Molecular Mechanisms behind Safranal’s Toxicity to HepG2 Cells from Dual Omics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Growth
2.2. RNA Extraction and Sequencing
2.3. Metabolite Extraction
2.4. LC/MS-QToF
2.5. Computational, Quantitative, and Statistical Analyses
2.6. Differential Gene Expression Analysis
2.7. Pathway Analysis
3. Results
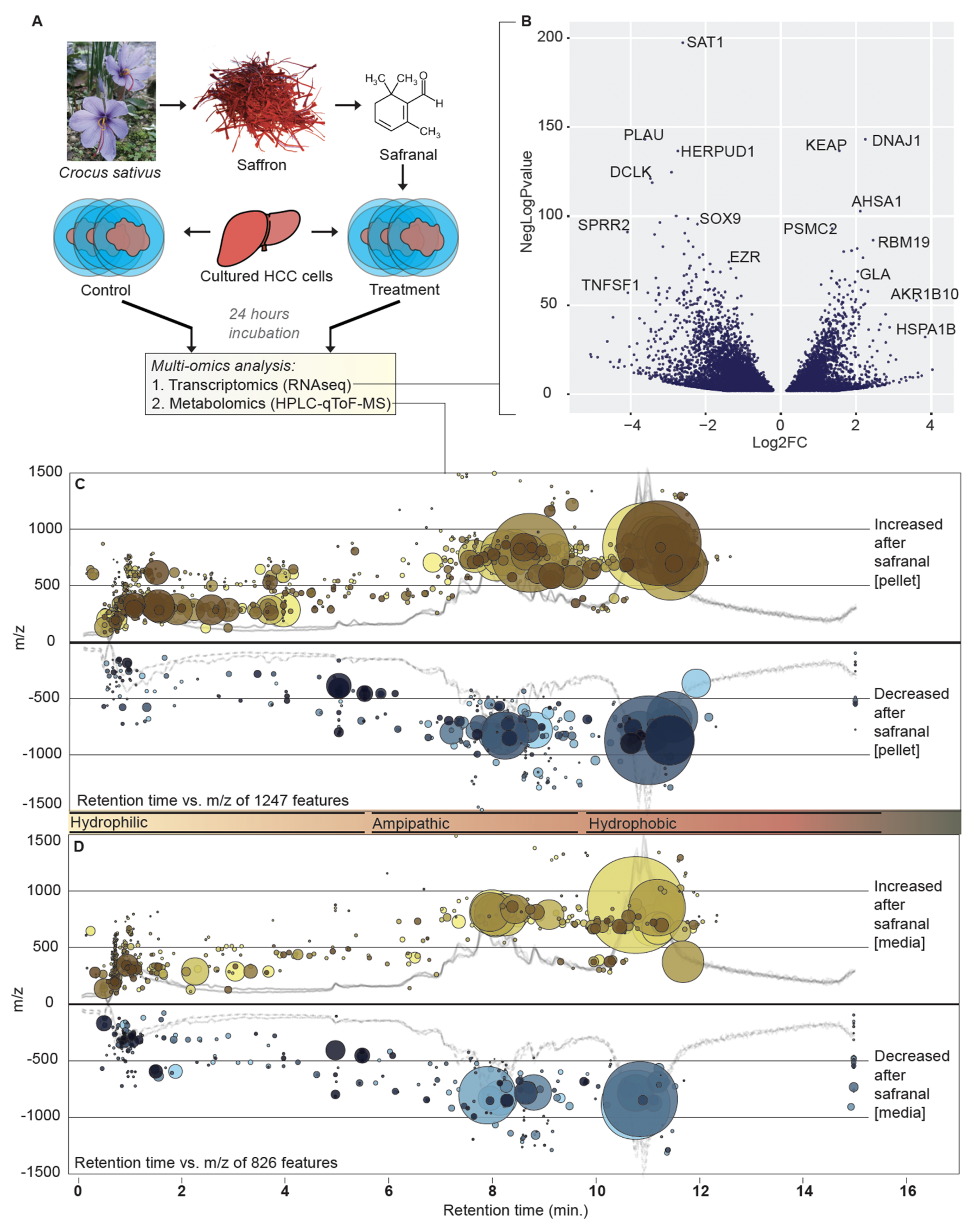
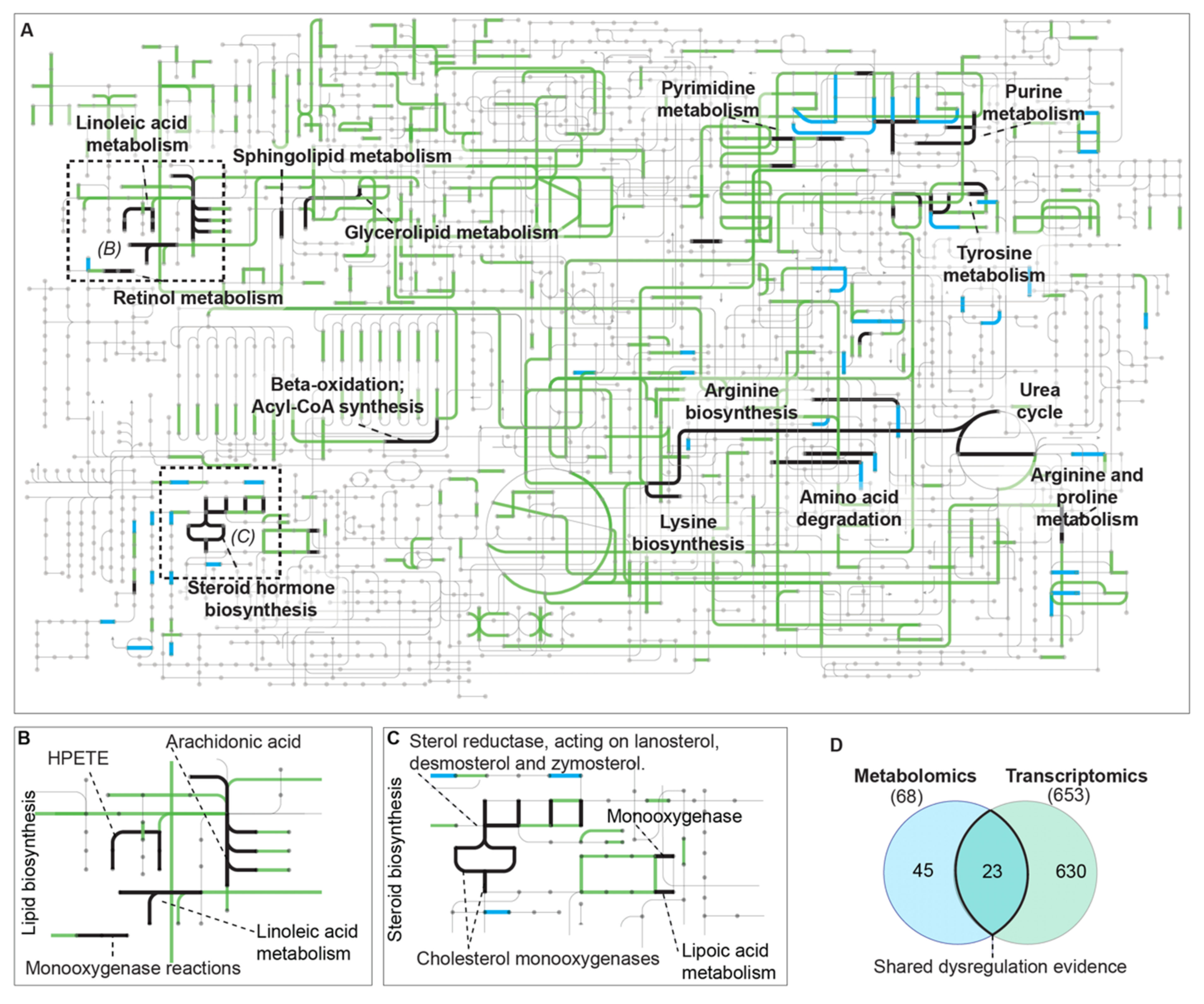
3.1. Dual Omics Systems Biology Resolves the HCC Safranal Response
3.2. Safranal Induces Shifts in the HCC Nucleoside Landscape
3.3. Safranal Induces ROS-Damage Biomarkers and Antioxidant Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jindal, A.; Thadi, A.; Shailubhai, K. Hepatocellular Carcinoma: Etiology and Current and Future Drugs. J. Clin. Exp. Hepatol. 2019, 9, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Zhu, R.X.; Seto, W.-K.; Lai, C.-L.; Yuen, M.-F. Epidemiology of Hepatocellular Carcinoma in the Asia-Pacific Region. Gut Liver 2016, 10, 332–339. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.C.; Chen, K.-F.; Chen, P.-J. Treatment of liver cancer. Cold Spring Harb. Perspect. Med. 2015, 5, a021535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, Z.; Bhandari, A.; Wang, Y.; Pan, Y.; Yang, F.; Chen, R.; Xia, E.; Wang, O. Dihydroartemisinin potentiates antitumor activity of 5-fluorouracil against a resistant colorectal cancer cell line. Biochem. Biophys. Res. Commun. 2018, 501, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Tarantilis, P.A.; Polissiou, M.; Manfait, M. Separation of picrocrocin, cis-trans-crocins and safranal of saffron using high-performance liquid chromatography with photodiode-array detection. J. Chromatogr. A 1994, 664, 55–61. [Google Scholar] [CrossRef]
- Hoshyar, R.; Mollaei, H. A comprehensive review on anticancer mechanisms of the main carotenoid of saffron, crocin. J. Pharm. Pharmacol. 2017, 69, 1419–1427. [Google Scholar] [CrossRef] [Green Version]
- Bathaie, S.Z.; Bolhassani, A.; Tamanoi, F. Anticancer Effect and Molecular Targets of Saffron Carotenoids. Enzymes 2014, 36, 57–86. [Google Scholar] [CrossRef] [PubMed]
- Bajbouj, K.; Schulze-Luehrmann, J.; Diermeier, S.; Amin, A.; Schneider-Stock, R. The anticancer effect of saffron in two p53 isogenic colorectal cancer cell lines. BMC Complement. Altern. Med. 2012, 12, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amin, A.; Hamza, A.A.; Bajbouj, K.; Ashraf, S.S.; Daoud, S. Saffron: A potential candidate for a novel anticancer drug against hepatocellular carcinoma. Hepatology 2011, 54, 857–867. [Google Scholar] [CrossRef]
