Selenium Nanoparticles (SeNPs) Immunomodulation Is More Than Redox Improvement: Serum Proteomics and Transcriptomic Analyses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selenium Nanoparticles (SeNPs) Preparation and Characterization
2.2. SeNPs Diet and Trolox Diet Preparation
2.3. Fish Maintenance
2.4. Comparison of Immunomodulation Effects with SeNP and Trolox Diet
2.5. Comparison of Antioxidation Effects with SeNP and Trolox Diet
2.6. Serum Collection and Proteomic Sample Preparation
2.7. Serum Proteomics Analysis
2.8. Transcriptome Analysis
2.9. ICP-MS Analysis
2.10. Statistical Analyses
3. Results
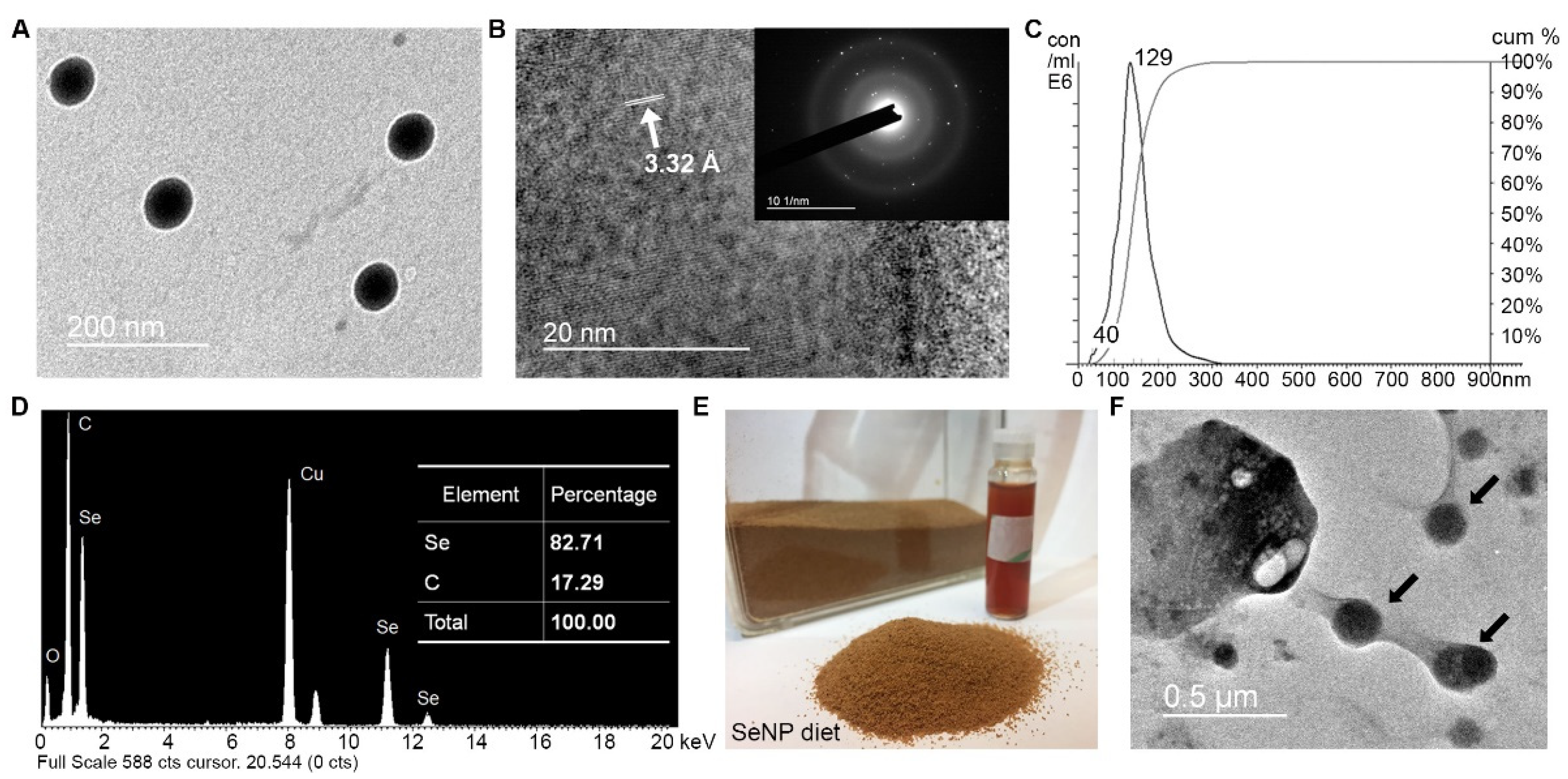
3.1. SeNPs and SeNP Diet Characterization
