Nrfs/ARE/Prdx6 activity during aging and oxidative stress. Sci. Rep. 2017, 7, 14130. [Google Scholar] [CrossRef] [Green Version]
- Clarke, J.D.; Hsu, A.; Williams, D.E.; Dashwood, R.H.; Stevens, J.F.; Ho, E. Metabolism and tissue distribution of sulforaphane in Nrfs Knockout and wild-type mice. Pharm. Res. 2011, 28, 3171–3179. [Google Scholar] [CrossRef] [Green Version]
- Thimmulappa, R.K.; Mai, K.H.; Srisuma, S.; Kensle, T.W.; Yamamoto, M.; Siswal, S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002, 62, 5196–5203. [Google Scholar] [PubMed]
- Chung, F.L.; Conaway, C.C.; Rao, C.V.; Reddy, B.S. Chemoprevention of colonic aberrant crypt foci in Fischer rats by sulforaphane and phenethyl isothiocyanate. Carcinogenesis 2000, 21, 2287–2291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Q.; Tian, X.; Li, W.; Chen, L.; Zhou, M.; Xu, C.; Ru, Q. Sulforaphane alleviates methamphetamine-induced oxidative damage and apoptosis via the Nrf2-mediated pathway in vitro and in vivo. Food Agric. Immunol. 2020, 31, 859–880. [Google Scholar] [CrossRef]
- Morimitsu, Y.; Nakagawa, Y.; Hayashi, K.; Fujii, H.; Kumagai, T.; Nkamura, Y.; Osawa, T.; Horio, F.; Itoh, K.; Iida, K.; et al. A sulforaphane analogue that potently activates the Nrf2-dependent detoxification pathway. J. Biol. Chem. 2002, 277, 3456–3463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.-H.; Jang, H.-J.; Cho, W.-Y.; Yeon, S.J.; Lee, C.-H. In vitro antioxidant actions of sulfur-containing amino acids. Arab. J. Chem. 2020, 13, 1678–1684. [Google Scholar] [CrossRef]
- Guo, D.; Yang, J.; Ling, F.; Tu, L.; Li, J.; Chen, Y.; Zou, K.; Zhu, L.; Hou, X. Elemental diet enriched with amino acids alleviates mucosal inflammatory response and prevents colonic epithelial barrier dysfunction in mice with DSS-induced chronic colitis. J. Immunol. Res. 2020, 2020, 9430763. [Google Scholar] [CrossRef]
- Kimura, Y.; Koike, S.; Shibuya, N.; Lefer, D.; Ogasawara, Y.; Kimura, H. 3-Mercaptopyruvate sulfurtransferase produces potential redox regulators cysteine- and glutathione-persulfide (Cys-SSH and GSSH) together with signaling molecules H2S2, H2S3 and H2S. Sci. Rep. 2017, 7, 10459. [Google Scholar] [CrossRef] [Green Version]
- Koike, S.; Ogasawara, Y.; Shibuya, N.; Kimura, H.; Ishii, K. Polysulfide exerts a protective effect against cytotoxicity caused by t-buthylhydroperoxide through Nrf2 signaling in neuroblastoma cells. FEBS Lett. 2013, 587, 3548–3555. [Google Scholar] [CrossRef] [Green Version]
- Tesseraud, S.; Coustrard, S.M.; Collin, A.; Seiliez, I. Role of sulfur amino acids in controlling nutrient metabolism and cell functions: Implications for nutrition. Br. J. Nutr. 2009, 101, 1132–1139. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.C.; Gladyshev, V.N. The biological significance of methionine sulfoxide stereochemistry. Free Radic. Biol. Med. 2011, 50, 271–277. [Google Scholar] [CrossRef] [Green Version]
