Abstract
Recent studies in mice and humans demonstrated the relevance of H2S synthesising enzymes, such as CTH, CBS, and MPST, in the physiology of adipose tissue and the differentiation of preadipocyte into adipocytes. Here, our objective was to investigate the combined role of CTH, CBS, and MPST in the preservation of adipocyte protein persulfidation and adipogenesis. Combined partial CTH, CBS, and MPST gene knockdown was achieved treating fully human adipocytes with siRNAs against these transcripts (siRNA_MIX). Adipocyte protein persulfidation was analyzed using label-free quantitative mass spectrometry coupled with a dimedone-switch method for protein labeling and purification. Proteomic analysis quantified 216 proteins with statistically different levels of persulfidation in KD cells compared to control adipocytes. In fully differentiated adipocytes, CBS and MPST mRNA and protein levels were abundant, while CTH expression was very low. It is noteworthy that siRNA_MIX administration resulted in a significant decrease in CBS and MPST expression, without impacting on CTH. The combined partial knockdown of the CBS and MPST genes resulted in reduced cellular sulfide levels in parallel to decreased expression of relevant genes for adipocyte biology, including adipogenesis, mitochondrial biogenesis, and lipogenesis, but increased proinflammatory- and senescence-related genes. It should be noted that the combined partial knockdown of CBS and MPST genes also led to a significant disruption in the persulfidation pattern of the adipocyte proteins. Although among the less persulfidated proteins, we identified several relevant proteins for adipocyte adipogenesis and function, among the most persulfidated, key mediators of adipocyte inflammation and dysfunction as well as some proteins that might play a positive role in adipogenesis were found. In conclusion, the current study indicates that the combined partial elimination of CBS and MPST (but not CTH) in adipocytes affects the expression of genes related to the maintenance of adipocyte function and promotes inflammation, possibly by altering the pattern of protein persulfidation in these cells, suggesting that these enzymes were required for the functional maintenance of adipocytes.
1. Introduction
Functional adipocytes are characterized by an increased capacity to store excess fuel and produce beneficial adipokines, increased expression of adipogenic genes, but decreased expression of proinflammatory- and cellular senescence-related genes [1,2,3,4]. Optimal adipocyte function is required to maintain adipose tissue physiology and prevents obesity-associated metabolic disturbances [3,4,5,6].
Recent studies in mice and humans demonstrated the relevance of H2S-synthesising enzymes, such as CTH, CBS, and MPST, to preserve adipogenesis of adipose tissue, healthy fat mass expansion and insulin action [7,8]. These enzymes play a crucial role in the differentiation of adipocytes. Specifically, gene knockdown experiments in mouse 3T3-L1 cells and human preadipocytes demonstrated the relevance of CTH, CBS and MPST in adipogenesis [7,8,9]. Interestingly, increased persulfidation in relevant adipogenic proteins has also been reported in human adipocytes [8]. Since persulfidation often increases the reactivity of target proteins [10], increased persulfidation in adipogenic enzymes suggests a possible role of these post-translational modifications in the promotion of adipocyte differentiation and in the maintenance of adipogenic status. Despite these intriguing studies, to the best of our knowledge, the combined role of these enzymes in the maintenance of adipocyte function has not been yet examined.
In the present study, our objective was to investigate the combined role of CTH, CBS, and MPST in the preservation of adipocyte adipogenesis and function and to investigate how the combined partial knockdown of these enzymes might impact the persulfidation of the adipocyte proteins.
2. Materials and Methods
2.1. Human Preadipocyte Differentiation and Adipocyte Maintenance
Primary human subcutaneous preadipocytes from a non-diabetic Caucasian female with BMI < 30 kg/m2 (Zen-Bio Inc., Research Triangle Park, Durham, NC, USA) were cultured with Preadipocyte Medium (PM-1, Zen-Bio Inc.) in a humidified incubator at 37 °C with 5% CO2. Twenty-four hours after plating, cells were checked for confluence. Once reached, differentiation started with commercially available Differentiation Medium (DM-2, Zen-Bio Inc.) and Maintenance Medium (AM-1 Zen-Bio Inc.), following the manufacturer’s instructions. Two weeks after initiating differentiation, differentiated cells appeared rounded with large lipid droplets apparent in the cytoplasm. The cells were then considered mature adipocytes. Forward siRNA transfections using siRNA_MIX, which included siRNAs against MPST, CBS, and CTH gene expression, were performed at the end of the differentiation process.
2.2. siRNA-Induced Gene Knockdown in Fully Differentiated Adipocytes
Primary human subcutaneous adipocytes were forward transfected with siRNAs at the end of differentiation (on day 11). Briefly, siRNA (Sigma-Aldrich, St. Louis, MO, USA) against CTH (NM_001190463), CBS (NM_000071), and MPST (NM_001130517) and Lipofectamine RNAiMAX (Life Technologies, Darmstadt, Germany) were diluted separately with Opti-MEM I Reduced Serum Medium (Life Technologies, Darmstadt, Germany) and mixed by pipetting afterwards. The siRNA-RNAiMAX complexes were left to incubate for 20 min at room temperature and then added to the top of the adherent cells drop-wise. The final concentrations of Lipofectamine RNAiMAX and siRNAs were 1.6 μL/cm2 and 100 nM, respectively, in 12-well cell culture plates, and the final amount of medium per well was 1 mL. The transfection conditions included combined silencing of CTH, CBS and MSPT (100 nM), and a non-targeting siRNA (100 nM) for the vehicle. Adipocytes were harvested on day 14 of differentiation, i.e., 72 h after transfection without changing cell culture medium. Transfection efficiency was assessed by real-time PCR and Western blot. The MISSION® siRNAs (Sigma-Aldrich) used were MISSION® esiRNA targeting human MPST (EHU086511), CBS (EHU099041), and CTH (EHU078251). The MISSION® siRNA Universal Negative Control #1 (Sigma-Aldrich, SIC001) was used as a control in all experiments (siRNA_SCR).
2.3. Gene Expression
RNA was extracted from cells using the RNeasy Lipid Tissue Mini Kit (Qiagen, Germantown, MD, USA). Total RNA was quantified using a spectrophotometer (GeneQuant; GE Health Care, Waukesha, WI, USA). RNA was reverse transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Life Technology, Darmstadt, Germany) according to standard procedures. Commercially available Taq-Man primer and probe sets were used for gene expression. Expression was assessed by real-time PCR using the LightCycler 480 Real-Time PCR System (Roche Diagnostics, Barcelona, Spain).
The commercially available and pre-validated TaqMan® primer/probe sets used were as follows: Peptidylprolyl isomerase A (cyclophilin A) (4333763, PPIA as endogenous control), cystathionine γ-lyase (CTH, Hs00542284_m1), cystathionine β-synthase (CBS, Hs00163925_m1), mercaptopyruvate sulfurtransferase (MPST, Hs00560401_m1), adiponectin (ADIPOQ, Hs00605917_m1), fatty acid binding protein 4, adipocyte (FABP4, Hs01086177_m1), peroxisome proliferator-activated receptor gamma (PPARG, Hs00234592_m1), perilipin 1 (PLIN1, Hs00160173_m1), PPARG coactivator 1 alpha (PPARGC1A, Hs00173304_m1), fatty acid synthase (FASN, Hs00188012_m1), acetyl-CoA carboxylase alpha (ACACA, Hs00167385_m1), solute carrier family 2 member 4 (SLC2A4 or GLUT4, Hs00168966_m1), interleukin 6 (interferon, beta 2) (IL6, Hs00985639_m1), tumor necrosis factor (TNF, Hs00174128_m1), C-X-C motif chemokine ligand 8 (CXCL8 or IL8, Hs00174103_m1), C–C motif chemokine ligand 2 (CCL2 or MCP1, Hs00234140_m1), and tumor protein p53 (TP53, Hs01034249_m1).
2.4. Western Blot
The proteins were extracted from cells using radioimmunoprecipitation assay (RIPA) buffer (0.1% SDS, 0.5% sodium deoxy-cholate, 1% Nonidet P-40, 150 mmol/L NaCl, and 50 mmol/L Tris-HCl, pH 8.0) supplemented with a commercially available cocktail of protease inhibitors (Millipore, Sigma, Madrid, Spain). Cellular debris and lipids were eliminated by centrifugation of the solubilized samples at 12,000× g for 10 min at 4 °C, recovering the soluble fraction. The protein concentration was determined using the RC/DC Protein Assay (Bio-Rad Laboratories, Hercules, CA, USA). Protein extracts were processed in SDS-PAGE and transferred to nitrocellulose membranes by conventional procedures. After blocking with 5% BSA in TBS-Tween, the membranes were incubated with primary antibodies, followed by incubation with horseradish peroxidase conjugated goat anti-rabbit antibody (Cell Signaling, 7074). Protein signal was detected by chemiluminescence ChemiDoc™ Gel Imaging System (Bio-Rad, California, USA). Actin was used as a housekeeping gene-coded protein. All primary antibodies were used at 1:1000 dilutions and were the following: CBS (sc-133154, Santa Cruz Biotechnology, Santa Cruz, CA, USA), CTH (sc-365382, Santa Cruz Biotechnology), MPST (sc-376168, Santa Cruz Biotechnology), FAS (C20G5, Cell Signaling), and β-actin (sc-47778, Santa Cruz Biotechnology).