3.2. Comparison of Immunomodulation Effects with SeNP and Trolox Diet
3.3. Comparison of Antioxidation Effects with SeNP and Trolox Diet
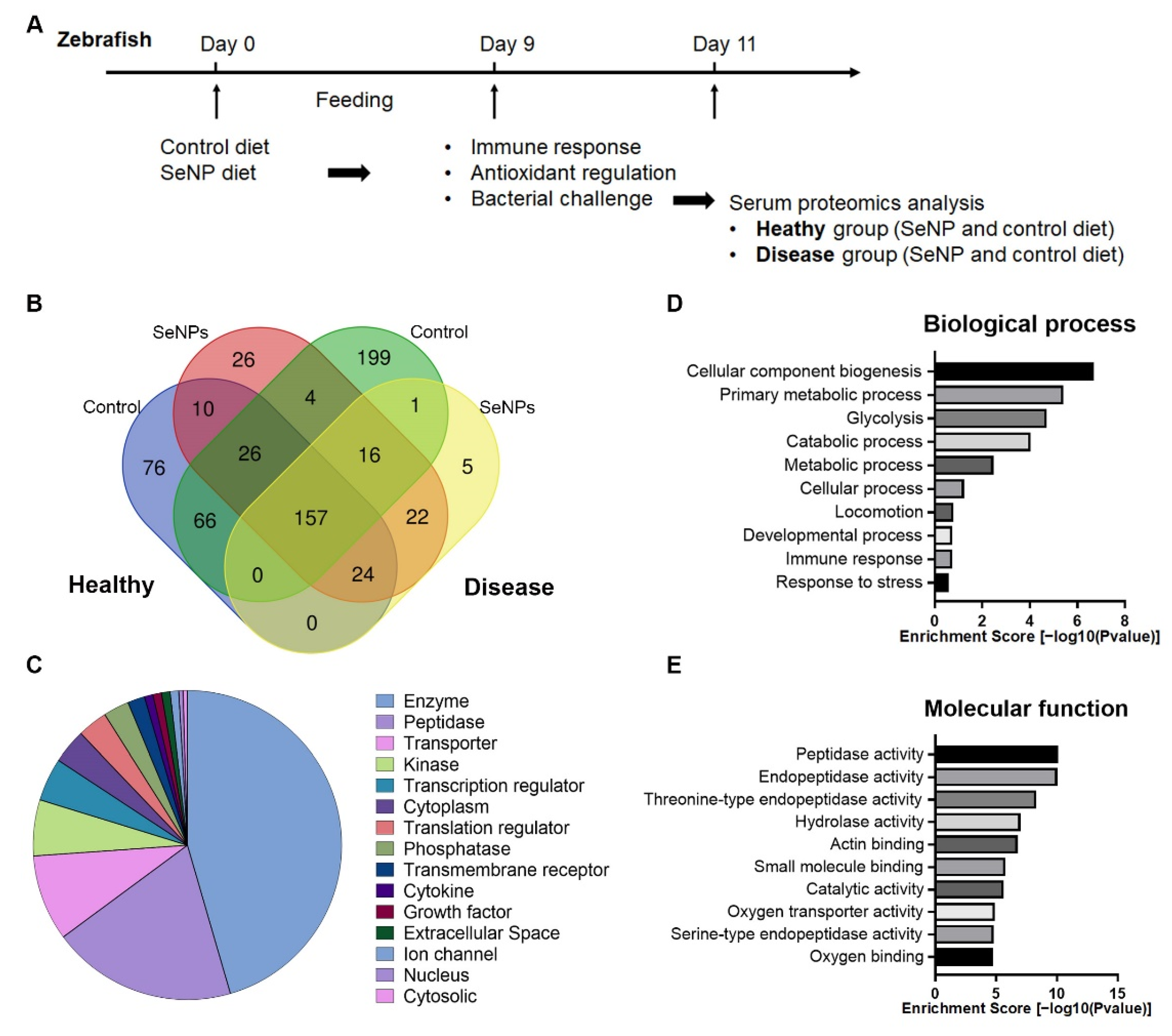
3.4. Serum Proteomic Analysis
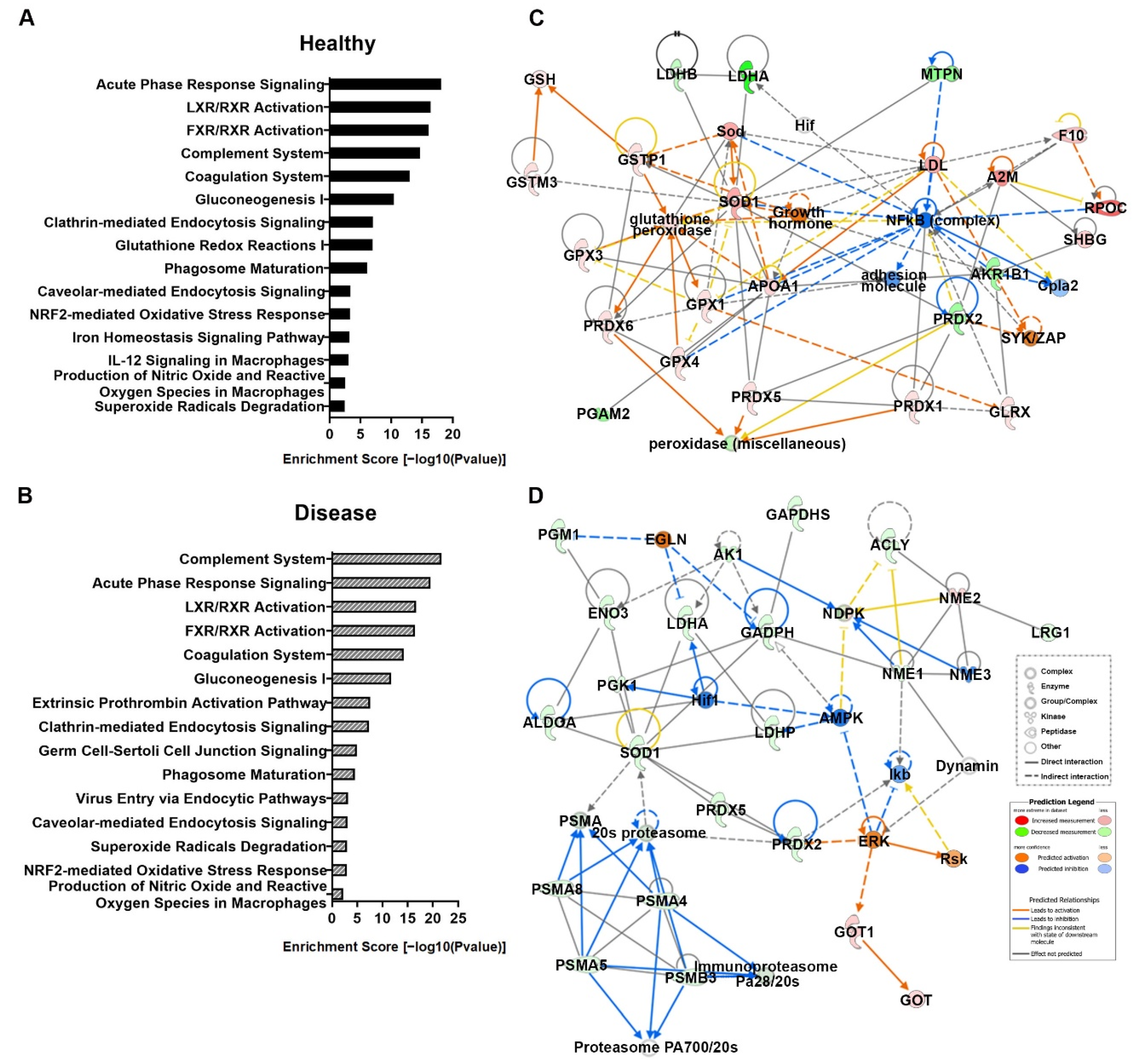
3.5. Canonical Pathway and Molecular Network Analyses from Serum Proteomics
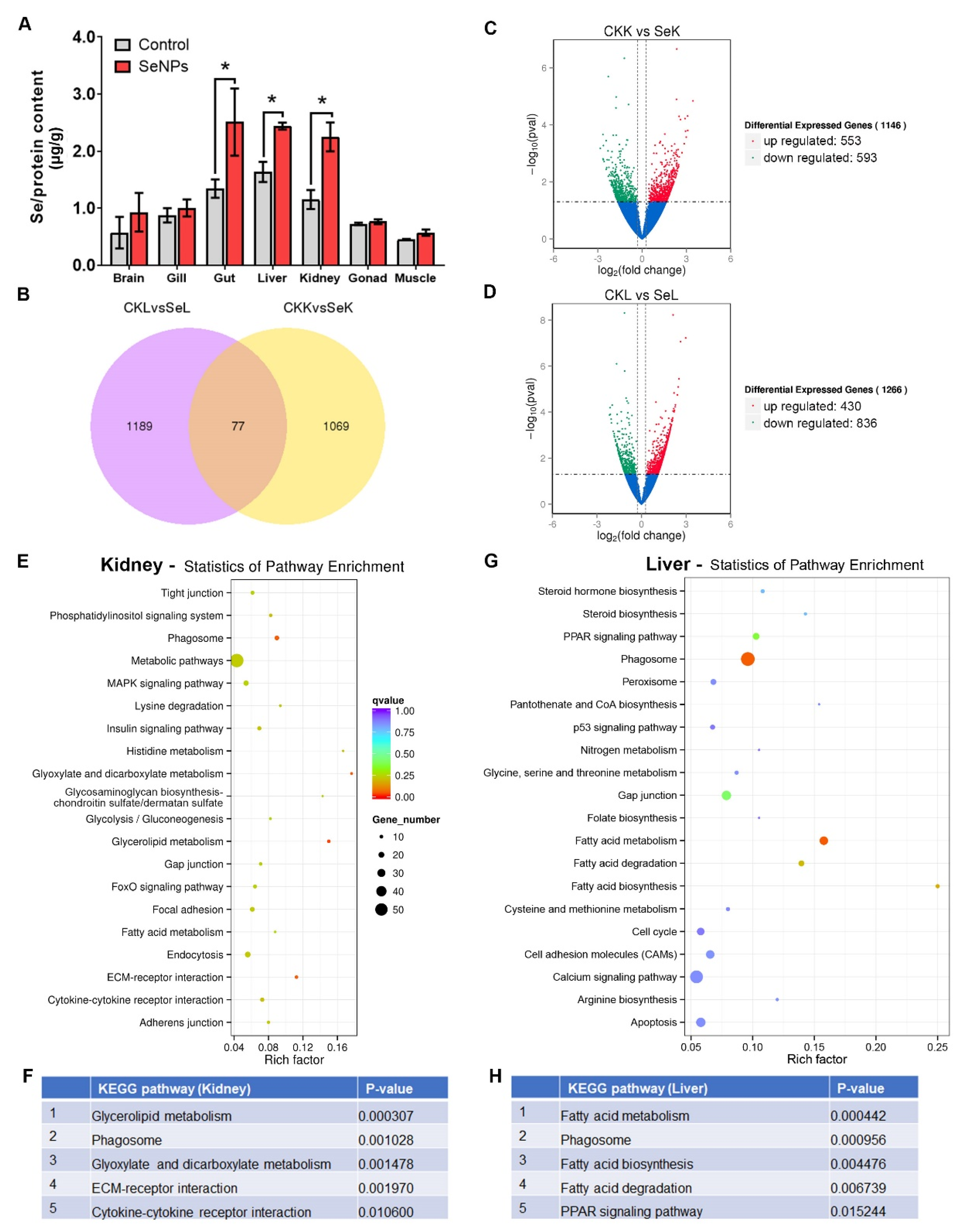
3.6. Uptake of Se and Transcriptomic Response of Kidney and Liver
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Short, S.P.; Williams, C.S. Selenoproteins in tumorigenesis and cancer progression. Adv. Cancer Res. 2017, 136, 49–83. [Google Scholar] [PubMed] [Green Version]