- Forman, H.J.; Zhang, H.; Rinna, A. Glutathione: Overview of its protective roles, measurement, and biosynthesis. Mol. Asp. Med. 2009, 30, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Chen, H.; Liu, P.; Yang, M.; Zou, L.; Xiao, D. Antioxidant Function and Metabolomics Study in Mice after Dietary Supplementation with Methionine. BioMed Res. Int. 2020, 2020, 9494528. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Coleman, D.N.; Hu, L.; Martinez-cortes, I.; Wang, W.; Parys, C.; Shen, X.; Loor, J.J. Methionine and arginine supplementation alter inflammatory and oxidative stress responses during lipopolysaccharide challenge in bovine mammary epithelial cells in vitro. J. Dairy Sci. 2020, 103, 676–689. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liang, M.; Li, H.; Cai, L.; He, H.; Wu, Q.; Yang, L. L-methionine activates Nrf2-ARE pathway to induce endogeneous antioxidant activity for depressing ROS- derived oxidative stress in growing rats. J. Sci. Food Agric. 2019, 99, 4849–4862. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cai, L.; Liang, M.; Wang, Z.; Zhang, Y.; Wu, Q.; Yang, L. Methionine augments endogeneous antioxidant capacity of rice protein through stimulating MSR antioxidant system and activating Nrf2-ARE pathway in growing and adult rats. Eur. Food Res. Technol. 2020, 246, 1051–1063. [Google Scholar] [CrossRef]
- Liu, R.; Diao, Q.; Cui, K. Effect of dietary methionine deficiency followed by a re-feeding phase on the hepatic antioxidants activities of lambs. Animals 2021, 11, 7. [Google Scholar] [CrossRef]
- Wanders, D.; Stone, K.P.; Forney, L.A.; Cortez, C.C.; Dille, K.N.; Simon, J.; Xu, M.; Hotard, E.C.; Nikonorova, I.A.; Pettit, A.P.; et al. Role of GCN2-Independent Signaling Through a Noncanonical PERK/NRF2 Pathway in the Physiological Responses to Dietary Methionine Restriction. Diabetes 2016, 65, 1499–1510. [Google Scholar] [CrossRef] [Green Version]
- Schuller-Levis, G.B.; Park, E. Taurine: New implications for an old amino acid. FEMS Microbiol. Lett. 2003, 226, 195–202. [Google Scholar] [CrossRef]
- Kosswig, K. Sulfonic acids, aliphatic. In Ullman’s Encyclopedia of Industrial Chemistry; Willey-VCH: Weinheim, Germany, 2000. [Google Scholar]
- Jurkowska, H.; Stipanuk, M.H.; Hirschberger, L.L.; Roman, H.B. Propargylglycine inhibits hypotaurine/taurine synthesis and elevates cystathionine and homocysteine concentrations in primary mouse hepatocytes. Amino Acids 2015, 47, 1215–1223. [Google Scholar] [CrossRef] [Green Version]