- Samarghandian, S.; Boskabady, M.H.; Davoodi, S. Use of in vitro assays to assess the potential antiproliferative and cytotoxic effects of saffron (Crocus sativus L.) in human lung cancer cell line. Pharmacogn. Mag. 2010, 6, 309–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farahzad, J.A.; Samarghandian, S.; Shoshtari, M.E.; Sargolzaei, J.; Hossinimoghadam, H. Anti-tumor activity of safranal against neuroblastoma cells. Pharmacogn. Mag. 2014, 10 (Suppl. 2), S419–S424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Hrout, A.; Chaiboonchoe, A.; Khraiwesh, B.; Murali, C.; Baig, B.; El-Awady, R.; Tarazi, H.; Alzahmi, A.; Nelson, D.R.; Greish, Y.E.; et al. Safranal induces DNA double-strand breakage and ER-stress-mediated cell death in hepatocellular carcinoma cells. Sci. Rep. 2018, 8, 16951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, D.R.; Chaiboonchoe, A.; Fu, W.; Hazzouri, K.M.; Huang, Z.; Jaiswal, A.K.; Daakour, S.; Mystikou, A.; Arnoux, M.; Sultana, M.; et al. Potential for Heightened Sulfur-Metabolic Capacity in Coastal Subtropical Microalgae. iScience 2019, 11, 450–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castro-Perez, J.M.; Kamphorst, J.; DeGroot, J.; Lafeber, F.; Goshawk, J.; Yu, K.; Shockcor, J.P.; Vreeken, R.J.; Hankemeier, T. Comprehensive LC−MS E Lipidomic Analysis using a Shotgun Approach and Its Application to Biomarker Detection and Identification in Osteoarthritis Patients. J. Proteome Res. 2010, 9, 2377–2389. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing Mass Spectrometry Data for Metabolite Profiling Using Nonlinear Peak Alignment, Matching, and Identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Shi, Y.; Guo, T.; Wang, Y.; Cong, W.; Zhu, J. Metabolite discovery of helicidum in rat urine with XCMS based on the data of ultra performance liquid chromatography coupled to time-of-flight mass spectrometry. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2012, 907, 146–153. [Google Scholar] [CrossRef]
- Huan, T.; Forsberg, E.M.; Rinehart, D.; Johnson, C.H.; Ivanisevic, J.; Benton, H.P.; Fang, M.; Aisporna, A.; Hilmers, B.; Poole, F.L.; et al. Systems biology guided by XCMS Online metabolomics. Nat. Methods 2017, 14, 461–462. [Google Scholar] [CrossRef]
- Gowda, H.; Ivanisevic, J.; Johnson, C.H.; Kurczy, M.E.; Benton, H.P.; Rinehart, D.; Nguyen, T.; Ray, J.; Kuehl, J.; Arevalo, B.; et al. Interactive XCMS Online: Simplifying Advanced Metabolomic Data Processing and Subsequent Statistical Analyses. Anal. Chem. 2014, 86, 6931–6939. [Google Scholar] [CrossRef] [PubMed]