2.5. Quantification of Hydrogen Sulfide in Biological Matrices by LC-MS/MS
Hydrogen sulfide was quantified following a previously described method [11]. A 50 μL sample (cell lysate) was spiked with 70 μL of MBB solution (20 mM). The mixture was vortexed, incubated for 60 min at room temperature, and the derivatization reaction stopped by adding 10 μL 20% formic acid. The mixture was subjected to centrifugation at 12,000 × rpm for 10 min. The derivative sulfide dibimane was analyzed by LC-MS/MS using the ExionLC™ UPLC system (Sciex, Framingham, MA, USA) with a reversed-phase column (100 mm × 4.6 mm × 100 Å particle, Kinetex XB-C18, Phenomenex, Torrance, CA, USA). The mobile phases were 0.1% formic acid in H2O (Buffer A, HPLC/LCMS grade) and 0.1% formic acid in acetonitrile (Buffer B, HPLC/LC-MS grade). A 5 µL sample was loaded per injection, and the gradient, applied at a flow rate of 600 µL min−1, was as follows: 1.7 min 80% A, from 1.7 to 3 min linear gradient from 80% A to 20% A, 1 min isocratic 20% A, 1 min linear gradient from 20% A to 80% A, and hold 3 min at 80% A to re-equilibrate the column. Mass spectra were acquired using a QTRAP 6500 + triple quadrupole (Sciex, Framingham, MA, USA) equipped with an electrospray ionization source operating in negative ionization mode using an ion spray voltage of −4500 V. The other ESI parameters were as follows: curtain gas, 35 psi; collision gas, medium; temperature, 500 °C; nebulizer gas (GS1), 60 psi; and heater gas (GS2), 60 psi. Data were acquired with Analyst® 1.7 software in the multiple reaction monitoring (MRM) mode with a detection window of 60 s. The measured ionization adducts [M-H]− selected for identification and quantification were 412.9 m/z (Q1) and 190.9 m/z (Q3) and optimized declustering potential (DP − 45 V) and collision energy (CE − 24 V). Data were processed with Sciex OS® software (Sciex, Redwood City, CA, USA) for peak integration and quantification.
2.6. Protein Persulfidation
A total of 500 µg of proteins from adipocyte lysates in 50 mM Tris-HCl, pH 8, supplemented with 1% protease inhibitor (cOmplete™, SigmaAldrich) and 2% SDS was incubated with 5 mM 4-chloro-7-nitrobenzofurazan (Cl-NBF) at 37 °C for 30 min, protected from light. A methanol/chloroform precipitation was performed to eliminate excess Cl-NBF, and protein pellets obtained were washed with cold methanol, dried, and dissolved in 50 mM Tris-HCl, pH 8, with 2% SDS, as previously described [12,13]. The proteins were then incubated with 100 µM DCP-Bio1 at 37 °C for 1.5 h. Afterwards, proteins were precipitated with methanol/chloroform and dissolved in 50 mM Tris-HCl, pH 8, supplemented with 0.1% SDS. Proteins were incubated with Sera-Mag™ Magnetic Streptavidin (Cytiva, Madrid, Spain) at 4 °C overnight with agitation and then, beads were washed with 8 column volumes of 50 mM Tris-HCl, pH 8, with 0.001% Tween, 2 column volumes of 50 mM Tris-HCl, pH 8, and 1 column volume of pure HPLC-quality water. After washing, beads were incubated 5 h with agitation with 2.25 M ammonium hydroxide at RT. The sample was then neutralized with formic acid and the protein concentration was determined. A total of 50 µg of proteins were trypsinized (GOLD, Promega) following the filter-aided sample preparation (FASP) method [14]. Trypsin was added to a trypsin:protein ratio of 1:20, and the mixture was incubated overnight at 37 °C, dried out in a RVC2 25 speedvac concentrator (Christ), and resuspended in 0.1% FA. Peptides were desalted and resuspended in 0.1% FA using C18 stage tips (Millipore). The samples were analyzed in a timsTOF Pro with PASEF (Bruker Daltonics, Billerica, MA, USA) coupled online to an Evosep ONE liquid chromatograph (Evosep). Hence, 200 ng were directly loaded onto the Evosep ONE and resolved using the 30 samples-per-day protocol. The processed data were analyzed with the MQ (Max Quant) search engine and the label-free quantification was performed using PEAKS Studio (BSI, Mississauga, Canada) [15]. Protein identification and quantification were carried out using PEAKS X software (Bioinformatics solutions). The searches were carried out against a database consisting of Homo sapiens (Uniprot/Swissprot), with precursor and fragment tolerances of 20 ppm and 0.05 Da. Only proteins identified with at least two peptides at FDR < 1% were considered for further analysis. Protein abundances inferred from PEAKS were loaded onto Perseus computational platform [16], log2 transformed, and imputated. A t-test was used to address significant differences in protein abundances within each sample group under analysis.
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium [17] via the PRIDE partner repository with the dataset identifier PXD032370 and 10.6019/PXD032370.
Functional protein association networks were explored using the STRING database (STRING: functional protein association networks (string-db.org) access date 26 February 2022).
2.7. Statistical Analysis
Statistical analyses were performed using SPSS 12.0 software. The unpaired t-test and nonparametric test (Mann–Whitney test) were used to analyze in vitro experimental data. The levels of statistical significance were set at p < 0.05.
3. Results and Discussion
3.1. The Relevance of CTH, CBS and MPST on Human Adipocytes
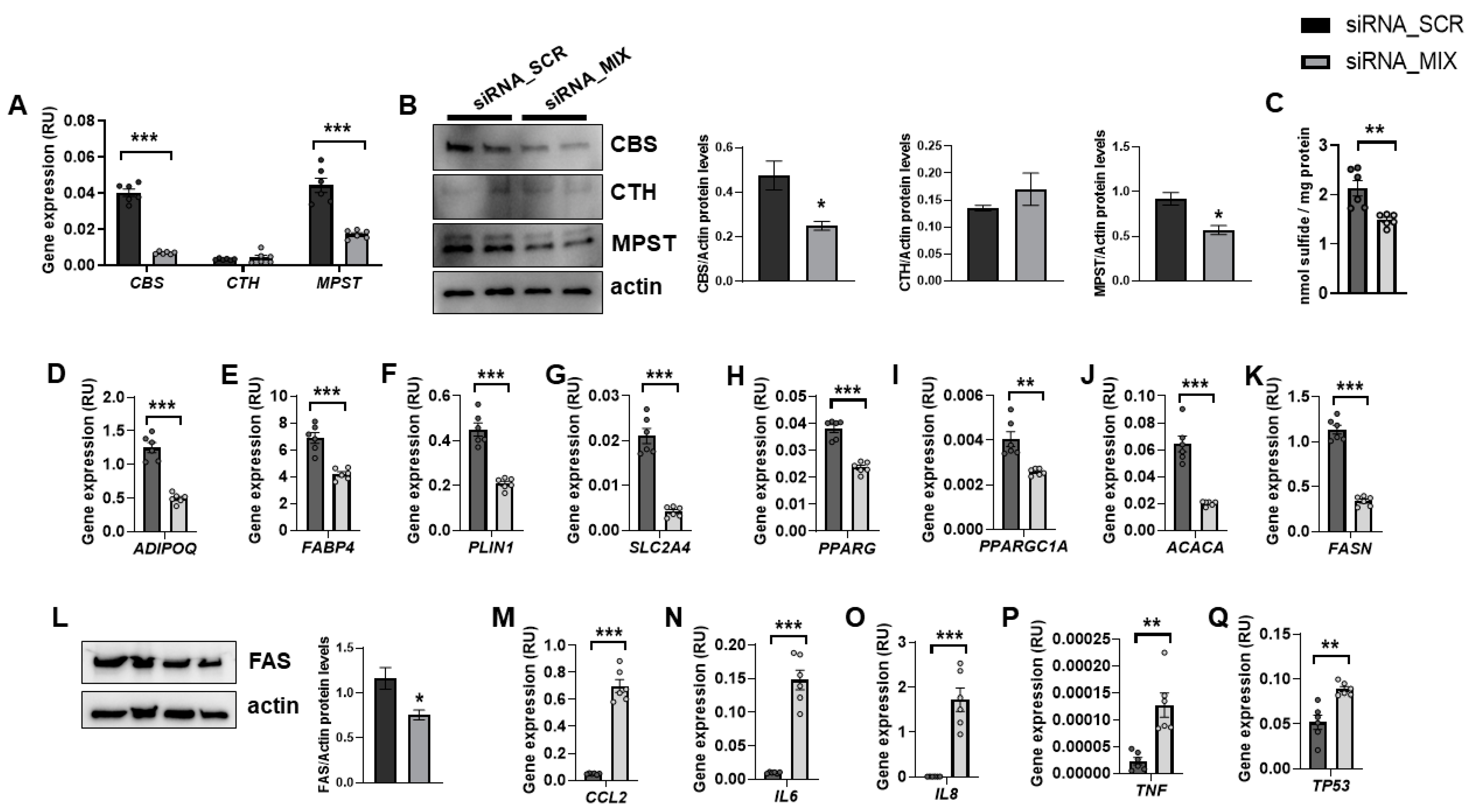
In fully differentiated adipocytes, CBS and MPST mRNA and protein levels were abundant, whereas CTH expression was very low (Figure 1A,B). It should be noted that siRNA_MIX administration resulted in a significant decrease in CBS and MPST expression, without impacting on CTH (Figure 1A,B). Possibly, the very low transcription and turnover of CTH make it resistant to the effect of siRNA. The combined partial knockdown of CBS and MPST genes led to a significant reduction in cellular sulfide levels (Figure 1C), indicating that H2S biosynthesis was attenuated in these cells. Interestingly, the combined partial CBS and MPST gene knockdown also resulted in decreased expression of relevant genes for adipocyte biology, including adipogenesis (ADIPOQ, FABP4, PLIN1, SLC2A4, PPARG), mitochondrial biogenesis (PPARGC1A), and lipogenesis (ACACA and FASN mRNA, and FAS protein) (Figure 1D–L), but increased proinflammatory- and senescence-related genes (TP53) (Figure 1M–Q). Importantly, these data indicated that MPST and CBS were not only required during adipocyte differentiation of mouse 3T3-L1 cells [9] or human preadipocytes [8], but these proteins were also necessary for the functional maintenance of adipocytes. In contrast to that observed during adipocyte differentiation or in mouse 3T3-L1 adipocytes [7,18,19], in fully differentiated human adipocytes, the expression of CTH was much lower than CBS and MPST.