- Maleki, V.; Mahdavi, R.; Hajzadeh-sharafabad, F.; Alizadeh, M. The effects of taurine supplementation on oxidative stress indices and inflammation biomarkers in patients with type 2 diabetes: A randomized, double blind, placebo-controlled trial. Diabetol. Metab. Syndr. 2020, 12, 9. [Google Scholar] [CrossRef]
- Quaradakhi, T.; Gadance, L.K.; McSweeney, K.R.; Abraham, J.R.; Apostolopoulos, V.; Zulli, A. The anti-inflammatory effect of taurine on cardiovascular disease. Nutrients 2020, 12, 2847. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.; Eraqi, M.M.; Alfaiz, F. Therapeutic role of taurine as antioxidant in reducing hypertension risks in rats. Heliyon 2020, 6, e03209. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Huang, J.; Xiao, B.; Liu, Y.; Zhu, Y.; Wang, F.; Sun, S. Taurine protects mouse spermatocytes from ionizing radiation-induced damage through activation of Nrf2-HO-1 signaling. Cell. Physiol. Biochem. 2017, 44, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- Agca, C.A.; Tuzcu, M.; Hayirli, A.; Sahin, K. Taurine ameliorates neuropathy via regulating NF-kB and Nrf2/HO-1 signaling cascades in diabetic rats. Food Chem. Toxicol. 2014, 711, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Aparna, P.; Betigeri, A.M.; Pasupathi, P. Homocysteine and oxidative stress markers and inflammation in patients with coronary artery disease. Int. J. Biol. Med. Res. 2010, 1, 125–129. [Google Scholar]
- Selhub, J. Homocysteine metabolism. Annu. Rev. Nutr. 1999, 19, 217–246. [Google Scholar] [CrossRef] [Green Version]
- Sbodio, J.I.; Snyder, S.H.; Paul, B.D. Regulators of the transsulfuration pathway. Br. J. Pharmacol. 2019, 176, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Zhao, K.; Ju, Y.; Mani, S.; Cao, Q.; Puukila, S.; Khaper, N.; Wu, L.; Wang, R. Hydrogen sulfide protects against cellular senescence via S-sulfhydration of Keap1 and activation of Nrf2. Antioxid. Redox Signal. 2013, 18, 1906–1919. [Google Scholar] [CrossRef] [PubMed]
- Ida, T.; Sawa, T.; Ihara, H.; Tsuchiya, Y.; Watanabe, Y.; Kumagai, Y.; Suematsu, M.; Motohashi, H.; Fujii, S.; Matsunaga, T.; et al. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 7606–7611. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.O.; Pearson, R.G. The factors determining nucleophilic reactivities. J. Am. Chem. Soc. 1962, 84, 16–24. [Google Scholar] [CrossRef]