- Forsberg, E.M.; Huan, T.; Rinehart, D.; Benton, H.P.; Warth, B.; Hilmers, B.; Siuzdak, G. Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS Online. Nat. Protoc. 2018, 13, 633–651. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Alborzi, S.Z.; Devignes, M.-D.; Ritchie, D.W. ECDomainMiner: Discovering hidden associations between enzyme commission numbers and Pfam domains. BMC Bioinform. 2017, 18, 107. [Google Scholar] [CrossRef] [PubMed]
- Save, S.S.; Rachineni, K.; Hosur, R.V.; Choudhary, S. Natural compound safranal driven inhibition and dis-aggregation of α-synuclein fibrils. Int. J. Biol. Macromol. 2019, 141, 585–595. [Google Scholar] [CrossRef]
- Stewart, G.R.; Robertson, B.D.; Young, D.B. Analysis of the function of mycobacterial DnaJ proteins by overexpression and microarray profiling. Tuberculosis 2004, 84, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Jia, J.; Zhang, J.; Sethuraman, V.; Yi, X.; Zhong, G. DnaJ homolog subfamily A member1 (DnaJ1) is a newly discovered anti-apoptotic protein regulated by azadirachtin in Sf9 cells. BMC Genom. 2018, 19, 413. [Google Scholar] [CrossRef] [Green Version]
- Stark, J.L.; Mehla, K.; Chaika, N.; Acton, T.B.; Xiao, R.; Singh, P.K.; Montelione, G.T.; Powers, R. Structure and Function of Human DnaJ Homologue Subfamily A Member 1 (DNAJA1) and Its Relationship to Pancreatic Cancer. Biochemistry 2014, 53, 1360–1372. [Google Scholar] [CrossRef]
- Stark, J.; Mercier, K.A.; Mueller, G.; Acton, T.B.; Xiao, R.; Montelione, G.T.; Powers, R. Solution structure and function of YndB, an AHSA1 protein from Bacillus subtilis. Proteins 2010, 78, 3328–3340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheehan-Rooney, K.; Swartz, M.E.; Zhao, F.; Liu, D.; Eberhart, J.K. Ahsa1 and Hsp90 activity confers more severe craniofacial phenotypes in a zebrafish model of hypopar-athyroidism, sensorineural deafness and renal dysplasia (HDR). Dis. Models Mech. 2013, 6, 1285–1291. [Google Scholar]
- Shao, J.; Wang, L.; Zhong, C.; Qi, R.; Li, Y. AHSA1 regulates proliferation, apoptosis, migration, and invasion of osteosarcoma. Biomed. Pharmacother. 2016, 77, 45–51. [Google Scholar] [CrossRef]
- Woodford, M.R.; Sager, R.A.; Marris, E.; Dunn, D.M.; Blanden, A.R.; Murphy, R.L.; Rensing, N.; Shapiro, O.; Panaretou, B.; Prodromou, C.; et al. Tumor suppressor Tsc1 is a new Hsp90 co-chaperone that facilitates folding of kinase and non-kinase clients. EMBO J. 2017, 36, 3650–3665. [Google Scholar] [CrossRef] [PubMed]