Figure 1.
The impact of the combined partial knockdown of CBS and MPST gene on relevant genes for adipocyte biology. The effect of siRNA_MIX administration on CBS, CTH and MPST mRNA (A) and protein (B) levels, on intracellular sulfide levels (C), and on expression of adipogenic (ADIPOQ, FABP4, PLIN1, SLC2A4, PPARG, PPARGC1A)-, lipogenic (ACACA, FASN)-, proinflammatory (CCL2, IL6, IL8, TNF)- and cellular senescence (TP53)-related genes and on FAS protein levels (D–Q). * p < 0.05, ** p < 0.01 and *** p < 0.001 compared to siRNA_SCR.
No visual differences in lipid droplets were observed between control and combined partial knockdown cells (data not shown). Considering that adipocyte dedifferentiation becomes morphologically evident at least one week after the start of the process [20], longer gene knockdown experiments should be required to appreciate significant changes in intracellular lipid accumulation.
3.2. Impact of the Combined Partial Knockdown of MPST and CBS Genes on Adipocyte Protein Persulfidation
The possible link between CTH, CBS, and MPST activities and protein persulfidation during human adipocyte differentiation [8] suggests that these enzymes might modulate persulfidation in several adipogenic proteins, affecting the physiology of adipocytes. To test this hypothesis, we examined whether the reduction of adipogenic markers observed in human adipocytes with the combined partial knockdown of MPST and CBS genes was linked to altered persulfidation in key adipogenic proteins. For this purpose, we used a mass spectrometry label-free quantitative proteomic approach combined with the dimedone-switch method to measure the protein profile of persulfidation. Proteins from three biological replicates from fully differentiated human adipocytes, control, and the combined partial MPST and CBS gene knockdown were isolated and subjected to the dimedone-switch procedure for isolation of the persulfidated proteins. The proteomic analysis quantified 1834 proteins with a FDR threshold of 1% (Supplementary Dataset S1) of which 216 showed statistically different levels of persulfidation in KD cells compared to control adipocytes (Student t-test, p < 0.05) (Supplementary Dataset S2). From these proteins, 136 proteins were less persulfidated in KD cells, and 80 proteins were more persulfidated.
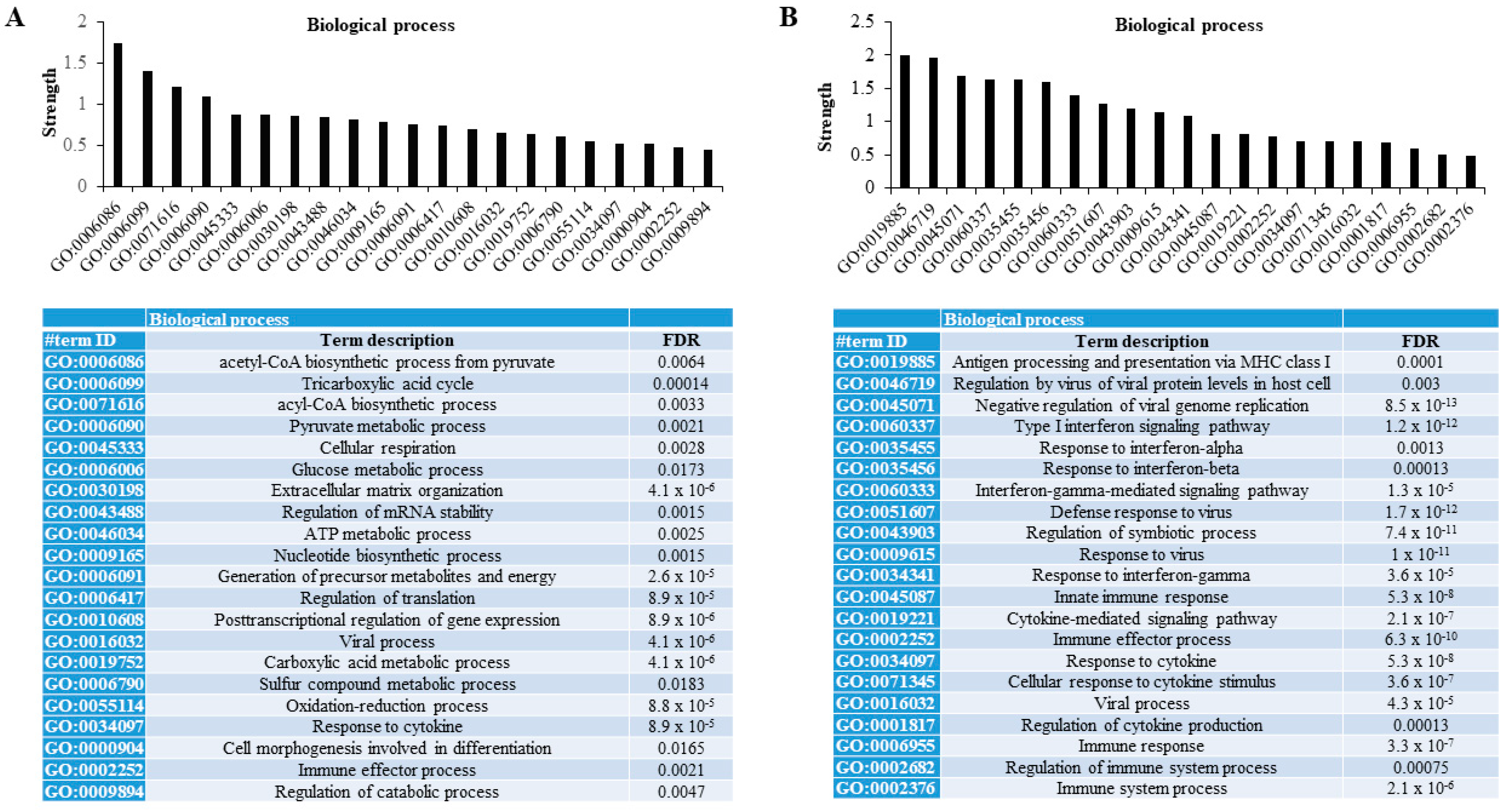
When functional protein association networks were explored using the STRING database, we found that those differentially less persulfidated proteins in human adipocytes with the combined partial knockdown of MPST and CBS genes were associated with relevant metabolic processes for adipocytes. These processes included the biosynthetic process of acetyl-CoA and acyl-CoA, tricarboxylic acid cycle, the metabolic process of pyruvate and glucose, extracellular matrix organization, the regulation of the mRNA stability, metabolic process of sulfur compound, the oxidation-reduction process, and the regulation of catabolic process (FDR < 0.02, Figure 2A). Otherwise, differentially increased persulfidated proteins in human adipocytes with the combined partial knockdown of MPST and CBS genes were mainly associated with processes related to immune response- and cytokine-mediated signaling pathways (FDR < 0.005, Figure 2B).
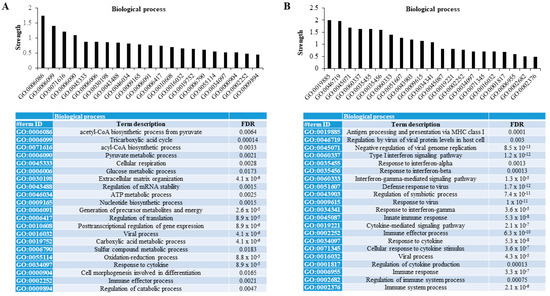
Figure 2.
Proteomic persulfidation networks analysis. Analysis of functional protein association networks using STRING database of differentially decreased (A) and increased (B) persulfidated proteins in siRNA_MIX- compared to siRNA_SCR-treated cells. FDR, false discovery rate adjusted p-value.