- Zuhra, K.; Tomé, C.S.; Forte, E.; Vicente, J.B.; Giuffrè, A. The multifaceted roles of sulfane sulfur species in cancer-associated processes. Biochim. Biophys. Acta Bioenerg. 2021, 1862, 148338. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Gu, Y.; Wen, M.; Zhao, S.; Wang, W.; Ma, Y.; Meng, G.; Han, Y.; Wang, Y.; Liu, G.; et al. Hydrogen Sulfide Induces Keap1 S-sulfhydration and Suppresses Diabetes-Accelerated Atherosclerosis via Nrf2 Activation. Diabetes 2016, 65, 3171–3184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panieri, E.; Buha, A.; Telkoparan-Akillilar, P.; Cevik, D.; Kouretas, D.; Veskoukis, A.; Skaperda, Z.; Tsatsakis, A.; Wallace, D.; Suzen, S.; et al. Potential Applications of NRF2 Modulators in Cancer Therapy. Antioxidants 2020, 9, 193. [Google Scholar] [CrossRef] [Green Version]
- Pirchl, M.; Ullrich, C.; Sperner-Unterweger, B.; Humpel, C. Homocysteine has anti-inflammatory properties in a hypercholesterolemic rat model in vivo. Mol. Cell. Neurosci. 2012, 49, 456–463. [Google Scholar] [CrossRef]
- Pirchl, M.; Ullrich, C.; Humpel, C. Differential effects of short- and long-term hyperhomocysteinaemia on cholinergic neurons, spatial memory and microbleedings in vivo in rats. Eur. J. Neurosci. 2010, 32, 1516–1527. [Google Scholar] [CrossRef] [PubMed]
- Mani, M.; Golmohammadi, T.; Khaghani, S.; Zamani, Z.; Azadmanesh, K.; Meshkani, R.; Pasalar, P. Homocysteine induces heme oxygenase-1expression via transcription factor Nrf2 activation in HepG2 cells. Iran. Biomed. J. 2013, 17, 93–100. [Google Scholar]
- Mani, M.; Khaghani, S.; Mohammadi, T.G.; Zamani, Z.; Azadmanesh, K.; Meshkani, R.; Pasalar, P.; Mostafavi, E. Activation of Nrf2-antioxidant response element mediated glutamate cysteine ligase expression in hepatoma cell line by homocysteine. Hepat. Mon. 2013, 13, e8394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Dong, J.-L.; Chen, Y.-L.; Liu, Y.; Huaug, S.-S.; Zhong, X.-L.; Cheng, Y.-H.; Wang, Z.-G. Nrf2 mediates the protective effects of homocysteine by increasing. Mol. Med. Rep. 2017, 16, 597–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navneet, S.; Cui, X.; Zhao, J.; Wang, J.; Kaidery, N.A.; Thomas, B.; Bollinger, K.E.; Yoon, Y.; Smith, S.B. Excess homocysteine upregulates the Nrf2-antioxidant pathway in retinal muller glial cells. Exp. Eye Res. 2019, 178, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Zed, P.J.; Krenzelok, E.P. Treatment of acetaminophen overdose. Am. J. Health Syst. Pharm. 1999, 56, 1081–1093. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Puyo, C.A. N-Acetylcysteine to Combat COVID-19: An Evidence Review. Ther. Clin. Risk Manag. 2020, 16, 1047–1055. [Google Scholar] [CrossRef]
- Cazzola, M.; Calzetta, L.; Facciolo, F.; Rogliani, P.; Matera, M.G. Pharmacological investigation on the antioxidant and anti-inflammatory activity of N-acetylcysteine in an ex vivo model of COPD exacerbation. Respire Res. 2017, 18, 26. [Google Scholar] [CrossRef] [PubMed]