- Cheriyamundath, S.; Choudhary, S.; Lopus, M. Safranal Inhibits HeLa Cell Viability by Perturbing the Reassembly Potential of Microtubules. Phytotherapy Res. 2018, 32, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Cadenas, S.; Aragonés, J.; Landázuri, M.O. Mitochondrial reprogramming through cardiac oxygen sensors in ischaemic heart disease. Cardiovasc. Res. 2010, 88, 219–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, J.; Han, D.; Sancheti, H.; Yap, L.-P.; Kaplowitz, N.; Cadenas, E. Regulation of Mitochondrial Glutathione Redox Status and Protein Glutathionylation by Respiratory Substrates. J. Biol. Chem. 2010, 285, 39646–39654. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Amini, A.; Ahmad, Z. Safranal and its analogs inhibit Escherichia coli ATP synthase and cell growth. Int. J. Biol. Macromol. 2017, 95, 145–152. [Google Scholar] [CrossRef] [Green Version]
- Pang, B.; McFaline, J.L.; Burgis, N.E.; Dong, M.; Taghizadeh, K.; Sullivan, M.R.; Elmquist, C.E.; Cunningham, R.P.; Dedon, P.C. Defects in purine nucleotide metabolism lead to substantial incorporation of xanthine and hypoxanthine into DNA and RNA. Proc. Natl. Acad. Sci. USA 2012, 109, 2319–2324. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.J.; Ryu, H.M.; Choi, J.Y.; Cho, J.H.; Kim, C.D.; Park, S.H.; Kim, Y.L. Hypoxanthine causes endothelial dysfunction through oxidative stress-induced apoptosis. Biochem. Biophys. Res. Commun. 2017, 482, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Clerici, S.; Boletta, A. Role of the KEAP1-NRF2 Axis in Renal Cell Carcinoma. Cancers 2020, 12, 3458. [Google Scholar] [CrossRef]
- Fakhri, S.; Pesce, M.; Patruno, A.; Moradi, S.Z.; Iranpanah, A.; Farzaei, M.H.; Sobarzo-Sánchez, E. Attenuation of Nrf2/Keap1/ARE in Alzheimer’s Disease by Plant Secondary Metabolites: A Mechanistic Review. Molecules 2020, 25, 4926. [Google Scholar] [CrossRef]
- Moretti, D.; Tambone, S.; Cerretani, M.; Fezzardi, P.; Missineo, A.; Sherman, L.T.; Munoz-Sajuan, I.; Harper, S.; Dominquez, C.; Pacifici, R.; et al. NRF2 activation by reversible KEAP1 binding induces the antioxidant response in primary neurons and astrocytes of a Huntington’s disease mouse model. Free Radic. Biol. Med. 2020, 162, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Thanas, C.; Ziros, P.G.; Chartoumpekis, D.V.; Renaud, C.O.; Sykiotis, G.P. The Keap1/Nrf2 Signaling Pathway in the Thyroid-2020 Update. Antioxidants 2020, 9, 1082. [Google Scholar] [CrossRef]