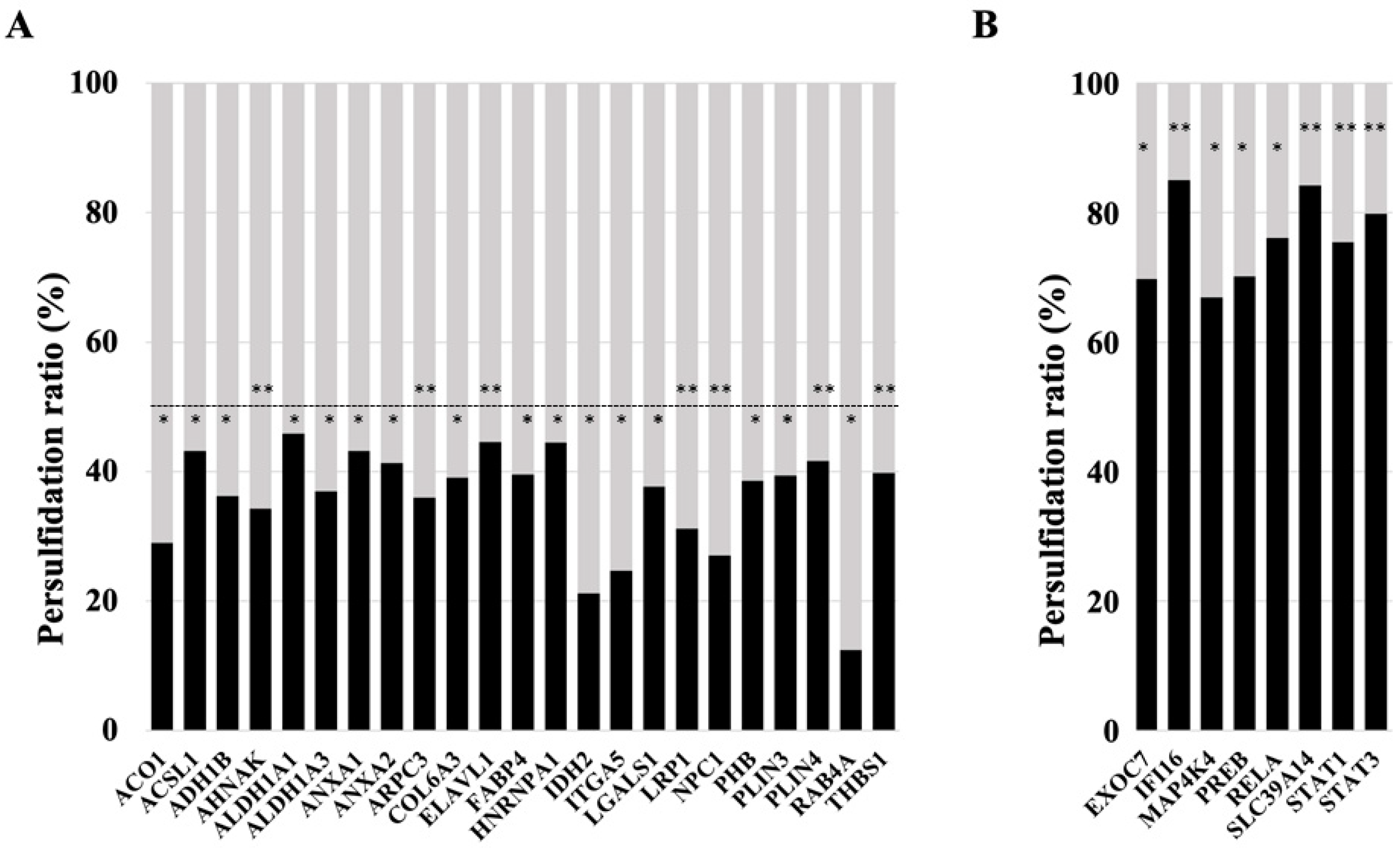
Specifically, among those proteins found less persulfidated, several relevant proteins for the maintenance of adipocyte adipogenesis and function were identified, including RAB4A, IDH2, ITGA5, NPC1, ACO1, LRP1, AHNAK, ARPC3, ADH1B, ALDH1A3, LGALS1, PHB, COL6A3, PLIN3, FABP4, THBS1, ANXA2, PLIN4, ACSL1, ANXA1, ELAVL1, ALDH1A1, and HNRNPA1 (Figure 3A, all with p < 0.05). Previous studies supported the relevance of these proteins in adipocyte biology. RAB4A is a small GTPase that participates in adipocyte GLUT4 trafficking, exerting an important role in glucose uptake [21,22]. IDH2 attenuates adipocyte inflammation through the induction of α-ketoglutarate production [23] and is required for lipogenesis in these cells [24]. ITGA5 promotes fibrosis of adipose tissue [25] and inhibits adipocyte differentiation in human adipose tissue-derived stem cells [26]. Decreased expression of the NPC1 gene or the partial deletion of this gene promoted weight gain and fat mass expansion [27,28,29], suggesting a relevant role of NPC1 in the prevention of obesity and adipocyte hypertrophy, possibly increasing lipolysis and beta oxidation and decreasing lipogenesis and triglyceride accumulation in adipocytes [30]. NPC1 is increased in adipocytes [31], but it was not associated with adipogenesis [32]. ACO1 is important to maintain adipogenesis in adipocytes, possibly modulating cytosolic NADPH levels and intracellular iron homeostasis, both processes required for adipocyte biology [33]. LRP1 is required for adipocyte differentiation and to maintain adipocyte adipogenesis [34], possibly in part due to its interaction with ShcA [35], or acting as APOA4 receptor, and favoring APOA4-induced glucose uptake [36]. Several studies have demonstrated that AHNAK is required for adipocyte differentiation [37,38,39]. Mechanistically, a recent study demonstrated that AHNAK gene knockdown decreased adipogenesis through the suppression of Bmpr1α expression and decreased BMP4/ Bmpr1α signaling [39]. ARPC3 is another protein required for adipocyte differentiation, exerting a relevant role in the late stage of the process, participating in GLUT4 exocytosis and insulin signal transduction, and suggesting a relevant role in maintaining insulin action in adipocytes [40]. ADH1B modulates intracellular lipid accumulation, without impacting adipogenesis [41], and preserved insulin-stimulated glucose uptake in adipocytes [42]. Inhibition of ALDH1A1 reduced visceral fat [43], negatively impacting adipogenesis in visceral adipose tissue [44]. LGALS1 (galectin 1) is an adipocyte secreted factor [45] that enhances the transcriptional activity of PPARG, increasing adipogenesis and accumulation of lipids in adipocytes [46]. Gain–loss in vivo studies demonstrated that this protein is crucial for adipose tissue expansion and adiposity [46]. LGALS1 expression in subcutaneous adipose tissue has been identified as a relevant factor to confer the risk of weight regain [47]. Pharmacological inhibition of LGALS1 displayed important anti-obesogenic and anti-adipogenic effects [48,49], strengthening the role of this protein in the development of fat mass and obesity. Otherwise, circulating galectin-1 levels were negatively associated with type 2 diabetes [50,51], indicating that it might be associated with healthy adiposity. PHB is induced during adipocyte differentiation, in response to insulin and PPARγ agonist, and its overexpression in 3T3-L1 fibroblasts was sufficient to induce adipogenesis [52], but its gene knockdown reduces the expression of adipogenic genes and lipid accumulation and impairs mitochondrial dysfunction [53,54]. In adipocytes, prohibitin is required for fatty acid uptake [55]. COL6A3 gene knockdown increased the triglyceride content, lipolysis, insulin-induced Akt phosphorylation, and the expression of adipogenic genes (peroxisome proliferator-activated receptor-γ, glucose transporter, adiponectin, and fatty acid binding protein), indicating improved adipocyte function and insulin sensitivity [56]. In addition, COL6A3 knockdown also decreased basal adipocyte MCP1 mRNA expression, reduced secreted protein levels, and attenuated TNFα- and LPS-induced MCP1 gene expression [57]. In contrast, increased COL6A3 mRNA in adipocytes is associated with obesity, insulin resistance, and inflammation of adipose tissue [58,59], supporting the negative impact of this protein on adipocytes and adipose tissue. Annexin A1 (AnxA1) is an endogenous glucocorticoid regulated protein that modulates systemic anti-inflammatory processes. AnxA1 gene expression and protein were significantly up-regulated during adipogenesis in human SGBS preadipocytes [60]. Epididymal fat mass was reduced by ANXA1 gene deletion, but adipocyte size was unchanged, suggesting that ANXA1 is required for maintaining adipocyte cell number [61]. Furthermore, the progressive accumulation of Annexin A2 (AnxA2) in the myofiber matrix causes muscle-resident fibro/adipogenic precursors (FAP) differentiation into adipocytes, and depletion of AnxA2 prevents FAP adipogenesis and muscle loss [62]. ACSL1 is required for beta oxidation in adipocytes, and it is associated to adipose insulin sensitivity, adipogenesis and adipocyte fatty acid uptake [63,64,65,66,67,68,69,70]. ELAVL1 (also named HuR) is an important repressor of adipogenesis. Knockdown and overexpression of HuR in primary adipocyte culture enhances and inhibits adipogenesis, respectively [71]. HuR also exerts an important role in the prevention of adipocyte hypertrophy and inflammation of adipose tissue [72]. However, previous studies in 3T3-L1 cells indicate that HuR is required in early events of adipogenesis, allowing C/EBPβ mRNA translocation into the cytosol, and its proper translation, a process required to activate CEBPα and PPARγ in mitotic clonal expansion [73,74]. Importantly, the reduction in HuR persulfidation, resulted in the stabilization of HuR-target mRNAs, and increased its translation and expression [75], suggesting that the decrease in HuR persulfidation improved HuR dimerization and its activity, and consequently, it has negative effects on adipocyte adipogenesis. Interestingly, perilipin persulfidation inhibits lipid mobilization and lipolysis in adipocytes, and contributes to adequate maintenance of lipid storage [76]. HNRNPA1 is a known regulator of INSR exon 11 splicing, which could have an impact on adipose insulin action [77].
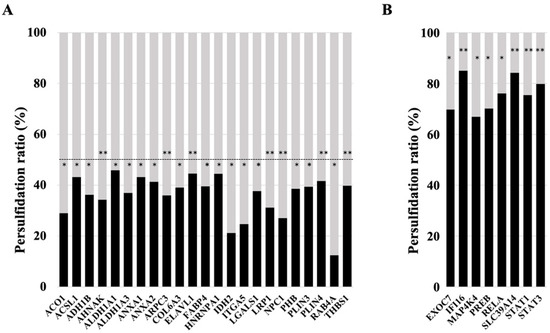
Figure 3.
Rate of persulfidation of relevant proteins involved in adipocyte function and the maintenance of adipogenesis. Level of protein persulfidation in siRNA_MIX samples (black boxes, in percentage) compare to the level in control samples (grey boxes). (A) ACO1, Cytoplasmic aconitate hydratase; ACSL1, Long-chain-fatty-acid—CoA ligase 1; ADH1B, All-trans-retinol dehydrogenase [NAD(+)] ADH1B; AHNAK, Neuroblast differentiation-associated protein AHNAK; ALDH1A1, Retinal dehydrogenase 1; ALDH1A3, Aldehyde dehydrogenase family 1 member A3; ANXA1, Annexin A1; ANXA2, Annexin A2; ARPC3, Actin-related protein 2/3 complex subunit 3; COL6A3, Collagen alpha-3(VI) chain; ELAVL1, ELAV-like protein 1; FABP4, Fatty acid-binding protein adipocyte; HNRNPA1, Heterogeneous nuclear ribonucleoprotein A1; IDH2, Isocitrate dehydrogenase [NADP] mitochondrial; ITGA5, Integrin alpha-5; LGALS1, Galectin-1; LRP1, Low-density lipoprotein receptor-related protein 1; NPC1, NPC intracellular cholesterol transporter 1; PHB, Prohibitin; PLIN3, Perilipin-3; PLIN4, Perilipin-4;RAB4A, Ras-related protein Rab-4A; THBS1, Thrombospondin-1. (B) EXOC7, Exocyst complex component 7; IFI16, Gamma-interferon-inducible protein 16; MAP4K4, Mitogen-activated protein kinase kinase kinase kinase 4; PREB, Prolactin regulatory element-binding protein; RELA, Transcription factor p65; SLC39A14, Metal cation symporter ZIP14; STAT1, Signal transducer and activator of transcription 1-alpha/beta; STAT3, Signal transducer and activator of transcription 3. Student t-test significance: * p < 0.05; ** p < 0.01.