- Jannatifar, R.; Parivar, K.; Roodbari, N.H.; Nasr-Esfahani, M.H. The effect of N-acetyl- cysteine on Nrf2 antioxidant gene expression in asthenoteratozoo-spermia men. A clinical trial study. Int. J. Fertil. Steril. 2020, 14, 171–175. [Google Scholar]
- Zhou, Y.; Wang, H.-D.; Zhou, X.-M.; Fang, J.; Zhu, L.; Ding, K. N-acetylcysteine amide provides. Neuroprotection via Nrf2-ARE pathway in a mouse model of traumatic brain injury. Drug Des. Dev. Ther. 2018, 12, 4117–4127. [Google Scholar] [CrossRef] [Green Version]
- Ezeriņa, D.; Takano, Y.; Hanaoka, K.; Urano, Y.; Dick, T.P. N-Acetyl Cysteine Functions as a Fast-Acting Antioxidant by Triggering Intracellular H2S and Sulfane Sulfur Production. Cell Chem. Biol. 2018, 25, 447–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuhra, K.; Tomé, C.S.; Masi, L.; Giardina, G.; Paulini, G.; Malagrinò, F.; Forte, E.; Vicente, J.B.; Giuffrè, A. N-Acetylcysteine Serves as Substrate of 3-Mercaptopyruvate Sulfurtransferase and Stimulates Sulfide Metabolism in Colon Cancer Cells. Cells 2019, 8, 828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maccio, A.; Madeddu, C.; Panzone, F.; Mantovani, G. Carbocysteine: Clinincal experience and new perspective in the treatment of chronic inflammatory diseases. Expert Opin. Pharmacother. 2009, 10, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Pace, E.; Ferraro, M.; Vincenzo, S.D.; Cipollina, C.; Gerbino, S.; Gjomarkaj, M. Comparative cytoprotective effects of carbocysteine and fluticasone propionate in cigarette smoke extract-stimulated bronchial epithelial cells. Cell Stress Chaperones 2013, 18, 733–743. [Google Scholar] [CrossRef] [Green Version]
- Borodina, I.; Kenny, L.C.; McCarthy, C.M.; Paramasivan, K.; Pretorius, E.; Roberts, T.J.; Van der Hoek, S.A.; Kell, D.B. The Biology of Ergothioneine an antioxidant nutraceutical. Nutr. Res. Rev. 2020, 33, 190–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, G.W.; Doule, S.; Fitzpatrick, D.A. The evolutionary history of the genes involved in the biosynthesis of the antioxidant ergothioneine. Gene 2014, 549, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Salama, S.A.; Abd-Allah, G.M.; Mohamadin, A.M.; Elshafey, M.M.; Gad, H.S. Ergothioneine mitigates cisplation-evoked nephrotoxicity via targeting Nrf2, NF-KB, an apoptotic signaling and inhibiting γ-glutanyle transpeptidase. Life Sci. 2021, 278, 119572. [Google Scholar] [CrossRef] [PubMed]
- Hseu, Y.C.; Lo, H.W.; Lorivi, M.; Tsai, Y.C.; Tang, M.J.; Yang, H.L. Dermato-protective properties of Ergothioneine through induction of Nrf2/ARE-mediated antioxidant genes in UVA-irradiated human keratinocytes. Free Radic. Biomed. 2015, 86, 102–117. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.K.; Kim, J.; Ahn, M.; Kim, J.H.; Lee, G.S.; Shin, T. Ergothionine alleviates senescence of fibroblasts induced by UVB damage of keratinocytes via activation of the Nrf2/H 0-1 pathway and HSP70 in keratinocytes. Exp. Cell Res. 2021, 400, 112516. [Google Scholar]