- Mandal, S.; Mandal, A.; Park, M.H. Depletion of the polyamines spermidine and spermine by overexpression of spermi-dine/spermine N(1)-acetyltransferase 1 (SAT1) leads to mitochondria-mediated apoptosis in mammalian cells. Biochem. J. 2015, 468, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, T.; Komoda, Y.; Nakajima, H. Biliverdin-IX alpha reductase and biliverdin-IX beta reductase from human liver. Purification and characterization. J. Biol. Chem. 1994, 269, 24343–24348. [Google Scholar] [CrossRef]
- Nikolaidis, A.; Kramer, R.; Ostojic, S. Nitric Oxide: The Missing Factor in COVID-19 Severity? Med. Sci. 2021, 10, 3. [Google Scholar] [CrossRef]
- Peluffo, R.D. Cationic amino acid transporters and their modulation by nitric oxide in cardiac muscle cells. Biophys. Rev. 2021, 13, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Karami, M.; Zarrindast, M.R. The regulatory role of nitric oxide in morphine-induced analgesia in the descending path of pain from the dorsal hippocampus to the dorsolateral periaqueductal gray. Eur. J. Pain 2022, 26, 888–901. [Google Scholar] [CrossRef] [PubMed]
- Farahmand, S.K.; Samini, F.; Samini, M.; Samarghandian, S. Safranal ameliorates antioxidant enzymes and suppresses lipid peroxidation and nitric oxide formation in aged male rat liver. Biogerontology 2013, 14, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Mardani, H.; Maninang, J.; Appiah, K.S.; Oikawa, Y.; Azizi, M.; Fujii, Y. Evaluation of Biological Response of Lettuce (Lactuca sativa L.) and Weeds to Safranal Allelochemical of Saffron (Crocus sativus) by Using Static Exposure Method. Molecules 2019, 24, 1788. [Google Scholar] [CrossRef] [Green Version]
- Barañano, D.E.; Rao, M.; Ferris, C.D.; Snyder, S.H. Biliverdin reductase: A major physiologic cytoprotectant. Proc. Natl. Acad. Sci. USA 2002, 99, 16093–16098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; de Montellano, P.R.O. The binding sites on human heme oxygenase-1 for cytochrome p450 reductase and biliverdin reductase. J. Biol. Chem. 2003, 278, 20069–20076. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Kang, H.T.; Choi, H.R.; Park, S.C. Biliverdin reductase A in the prevention of cellular senescence against oxidative stress. Exp. Mol. Med. 2011, 43, 15–23. [Google Scholar] [CrossRef] [Green Version]
- Kamatani, N.; Nelson-Rees, W.A.; Carson, D.A. Selective killing of human malignant cell lines deficient in methylthioadenosine phosphorylase, a purine metabolic enzyme. Proc. Natl. Acad. Sci. USA 1981, 78, 1219–1223. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, R.; Henkel, K. Über die Senkung der Körpertemperatur durch Adenylthiomethylpentose. Hoppe Seyler’s Z. Physiol. Chemie 1941, 269, 41–46. [Google Scholar] [CrossRef]