Among the most persulfidated proteins, some key mediators of adipocyte inflammation and dysfunction (IFI16, STAT3, RELA, STAT1, MAP4K4, PREB) were found, but also proteins that might have a positive role in adipogenesis (EXOC7, SLC39A14) (Figure 3B, all with p < 0.05). In mice and humans, increased IFI16 levels are associated with larger adipocytes, enhanced inflammatory state, and impaired insulin-stimulated glucose uptake in white adipose tissue [78]. STAT3 is a relevant transcription factor of proinflammatory genes in adipocytes that is associated with adipocyte inflammation [79,80]. RELA is a relevant transcription factor associated with adipocyte inflammation [81] that inhibits adipogenesis and accumulation of lipids in adipocytes [82]. The activation of STAT1 transcription factor inhibits adipogenesis and promotes adipocyte and adipose tissue inflammation [83,84,85,86,87,88]. Isolated mature adipocytes from obese individuals had increased expression of mitogen-activated protein 4 kinase 4 (MAP4K4), which is known to inhibit PPARγ transcriptional activity, adipogenesis, and insulin-stimulated glucose transport [89,90]. In line with this, MAP4K4 inhibits adipose lipogenesis via the suppression of Srebp-1 [91], and deletion of this kinase increases insulin sensitivity in adipose tissue [92]. Prolactin regulatory element-binding (PREB) is a negative regulator of adiponectin gene expression in adipocytes [93].
On the other hand, EXOC7 might participate in Glut4 trafficking [94,95], and SLC39A14 is a zinc transporter, which is rapidly induced in the early stages of differentiation, suggesting a possible role in adipogenesis [96]. In fact, depletion of SLC39A14 caused hypertrophy and inflammation of adipocytes [97].
These data indicated that the combined partial knockdown of the MPST and CBS genes disrupts the pattern of protein persulfidation in human adipocytes, suggesting a possible link between this disruption and adipocyte dysfunction, which is characterized by decreased adipogenesis but increased inflammation. This disruption included a significant decrease in persulfidation in key proteins for adipocyte function (RAB4A, IDH2, NPC1, ACO1, LRP1, AHNAK, ARPC3, ADH1B, ALDH1A1, LGALS1, PHB, ANXA1, ANXA2, ACSL1, PLIN3, PLIN4, FABP4, HNRNPA1), but also in proteins that negatively impact adipocytes and adipogenesis (ITGA5, COL6A3, ELAVL1). In addition, the combined partial knockdown of MPST and CBS genes in adipocytes resulted in increased persulfidation in relevant proinflammatory transcription factors (RELA, STAT1, STAT3) and anti-adipogenic proteins (IFI16, MAP4K4, PREB), but also in some proteins that might exert positive effects on adipogenesis, such as SLC39A14 and EXOC7. Persulfidation increases the reactivity of target proteins, modulating their biological activities [10]. However, it is important to note that modulation in persulfidation-induced protein activity can trigger activation or inhibition of the biological function of the persulfidated protein [98]. Other consequences of CBS and MPST gene knockdown, such as decreased H2S biosynthesis [8,18] or increased homocysteine levels [99], could also explain the negative impact on adipocyte function. Additional experiments using specific chemical inhibitors of H₂S biosynthesis and the administration of H2S donor molecules to CBS/MPST gene knockdown cells should be performed to further investigate the possible role of H2S in this model.
Selenium-binding protein 1 (SELENBP1), a fourth enzyme that participates in H2S biosynthesis, increased during differentiation of 3T3L1 adipocyte in association with lipogenesis and lipid accumulation, and it has been recently described as a marker of differentiated adipocytes [100]. SELENBP1 gene knockdown had a negative impact on adipocyte function in parallel to the decreased expression of the CBS, CTH, and MPST enzymes [101]. The contribution of other enzymes, such as SELENBP1 [100,101], or non-enzymatic pathways [102] in H2S production cannot be excluded in the current study.
4. Conclusions
In summary, these findings indicate that the combined partial knockdown of CBS and MPST (but not CTH) in adipocytes impairs the expression of genes related with the maintenance of adipocyte function and promotes inflammation, possibly by disrupting the pattern of protein persulfidation in these cells. The current study points to the fact that these enzymes were not only required during adipocyte differentiation [8,9], but they were also needed for the functional maintenance of adipocytes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antiox11061095/s1, Supplementary Dataset S1 and Supplementary Dataset S2.
Author Contributions
J.L. participated in this study conducting experiments, acquiring and analyzing data and reviewed the manuscript. A.A. and L.C.R. participated in this study acquiring and analyzing data, contributed to the discussion and reviewed the manuscript. J.M.F.-R. contributed to the discussion and reviewed the manuscript. J.M.M.-N. contributed to research study design, conducting experiments, acquiring and analyzing data, and writing the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by research grants PI16/01173, PI19/01712 and PI21/01361 from the Instituto de Salud Carlos III from Spain, Fondo Europeo de Desarrollo Regional (FEDER) funds, and VII Spanish Diabetes Association grants to Basic Diabetes Research Projects led by young researchers. This work was also partially supported by ERDF A way of making Europe, Junta de Andalucía (grant No. US-1255781).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available via ProteomeXchange with identifier PXD032370.
Acknowledgments
CIBEROBN Fisiopatología de la Obesidad y Nutrición is an initiative from the Instituto de Salud Carlos III from Spain.
Conflicts of Interest
The authors declared no conflict of interest.
References
- Maurizi, G.; Della Guardia, L.; Maurizi, A.; Poloni, A. Adipocytes properties and crosstalk with immune system in obesity-related inflammation. J. Cell. Physiol. 2018, 233, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Sakers, A.; De Siqueira, M.K.; Seale, P.; Villanueva, C.J. Adipose-tissue plasticity in health and disease. Cell 2022, 185, 419–446. [Google Scholar] [CrossRef] [PubMed]
- Virtue, S.; Vidal-Puig, A. Adipose tissue expandability, lipotoxicity and the Metabolic Syndrome-an allostatic perspective. Biochim. Biophys. Acta 2010, 1801, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Van De Wall, E.; Laplante, M.; Azzara, A.; Trujillo, M.E.; Hofmann, S.M.; Schraw, T.; Durand, J.L.; Li, H.; Li, G.; et al. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J. Clin. Investig. 2007, 117, 2621–2637. [Google Scholar] [CrossRef] [PubMed]
- Vishvanath, L.; Gupta, R.K. Contribution of adipogenesis to healthy adipose tissue expansion in obesity. J. Clin. Investig. 2019, 129, 4022–4031. [Google Scholar] [CrossRef]
- De Ferranti, S.; Mozaffarian, D. The perfect storm: Obesity, adipocyte dysfunction, and metabolic consequences. Clin. Chem. 2008, 54, 945–955. [Google Scholar] [CrossRef]
- Yang, G.; Ju, Y.; Fu, M.; Zhang, Y.; Pei, Y.; Racine, M.; Baath, S.; Merritt, T.J.S.; Wang, R.; Wu, L. Cystathionine gamma-lyase/hydrogen sulfide system is essential for adipogenesis and fat mass accumulation in mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 165–176. [Google Scholar] [CrossRef]
- Comas, F.; Latorre, J.; Ortega, F.; Arnoriaga Rodríguez, M.; Kern, M.; Lluch, A.; Ricart, W.; Blüher, M.; Gotor, C.; Romero, L.C.; et al. Activation of Endogenous H(2)S Biosynthesis or Supplementation with Exogenous H(2)S Enhances Adipose Tissue Adipogenesis and Preserves Adipocyte Physiology in Humans. Antioxid. Redox Signal. 2021, 35, 319–340. [Google Scholar] [CrossRef]
- Tsai, C.-Y.; Peh, M.T.; Feng, W.; Dymock, B.W.; Moore, P.K. Hydrogen sulfide promotes adipogenesis in 3T3L1 cells. PLoS ONE 2015, 10, e0119511. [Google Scholar] [CrossRef]