- Borlingaus, J.; Albrecht, F.; Gruhike, M.C.; Nwachukwu, I.D.; Slusarenko, A.J. Allicin: Chemistry and biological properties. Molecules 2014, 19, 12591–12618. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.Y.; Yuen, A.C.; Chan, R.Y.; Chan, S.W. A review of the cardiovascular benefits and antioxidant properties of allicin. Phytother. Res. 2013, 27, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Rabinkov, A.; Miron, T.; Konstantinovski, L.; Wilhek, M.; Mirelman, D.; Weiner, L. The mode of action of allicin: Trapping of radicals and interaction with thiol containing proteins. Biochim. Biophys. Acta 1998, 1379, 233–244. [Google Scholar] [CrossRef]
- Metwally, D.M.; Al-Olayan, E.M.; Alanazi, M.; Alzahrany, S.B.; Semlali, A. Antischistosomal and anti-inflammatory activity of garlic and allicin compared with that of praziquantel in vivo. BMC Complement. Altern. Med. 2018, 18, 135. [Google Scholar] [CrossRef] [Green Version]
- Bat-Chen, W.; Golan, T.; Peri, I.; Ludmer, Z.; Schwartz, B. Allicin purified from fresh garlic cloves induces apoptosis in colon cancer cells via Nrf2. Nutr. Cancer 2010, 62, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Pan, H.; Xu, Y.; Wnag, X.; Qiu, Z.; Jiang, L. Allicin decrease Lipopolysaccharide-induced oxidative stress and inflammation in human umbilical vein endothelial cells through suppression of mitochondrial dysfunction and activation of Nrf2. Cell. Physiol. Biochem. 2017, 41, 2255–2267. [Google Scholar] [CrossRef]
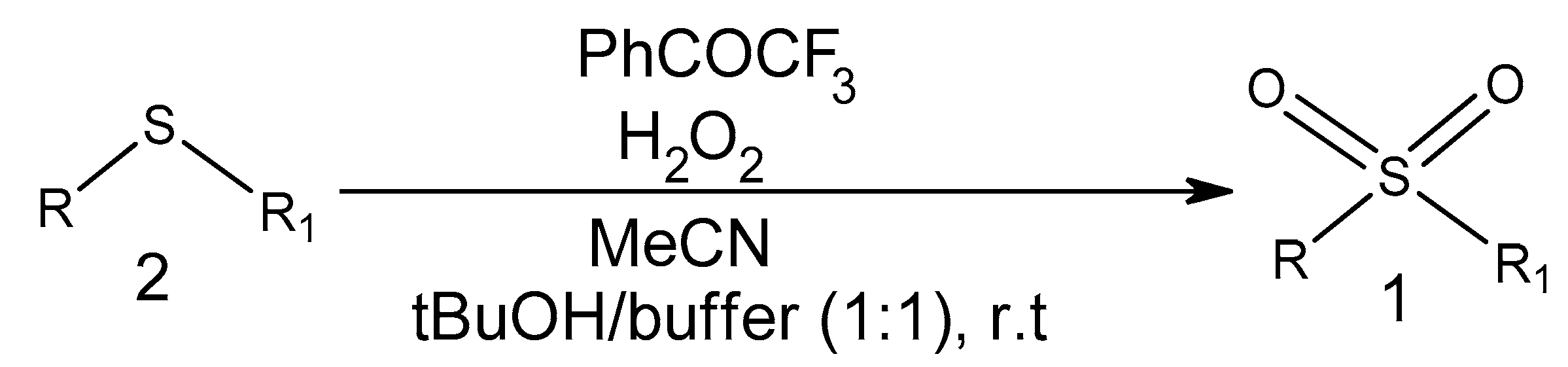

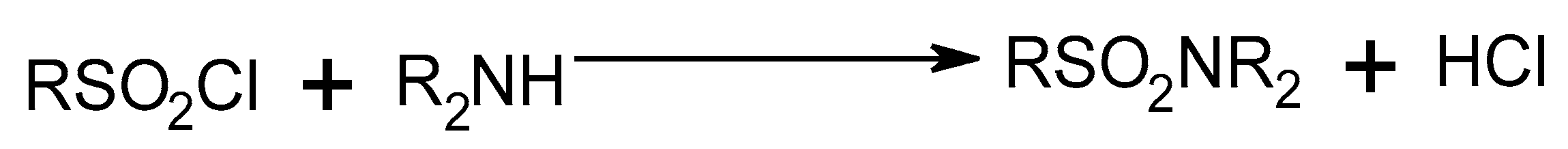


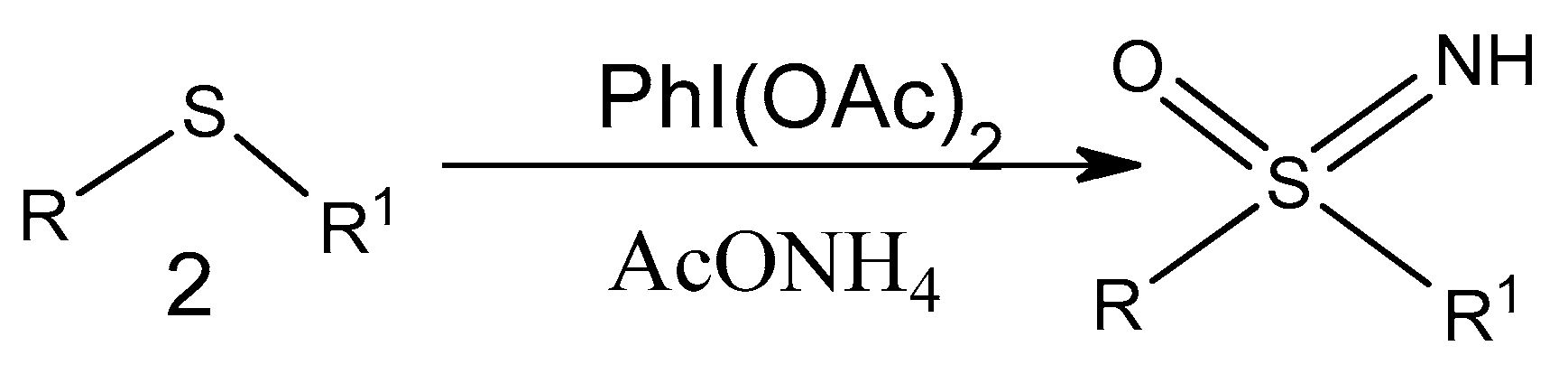
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Egbujor, M.C.; Petrosino, M.; Zuhra, K.; Saso, L. The Role of Organosulfur Compounds as Nrf2 Activators and Their Antioxidant Effects. Antioxidants 2022, 11, 1255. https://doi.org/10.3390/antiox11071255
Egbujor MC, Petrosino M, Zuhra K, Saso L. The Role of Organosulfur Compounds as Nrf2 Activators and Their Antioxidant Effects. Antioxidants. 2022; 11(7):1255. https://doi.org/10.3390/antiox11071255
Chicago/Turabian StyleEgbujor, Melford Chuka, Maria Petrosino, Karim Zuhra, and Luciano Saso. 2022. "The Role of Organosulfur Compounds as Nrf2 Activators and Their Antioxidant Effects" Antioxidants 11, no. 7: 1255. https://doi.org/10.3390/antiox11071255
APA StyleEgbujor, M. C., Petrosino, M., Zuhra, K., & Saso, L. (2022). The Role of Organosulfur Compounds as Nrf2 Activators and Their Antioxidant Effects. Antioxidants, 11(7), 1255. https://doi.org/10.3390/antiox11071255