- Lindberg, J.E.; Jacobsson, K.-G. Nitrogen and purine metabolism at varying energy and protein supplies in sheep sustained on intragastric infusion. Br. J. Nutr. 1990, 64, 359–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, L.; Dreyer, J.H.; Hartmann, D.; Barros, M.H.M.; Büttner-Herold, M.; Grittner, U.; Niedobitek, G. Tumor-associated macrophages in classical Hodgkin lymphoma: Hormetic relationship to outcome. Sci. Rep. 2020, 10, 9410. [Google Scholar] [CrossRef] [PubMed]

| Metabolites | Pathways Involved | Fold- Change | p-Value | m/z | Retention Time | Adduct |
|---|---|---|---|---|---|---|
| α-linolenate | eicosapentaenoate biosynthesis II | 32.2 | 2.2 × 10−7 | 261.2201 | 1.4 | M-H2O[1+] |
| α-linolenate | eicosapentaenoate biosynthesis II | 42.5 | 2 × 10−6 | 279.2305 | 1.39 | M+H[1+] |
| HPETE | leukotriene biosynthesis | 2.5 | 6 × 10−6 | 169.1215 | 0.85 | M+2H[2+] |
| S-3-methyl-2- oxopentanoate | isoleucine degradation | 11.1 | 4.4 × 10−7 | 113.0591 | 0.75 | M-H2O[1+] |
| 2-trans-hexadecenal | sphingosine metabolism | 15.8 | 1.2 × 10−5 | 261.2201 | 0.87 | M+Na[1+] |
| 2-trans-hexadecenal | sphingosine metabolism | 32.2 | 2.2 × 10−7 | 261.2201 | 1.4 | M+Na[1+] |
| 3-ureido-isobutyrate | Thymine degradation | 18.4 | 3.6 × 10−5 | 169.0579 | 1.06 | M+Na[1+] |
| 4-hydroxy-2-nonenal-glutathione conjugate | 4-hydroxy-2-nonenal detoxification | 15 | 1.2 × 10−6 | 465.2138 | 5.09 | M+H[1+] |
| glutathione disulfide | glutaredoxin ascorbate recycling | 236.6 | 1.6 × 10−5 | 308.0897 | 0.52 | M+2H[2+] |
| hypoxanthine | purine degradation | 42.3 | 7.7 × 10−5 | 119.0346 | 0.52 | M-H2O[1+] |
| hypoxanthine | purine degradation | 583.7 | 7.7 × 10−6 | 137.0451 | 0.53 | M+H[1+] |
| laurate | palmitate biosynthesis | 4.9 | 3 × 10−5 | 223.1667 | 0.79 | M+Na[1+] |
| methylhistamine | histamine degradation | 3.9 | 3.4 × 10−7 | 144.1374 | 0.77 | M+NH3[1+] |
| N-acetylputrescine | putrescine degradation III | 3 | 1.8 × 10−6 | 170.081 | 0.73 | M+K[1+] |
| oleate | oleate biosynthesis | 2 | 5.5 × 10−5 | 300.2884 | 3.19 | M+NH3[1+] |
| oleate | oleate biosynthesis | 10.2 | 4 × 10−5 | 153.1264 | 1.37 | M+H+Na |
| palmitate | aldehydesphingosine andsphingosine-1-phosphate metabolism | 43.8 | 8 × 10−7 | 263.2354 | 1.34 | M+Na[1+] |
| palmitate | palmitate biosynthesis-stearate biosynthesis | 42.5 | 2 × 10−6 | 279.2305 | 1.39 | M+Na[1+] |
| pyridoxine | pyridoxal 5′-phosphate salvage | 3 | 1.8 × 10−6 | 170.081 | 0.73 | M+H[1+] |
| S-methyl-5′- thioadenosine | wyosine biosynthesis | 43.4 | 7.2 × 10−5 | 298.0963 | 0.54 | M+H[1+] |
| urocanate | histidine degradation | 3.7 | 2 × 10−7 | 156.076 | 0.5 | M+NH3+ |