- Filipovic, M.R. Persulfidation (S-sulfhydration) and H2S. Handb. Exp. Pharmacol. 2015, 230, 29–59. [Google Scholar] [CrossRef]
- Tan, B.; Jin, S.; Sun, J.; Gu, Z.; Sun, X.; Zhu, Y.; Huo, K.; Cao, Z.; Yang, P.; Xin, X.; et al. New method for quantification of gasotransmitter hydrogen sulfide in biological matrices by LC-MS/MS. Sci. Rep. 2017, 7, 46278. [Google Scholar] [CrossRef] [PubMed]
- Aroca, A.; Yruela, I.; Gotor, C.; Bassham, D.C. Persulfidation of ATG18a regulates autophagy under ER stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2023604118. [Google Scholar] [CrossRef] [PubMed]
- Zivanovic, J.; Kouroussis, E.; Kohl, J.B.; Adhikari, B.; Bursac, B.; Schott-Roux, S.; Petrovic, D.; Miljkovic, J.L.; Thomas-Lopez, D.; Jung, Y.; et al. Selective Persulfide Detection Reveals Evolutionarily Conserved Antiaging Effects of S-Sulfhydration. Cell Metab. 2019, 30, 1152–1170.e13. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xin, L.; Shan, B.; Chen, W.; Xie, M.; Yuen, D.; Zhang, W.; Zhang, Z.; Lajoie, G.A.; Ma, B. PEAKS DB: De novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell. Proteomics 2012, 11, M111.010587. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Deutsch, E.W.; Wang, R.; Csordas, A.; Reisinger, F.; Ríos, D.; Dianes, J.A.; Sun, Z.; Farrah, T.; Bandeira, N.; et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [Google Scholar] [CrossRef]
- Cai, J.; Shi, X.; Wang, H.; Fan, J.; Feng, Y.; Lin, X.; Yang, J.; Cui, Q.; Tang, C.; Xu, G.; et al. Cystathionine γ lyase-hydrogen sulfide increases peroxisome proliferator-activated receptor γ activity by sulfhydration at C139 site thereby promoting glucose uptake and lipid storage in adipocytes. Biochim. Biophys. Acta 2016, 1861, 419–429. [Google Scholar] [CrossRef]
- Comas, F.; Latorre, J.; Cussó, O.; Ortega, F.; Lluch, A.; Sabater, M.; Castells-Nobau, A.; Ricart, W.; Ribas, X.; Costas, M.; et al. Hydrogen sulfide impacts on inflammation-induced adipocyte dysfunction. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2019, 131, 110543. [Google Scholar] [CrossRef]
- Sugihara, H.; Yonemitsu, N.; Miyabara, S.; Yun, K. Primary cultures of unilocular fat cells: Characteristics of growth in vitro and changes in differentiation properties. Differentiation 1986, 31, 42–49. [Google Scholar] [CrossRef]
- Kaddai, V.; Gonzalez, T.; Keslair, F.; Grémeaux, T.; Bonnafous, S.; Gugenheim, J.; Tran, A.; Gual, P.; Le Marchand-Brustel, Y.; Cormont, M. Rab4b is a small GTPase involved in the control of the glucose transporter GLUT4 localization in adipocyte. PLoS ONE 2009, 4, e5257. [Google Scholar] [CrossRef] [PubMed]
- Błaszczyk, M.; Gajewska, M.; Milewska, M.; Grzelkowska-Kowalczyk, K. Insulin-dependent cytoplasmic distribution of Rab4a in mouse adipocytes is inhibited by interleukin-6, -8, and -15. Cell Biol. Int. 2017, 41, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Gan, L.; Zhang, T.; Ren, Q.; Sun, C. Melatonin alleviates adipose inflammation through elevating α-ketoglutarate and diverting adipose-derived exosomes to macrophages in mice. J. Pineal Res. 2018, 64, e12455. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Kim, S.H.; Park, K.M.; Lee, J.H.; Park, J.-W. Increased obesity resistance and insulin sensitivity in mice lacking the isocitrate dehydrogenase 2 gene. Free Radic. Biol. Med. 2016, 99, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Liang, J.; Sun, Z.; Wu, Q.; Liu, Y.; Sun, C. The Mechanism of Leptin on Inhibiting Fibrosis and Promoting Browning of White Fat by Reducing ITGA5 in Mice. Int. J. Mol. Sci. 2021, 22, 2353. [Google Scholar] [CrossRef] [PubMed]
- Morandi, E.M.; Verstappen, R.; Zwierzina, M.E.; Geley, S.; Pierer, G.; Ploner, C. ITGAV and ITGA5 diversely regulate proliferation and adipogenic differentiation of human adipose derived stem cells. Sci. Rep. 2016, 6, 28889. [Google Scholar] [CrossRef] [PubMed]
- Jelinek, D.; Heidenreich, R.A.; Erickson, R.P.; Garver, W.S. Decreased Npc1 gene dosage in mice is associated with weight gain. Obesity 2010, 18, 1457–1459. [Google Scholar] [CrossRef]
- Liu, R.; Zou, Y.; Hong, J.; Cao, M.; Cui, B.; Zhang, H.; Chen, M.; Shi, J.; Ning, T.; Zhao, S.; et al. Rare Loss-of-Function Variants in NPC1 Predispose to Human Obesity. Diabetes 2017, 66, 935–947. [Google Scholar] [CrossRef]
- Jelinek, D.; Millward, V.; Birdi, A.; Trouard, T.P.; Heidenreich, R.A.; Garver, W.S. Npc1 haploinsufficiency promotes weight gain and metabolic features associated with insulin resistance. Hum. Mol. Genet. 2011, 20, 312–321. [Google Scholar] [CrossRef][Green Version]
- Castillo, J.J.; Jelinek, D.; Wei, H.; Gannon, N.P.; Vaughan, R.A.; Horwood, L.J.; Meaney, F.J.; Garcia-Smith, R.; Trujillo, K.A.; Heidenreich, R.A.; et al. The Niemann-Pick C1 gene interacts with a high-fat diet to promote weight gain through differential regulation of central energy metabolism pathways. Am. J. Physiol. Endocrinol. Metab. 2017, 313, E183–E194. [Google Scholar] [CrossRef]
- Bambace, C.; Dahlman, I.; Arner, P.; Kulyté, A. NPC1 in human white adipose tissue and obesity. BMC Endocr. Disord. 2013, 13, 5. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, F.; Landgraf, K.; Klöting, N.; Berthold, A.; Büttner, P.; Friebe, D.; Kiess, W.; Kovacs, P.; Blüher, M.; Körner, A. Functional relevance of genes implicated by obesity genome-wide association study signals for human adipocyte biology. Diabetologia 2013, 56, 311–322. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.; Ortega, F.; Xifra, G.; Ricart, W.; Fernández-Real, J.M.; Moreno-Navarrete, J.M. Cytosolic aconitase activity sustains adipogenic capacity of adipose tissue connecting iron metabolism and adipogenesis. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2015, 29, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Masson, O.; Chavey, C.; Dray, C.; Meulle, A.; Daviaud, D.; Quilliot, D.; Muller, C.; Valet, P.; Liaudet-Coopman, E. LRP1 receptor controls adipogenesis and is up-regulated in human and mouse obese adipose tissue. PLoS ONE 2009, 4, e7422. [Google Scholar] [CrossRef] [PubMed]
- Woldt, E.; Matz, R.L.; Terrand, J.; Mlih, M.; Gracia, C.; Foppolo, S.; Martin, S.; Bruban, V.; Ji, J.; Velot, E.; et al. Differential signaling by adaptor molecules LRP1 and ShcA regulates adipogenesis by the insulin-like growth factor-1 receptor. J. Biol. Chem. 2011, 286, 16775–16782. [Google Scholar] [CrossRef]
- Qu, J.; Fourman, S.; Fitzgerald, M.; Liu, M.; Nair, S.; Oses-Prieto, J.; Burlingame, A.; Morris, J.H.; Davidson, W.S.; Tso, P.; et al. Low-density lipoprotein receptor-related protein 1 (LRP1) is a novel receptor for apolipoprotein A4 (APOA4) in adipose tissue. Sci. Rep. 2021, 11, 13289. [Google Scholar] [CrossRef]
- Shin, S.; Seong, J.K.; Bae, Y.S. Ahnak stimulates BMP2-mediated adipocyte differentiation through Smad1 activation. Obesity 2016, 24, 398–407. [Google Scholar] [CrossRef]
- Shin, J.H.; Kim, I.Y.; Kim, Y.N.; Shin, S.M.; Roh, K.J.; Lee, S.H.; Sohn, M.; Cho, S.Y.; Lee, S.H.; Ko, C.-Y.; et al. Obesity Resistance and Enhanced Insulin Sensitivity in Ahnak-/- Mice Fed a High Fat Diet Are Related to Impaired Adipogenesis and Increased Energy Expenditure. PLoS ONE 2015, 10, e0139720. [Google Scholar] [CrossRef]
- Woo, J.K.; Shin, J.H.; Lee, S.H.; Park, H.-M.; Cho, S.Y.; Sung, Y.M.; Kim, I.Y.; Seong, J.K. Essential role of Ahnak in adipocyte differentiation leading to the transcriptional regulation of Bmpr1α expression. Cell Death Dis. 2018, 9, 864. [Google Scholar] [CrossRef]
- Yang, W.; Thein, S.; Lim, C.-Y.; Ericksen, R.E.; Sugii, S.; Xu, F.; Robinson, R.C.; Kim, J.B.; Han, W. Arp2/3 complex regulates adipogenesis by controlling cortical actin remodelling. Biochem. J. 2014, 464, 179–192. [Google Scholar] [CrossRef]
- Kerr, A.G.; Sinha, I.; Dadvar, S.; Arner, P.; Dahlman, I. Epigenetic regulation of diabetogenic adipose morphology. Mol. Metab. 2019, 25, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Morales, L.D.; Cromack, D.T.; Tripathy, D.; Fourcaudot, M.; Kumar, S.; Curran, J.E.; Carless, M.; Göring, H.H.H.; Hu, S.L.; Lopez-Alvarenga, J.C.; et al. Further evidence supporting a potential role for ADH1B in obesity. Sci. Rep. 2021, 11, 1932. [Google Scholar] [CrossRef] [PubMed]
- Haenisch, M.; Nguyen, T.; Fihn, C.A.; Goldstein, A.S.; Amory, J.K.; Treuting, P.; Brabb, T.; Paik, J. Investigation of an ALDH1A1-specific inhibitor for suppression of weight gain in a diet-induced mouse model of obesity. Int. J. Obes. 2021, 45, 1542–1552. [Google Scholar] [CrossRef] [PubMed]
- Reichert, B.; Yasmeen, R.; Jeyakumar, S.M.; Yang, F.; Thomou, T.; Alder, H.; Duester, G.; Maiseyeu, A.; Mihai, G.; Harrison, E.H.; et al. Concerted action of aldehyde dehydrogenases influences depot-specific fat formation. Mol. Endocrinol. 2011, 25, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Mariman, E.; Keijer, J.; Bouwman, F.; Noben, J.-P.; Robben, J.; Renes, J. Profiling of the secreted proteins during 3T3-L1 adipocyte differentiation leads to the identification of novel adipokines. Cell. Mol. Life Sci. 2004, 61, 2405–2417. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.-H.; Kim, D.-H.; Lee, J.; Kim, S.-J.; Chun, K.-H. Galectin-1 accelerates high-fat diet-induced obesity by activation of peroxisome proliferator-activated receptor gamma (PPARγ) in mice. Cell Death Dis. 2021, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Roumans, N.J.T.; Vink, R.G.; Bouwman, F.G.; Fazelzadeh, P.; van Baak, M.A.; Mariman, E.C.M. Weight loss-induced cellular stress in subcutaneous adipose tissue and the risk for weight regain in overweight and obese adults. Int. J. Obes. 2017, 41, 894–901. [Google Scholar] [CrossRef]
- Mukherjee, R.; Yun, J.W. Pharmacological inhibition of galectin-1 by lactulose alleviates weight gain in diet-induced obese rats. Life Sci. 2016, 148, 112–117. [Google Scholar] [CrossRef]
- Mukherjee, R.; Kim, S.W.; Park, T.; Choi, M.S.; Yun, J.W. Targeted inhibition of galectin 1 by thiodigalactoside dramatically reduces body weight gain in diet-induced obese rats. Int. J. Obes. 2015, 39, 1349–1358. [Google Scholar] [CrossRef]
- Fryk, E.; Strindberg, L.; Lundqvist, A.; Sandstedt, M.; Bergfeldt, L.; Mattsson Hultén, L.; Bergström, G.; Jansson, P.-A. Galectin-1 is inversely associated with type 2 diabetes independently of obesity—A SCAPIS pilot study. Metab. Open 2019, 4, 100017. [Google Scholar] [CrossRef]
- Acar, S.; Paketçi, A.; Küme, T.; Tuhan, H.; Gürsoy Çalan, Ö.; Demir, K.; Böber, E.; Abaci, A. Serum galectin-1 levels are positively correlated with body fat and negatively with fasting glucose in obese children. Peptides 2017, 95, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Ande, S.R.; Xu, Z.; Gu, Y.; Mishra, S. Prohibitin has an important role in adipocyte differentiation. Int. J. Obes. 2012, 36, 1236–1244. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Lin, Y.; Kang, T.; Huang, B.; Xu, W.; Garcia-Barrio, M.; Olatinwo, M.; Matthews, R.; Chen, Y.E.; Thompson, W.E. Mitochondrial dysfunction and adipogenic reduction by prohibitin silencing in 3T3-L1 cells. PLoS ONE 2012, 7, e34315. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.; Lu, W.; Xu, W.; Anderson, L.; Bacanamwo, M.; Thompson, W.; Chen, Y.E.; Liu, D. MicroRNA-27 (miR-27) targets prohibitin and impairs adipocyte differentiation and mitochondrial function in human adipose-derived stem cells. J. Biol. Chem. 2013, 288, 34394–34402. [Google Scholar] [CrossRef]
- Salameh, A.; Daquinag, A.C.; Staquicini, D.I.; An, Z.; Hajjar, K.A.; Pasqualini, R.; Arap, W.; Kolonin, M.G. Prohibitin/annexin 2 interaction regulates fatty acid transport in adipose tissue. JCI Insight 2016, 1, e86351. [Google Scholar] [CrossRef]
- Gesta, S.; Guntur, K.; Majumdar, I.D.; Akella, S.; Vishnudas, V.K.; Sarangarajan, R.; Narain, N.R. Reduced expression of collagen VI alpha 3 (COL6A3) confers resistance to inflammation-induced MCP1 expression in adipocytes. Obesity 2016, 24, 1695–1703. [Google Scholar] [CrossRef]
- Pardo, F.; Villalobos-Labra, R.; Sobrevia, B.; Toledo, F.; Sobrevia, L. Extracellular vesicles in obesity and diabetes mellitus. Mol. Aspects Med. 2018, 60, 81–91. [Google Scholar] [CrossRef]
- Dankel, S.N.; Svärd, J.; Matthä, S.; Claussnitzer, M.; Klöting, N.; Glunk, V.; Fandalyuk, Z.; Grytten, E.; Solsvik, M.H.; Nielsen, H.-J.; et al. COL6A3 expression in adipocytes associates with insulin resistance and depends on PPARγ and adipocyte size. Obesity 2014, 22, 1807–1813. [Google Scholar] [CrossRef]
- Pasarica, M.; Gowronska-Kozak, B.; Burk, D.; Remedios, I.; Hymel, D.; Gimble, J.; Ravussin, E.; Bray, G.A.; Smith, S.R. Adipose tissue collagen VI in obesity. J. Clin. Endocrinol. Metab. 2009, 94, 5155–5162. [Google Scholar] [CrossRef]
- Kosicka, A.; Cunliffe, A.D.; Mackenzie, R.; Zariwala, M.G.; Perretti, M.; Flower, R.J.; Renshaw, D. Attenuation of plasma annexin A1 in human obesity. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2013, 27, 368–378. [Google Scholar] [CrossRef]
- Warne, J.P.; John, C.D.; Christian, H.C.; Morris, J.F.; Flower, R.J.; Sugden, D.; Solito, E.; Gillies, G.E.; Buckingham, J.C. Gene deletion reveals roles for annexin A1 in the regulation of lipolysis and IL-6 release in epididymal adipose tissue. Am. J. Physiol. Endocrinol. Metab. 2006, 291, E1264–E1273. [Google Scholar] [CrossRef] [PubMed]
- Hogarth, M.W.; Defour, A.; Lazarski, C.; Gallardo, E.; Diaz Manera, J.; Partridge, T.A.; Nagaraju, K.; Jaiswal, J.K. Fibroadipogenic progenitors are responsible for muscle loss in limb girdle muscular dystrophy 2B. Nat. Commun. 2019, 10, 2430. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Zhao, Y.; He, L.; Zhao, J.; Gao, T.; Liu, F.; Qi, B.; Kang, F.; Wang, G.; Zhao, Y.; et al. AKAP1 Deficiency Attenuates Diet-Induced Obesity and Insulin Resistance by Promoting Fatty Acid Oxidation and Thermogenesis in Brown Adipocytes. Adv. Sci. 2021, 8, 2002794. [Google Scholar] [CrossRef] [PubMed]
- Joseph, R.; Poschmann, J.; Sukarieh, R.; Too, P.G.; Julien, S.G.; Xu, F.; Teh, A.L.; Holbrook, J.D.; Ng, K.L.; Chong, Y.S.; et al. ACSL1 Is Associated With Fetal Programming of Insulin Sensitivity and Cellular Lipid Content. Mol. Endocrinol. 2015, 29, 909–920. [Google Scholar] [CrossRef]
- Ellis, J.M.; Li, L.O.; Wu, P.-C.; Koves, T.R.; Ilkayeva, O.; Stevens, R.D.; Watkins, S.M.; Muoio, D.M.; Coleman, R.A. Adipose acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell Metab. 2010, 12, 53–64. [Google Scholar] [CrossRef]
- Lobo, S.; Wiczer, B.M.; Bernlohr, D.A. Functional analysis of long-chain acyl-CoA synthetase 1 in 3T3-L1 adipocytes. J. Biol. Chem. 2009, 284, 18347–18356. [Google Scholar] [CrossRef]
- Liu, Q.; Gauthier, M.-S.; Sun, L.; Ruderman, N.; Lodish, H. Activation of AMP-activated protein kinase signaling pathway by adiponectin and insulin in mouse adipocytes: Requirement of acyl-CoA synthetases FATP1 and Acsl1 and association with an elevation in AMP/ATP ratio. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2010, 24, 4229–4239. [Google Scholar] [CrossRef]
- Yi, X.; Liu, J.; Wu, P.; Gong, Y.; Xu, X.; Li, W. The key microRNA on lipid droplet formation during adipogenesis from human mesenchymal stem cells. J. Cell. Physiol. 2020, 235, 328–338. [Google Scholar] [CrossRef]
- Zhan, T.; Poppelreuther, M.; Ehehalt, R.; Füllekrug, J. Overexpressed FATP1, ACSVL4/FATP4 and ACSL1 increase the cellular fatty acid uptake of 3T3-L1 adipocytes but are localized on intracellular membranes. PLoS ONE 2012, 7, e45087. [Google Scholar] [CrossRef]
- Richards, M.R.; Harp, J.D.; Ory, D.S.; Schaffer, J.E. Fatty acid transport protein 1 and long-chain acyl coenzyme A synthetase 1 interact in adipocytes. J. Lipid Res. 2006, 47, 665–672. [Google Scholar] [CrossRef]
- Siang, D.T.C.; Lim, Y.C.; Kyaw, A.M.M.; Win, K.N.; Chia, S.Y.; Degirmenci, U.; Hu, X.; Tan, B.C.; Walet, A.C.E.; Sun, L.; et al. The RNA-binding protein HuR is a negative regulator in adipogenesis. Nat. Commun. 2020, 11, 213. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gong, L.; Liu, S.; Zhang, Y.; Zhang, C.; Tian, M.; Lu, H.; Bu, P.; Yang, J.; Ouyang, C.; et al. Adipose HuR protects against diet-induced obesity and insulin resistance. Nat. Commun. 2019, 10, 2375. [Google Scholar] [CrossRef] [PubMed]