| Metabolites | Pathways Involved | Fold- Change | p-Value | m/z | Retention Time | Adduct |
|---|---|---|---|---|---|---|
| 1-pyrroline-3-hydroxy-5- carboxylate | 4-hydroxyproline degradation | –3.7 | 1.3 × 10−5 | 130.0493 | 0.5 | M+H[1+] |
| S-dihydroorotate | UMP biosynthesis | –20.5 | 1.8 × 10−5 | 176.066 | 1.21 | M+NH3[1+] |
| 3-4-dihydroxy-5-all-trans- decaprenylbenzoate | ubiquinol-10 biosynthesis | –3 | 1.9 × 10−5 | 835.659 | 8.84 | M+H[1+] |
| 3-ureido-isobutyrate | thymine degradation | –3.7 | 1.3 × 10−5 | 130.0493 | 0.5 | M-NH3[1+] |
| 5S hydro peroxy-18R-hydroxy-eicosapentaenoate | resolvin E biosynthesis | –138.8 | 4 × 10−5 | 333.2044 | 1.57 | M-H2O[1+] |
| all-trans-retinoate | retinoate biosynthesis | –4.2 | 2.5 × 10−8 | 283.2043 | 2.22 | M-H2O[1+] |
| all-trans-retinoate | retinoate biosynthesis | –33.9 | 1.2 × 10−5 | 323.1968 | 1.11 | M+Na[1+] |
| all-trans-retinoate | retinoate biosynthesis | –141.3 | 4.2 × 10−6 | 323.1966 | 1.53 | M+Na[1+] |
| biliverdin-IX- | α-heme degradation | –3281.8 | 5.3 × 10−6 | 600.2797 | 1.28 | M+NH3[1+] |
| codeine | morphine biosynthesis | –3.7 | 8.2 × 10−5 | 318.1929 | 0.84 | M+NH3[1+] |
| D-sorbitol | sorbitol degradation I | –5 | 3.1 × 10−5 | 205.0675 | 1.16 | M+Na[1+] |
| N-formylkynurenine | tryptophan degradation | –4.3 | 5 × 10−5 | 255.1223 | 0.86 | M+NH3[1+] |
| resolvin E1 | resolvin E biosynthesis | –138.8 | 4 × 10−5 | 333.2044 | 1.57 | M-H2O[1+] |
| resolvin E2 | resolvin E biosynthesis | –8.8 | 2.6 × 10−5 | 317.2101 | 0.88 | M-H2O[1+] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nelson, D.R.; Hrout, A.A.; Alzahmi, A.S.; Chaiboonchoe, A.; Amin, A.; Salehi-Ashtiani, K. Molecular Mechanisms behind Safranal’s Toxicity to HepG2 Cells from Dual Omics. Antioxidants 2022, 11, 1125. https://doi.org/10.3390/antiox11061125
Nelson DR, Hrout AA, Alzahmi AS, Chaiboonchoe A, Amin A, Salehi-Ashtiani K. Molecular Mechanisms behind Safranal’s Toxicity to HepG2 Cells from Dual Omics. Antioxidants. 2022; 11(6):1125. https://doi.org/10.3390/antiox11061125
Chicago/Turabian StyleNelson, David Roy, Ala’a Al Hrout, Amnah Salem Alzahmi, Amphun Chaiboonchoe, Amr Amin, and Kourosh Salehi-Ashtiani. 2022. "Molecular Mechanisms behind Safranal’s Toxicity to HepG2 Cells from Dual Omics" Antioxidants 11, no. 6: 1125. https://doi.org/10.3390/antiox11061125
APA StyleNelson, D. R., Hrout, A. A., Alzahmi, A. S., Chaiboonchoe, A., Amin, A., & Salehi-Ashtiani, K. (2022). Molecular Mechanisms behind Safranal’s Toxicity to HepG2 Cells from Dual Omics. Antioxidants, 11(6), 1125. https://doi.org/10.3390/antiox11061125