- Gantt, K.; Cherry, J.; Tenney, R.; Karschner, V.; Pekala, P.H. An early event in adipogenesis, the nuclear selection of the CCAAT enhancer-binding protein {beta} (C/EBP{beta}) mRNA by HuR and its translocation to the cytosol. J. Biol. Chem. 2005, 280, 24768–24774. [Google Scholar] [CrossRef] [PubMed]
- Karschner, V.A.; Pekala, P.H. HuR involvement in mitotic clonal expansion during acquisition of the adipocyte phenotype. Biochem. Biophys. Res. Commun. 2009, 383, 203–205. [Google Scholar] [CrossRef][Green Version]
- Bibli, S.-I.; Hu, J.; Sigala, F.; Wittig, I.; Heidler, J.; Zukunft, S.; Tsilimigras, D.I.; Randriamboavonjy, V.; Wittig, J.; Kojonazarov, B.; et al. Cystathionine γ Lyase Sulfhydrates the RNA Binding Protein Human Antigen R to Preserve Endothelial Cell Function and Delay Atherogenesis. Circulation 2019, 139, 101–114. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, H.; Geng, B.; Xu, G. Sulfhydration of perilipin 1 is involved in the inhibitory effects of cystathionine gamma lyase/hydrogen sulfide on adipocyte lipolysis. Biochem. Biophys. Res. Commun. 2020, 521, 786–790. [Google Scholar] [CrossRef]
- Kaminska, D.; Hämäläinen, M.; Cederberg, H.; Käkelä, P.; Venesmaa, S.; Miettinen, P.; Ilves, I.; Herzig, K.-H.; Kolehmainen, M.; Karhunen, L.; et al. Adipose tissue INSR splicing in humans associates with fasting insulin level and is regulated by weight loss. Diabetologia 2014, 57, 347–351. [Google Scholar] [CrossRef]
- Stadion, M.; Schwerbel, K.; Graja, A.; Baumeier, C.; Rödiger, M.; Jonas, W.; Wolfrum, C.; Staiger, H.; Fritsche, A.; Häring, H.-U.; et al. Increased Ifi202b/IFI16 expression stimulates adipogenesis in mice and humans. Diabetologia 2018, 61, 1167–1179. [Google Scholar] [CrossRef]
- Valentino, R.; D’Esposito, V.; Passaretti, F.; Liotti, A.; Cabaro, S.; Longo, M.; Perruolo, G.; Oriente, F.; Beguinot, F.; Formisano, P. Bisphenol-A impairs insulin action and up-regulates inflammatory pathways in human subcutaneous adipocytes and 3T3-L1 cells. PLoS ONE 2013, 8, e82099. [Google Scholar] [CrossRef]
- Guo, T.; Gupta, A.; Yu, J.; Granados, J.Z.; Gandhi, A.Y.; Evers, B.M.; Iyengar, P.; Infante, R.E. LIFR-α-dependent adipocyte signaling in obesity limits adipose expansion contributing to fatty liver disease. iScience 2021, 24, 102227. [Google Scholar] [CrossRef]
- Berg, A.H.; Lin, Y.; Lisanti, M.P.; Scherer, P.E. Adipocyte differentiation induces dynamic changes in NF-kappaB expression and activity. Am. J. Physiol. Endocrinol. Metab. 2004, 287, E1178–E1188. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Zhang, J.; Yin, J.; Staszkiewicz, J.; Gawronska-Kozak, B.; Jung, D.Y.; Ko, H.J.; Ong, H.; Kim, J.K.; Mynatt, R.; et al. Uncoupling of inflammation and insulin resistance by NF-kappaB in transgenic mice through elevated energy expenditure. J. Biol. Chem. 2010, 285, 4637–4644. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Um, S.H.; Rhee, D.K.; Pyo, S. Interferon-alpha inhibits adipogenesis via regulation of JAK/STAT1 signaling. Biochim. Biophys. Acta 2016, 1860, 2416–2427. [Google Scholar] [CrossRef] [PubMed]
- Chernis, N.; Masschelin, P.; Cox, A.R.; Hartig, S.M. Bisphenol AF promotes inflammation in human white adipocytes. Am. J. Physiol. Cell Physiol. 2020, 318, C63–C72. [Google Scholar] [CrossRef] [PubMed]
- Annamalai, D.; Clipstone, N.A. Prostaglandin F2α inhibits adipogenesis via an autocrine-mediated interleukin-11/glycoprotein 130/STAT1-dependent signaling cascade. J. Cell. Biochem. 2014, 115, 1308–1321. [Google Scholar] [CrossRef]
- Antony, A.; Lian, Z.; Perrard, X.D.; Perrard, J.; Liu, H.; Cox, A.R.; Saha, P.; Hennighausen, L.; Hartig, S.M.; Ballantyne, C.M.; et al. Deficiency of Stat1 in CD11c(+) Cells Alters Adipose Tissue Inflammation and Improves Metabolic Dysfunctions in Mice Fed a High-Fat Diet. Diabetes 2021, 70, 720–732. [Google Scholar] [CrossRef]
- Chattopadhyay, D.; Das, S.; Guria, S.; Basu, S.; Mukherjee, S. Fetuin-A regulates adipose tissue macrophage content and activation in insulin resistant mice through MCP-1 and iNOS: Involvement of IFNγ-JAK2-STAT1 pathway. Biochem. J. 2021, 478, 4027–4043. [Google Scholar] [CrossRef]
- Cox, A.R.; Chernis, N.; Bader, D.A.; Saha, P.K.; Masschelin, P.M.; Felix, J.B.; Sharp, R.; Lian, Z.; Putluri, V.; Rajapakshe, K.; et al. STAT1 Dissociates Adipose Tissue Inflammation From Insulin Sensitivity in Obesity. Diabetes 2020, 69, 2630–2641. [Google Scholar] [CrossRef]
- Isakson, P.; Hammarstedt, A.; Gustafson, B.; Smith, U. Impaired preadipocyte differentiation in human abdominal obesity: Role of Wnt, tumor necrosis factor-alpha, and inflammation. Diabetes 2009, 58, 1550–1557. [Google Scholar] [CrossRef]
- Tang, X.; Guilherme, A.; Chakladar, A.; Powelka, A.M.; Konda, S.; Virbasius, J.V.; Nicoloro, S.M.C.; Straubhaar, J.; Czech, M.P. An RNA interference-based screen identifies MAP4K4/NIK as a negative regulator of PPARgamma, adipogenesis, and insulin-responsive hexose transport. Proc. Natl. Acad. Sci. USA 2006, 103, 2087–2092. [Google Scholar] [CrossRef]
- Danai, L.V.; Guilherme, A.; Guntur, K.V.; Straubhaar, J.; Nicoloro, S.M.; Czech, M.P. Map4k4 suppresses Srebp-1 and adipocyte lipogenesis independent of JNK signaling. J. Lipid Res. 2013, 54, 2697–2707. [Google Scholar] [CrossRef] [PubMed]
- Danai, L.V.; Flach, R.J.R.; Virbasius, J.V.; Menendez, L.G.; Jung, D.Y.; Kim, J.H.; Kim, J.K.; Czech, M.P. Inducible Deletion of Protein Kinase Map4k4 in Obese Mice Improves Insulin Sensitivity in Liver and Adipose Tissues. Mol. Cell. Biol. 2015, 35, 2356–2365. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Murao, K.; Imachi, H.; Yu, X.; Muraoka, T.; Kim, J.B.; Ishida, T. Prolactin regulatory element-binding protein involved in cAMP-mediated suppression of adiponectin gene. J. Cell. Mol. Med. 2010, 14, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Crisman, L.; Miller, J.; Datta, I.; Gulbranson, D.R.; Tian, Y.; Yin, Q.; Yu, H.; Shen, J. Inducible Exoc7/Exo70 knockout reveals a critical role of the exocyst in insulin-regulated GLUT4 exocytosis. J. Biol. Chem. 2019, 294, 19988–19996. [Google Scholar] [CrossRef]
- Lizunov, V.A.; Lisinski, I.; Stenkula, K.; Zimmerberg, J.; Cushman, S.W. Insulin regulates fusion of GLUT4 vesicles independent of Exo70-mediated tethering. J. Biol. Chem. 2009, 284, 7914–7919. [Google Scholar] [CrossRef]
- Tominaga, K.; Kagata, T.; Johmura, Y.; Hishida, T.; Nishizuka, M.; Imagawa, M. SLC39A14, a LZT protein, is induced in adipogenesis and transports zinc. FEBS J. 2005, 272, 1590–1599. [Google Scholar] [CrossRef]
- Troche, C.; Aydemir, T.B.; Cousins, R.J. Zinc transporter Slc39a14 regulates inflammatory signaling associated with hypertrophic adiposity. Am. J. Physiol. Endocrinol. Metab. 2016, 310, E258–E268. [Google Scholar] [CrossRef]
- Fu, L.; Liu, K.; He, J.; Tian, C.; Yu, X.; Yang, J. Direct Proteomic Mapping of Cysteine Persulfidation. Antioxid. Redox Signal. 2020, 33, 1061–1076. [Google Scholar] [CrossRef]
- Wang, Z.; Dou, X.; Yao, T.; Song, Z. Homocysteine inhibits adipogenesis in 3T3-L1 preadipocytes. Exp. Biol. Med. 2011, 236, 1379–1388. [Google Scholar] [CrossRef]
- Steinbrenner, H.; Micoogullari, M.; Hoang, N.A.; Bergheim, I.; Klotz, L.-O.; Sies, H. Selenium-binding protein 1 (SELENBP1) is a marker of mature adipocytes. Redox Biol. 2019, 20, 489–495. [Google Scholar] [CrossRef]
- Randi, E.B.; Casili, G.; Jacquemai, S.; Szabo, C. Selenium-Binding Protein 1 (SELENBP1) Supports Hydrogen Sulfide Biosynthesis and Adipogenesis. Antioxidants 2021, 10, 361. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Minkler, P.; Grove, D.; Wang, R.; Willard, B.; Dweik, R.; Hine, C. Non-enzymatic hydrogen sulfide production from cysteine in blood is catalyzed by iron and vitamin B6. Commun. Biol. 2019, 2, 88–97. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).