- Weekley, C.M.; Harris, H.H. Which form is that? The importance of selenium speciation and metabolism in the prevention and treatment of disease. Chem. Soc. Rev. 2013, 42, 8870–8894. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zhang, C.; Xia, I.F.; Cheung, S.T.; Wong, K.S.; Wong, K.H.; Au, D.W.T.; Hinton, D.E.; Kwok, K.W.H. Maternal dietary exposure to selenium nanoparticle led to malformation in offspring. Ecotoxicol. Environ. Saf. 2018, 156, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.L.; Li, X.L.; Liu, W.; Chen, T.F.; Li, Y.H.; Zheng, W.J.; Man, C.W.Y.; Wong, M.K.; Wong, K.H. Surface decoration of selenium nanoparticles by mushroom polysaccharides-protein complexes to achieve enhanced cellular uptake and antiproliferative activity. J. Mater. Chem. 2012, 22, 9602–9610. [Google Scholar] [CrossRef]
- Xia, I.F.; Cheung, J.S.; Wu, M.; Wong, K.S.; Kong, H.K.; Zheng, X.T.; Wong, K.H.; Kwok, K.W. Dietary chitosan-selenium nanoparticle (CTS-SeNP) enhance immunity and disease resistance in zebrafish. Fish Shellfish Immunol. 2019, 87, 449–459. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Azadmanesh, K.; Shokrgozar, M.A.; Journeay, W.S.; Laurent, S. Effect of nanoparticles on the cell life cycle. Chem. Rev. 2011, 111, 3407–3432. [Google Scholar] [CrossRef]
- Shamsi, M.M.; Chekachak, S.; Soudi, S.; Gharakhanlou, R.; Quinn, L.S.; Ranjbar, K.; Rezaei, S.; Shirazi, F.J.; Allahmoradi, B.; Yazdi, M.H.; et al. Effects of exercise training and supplementation with selenium nanoparticle on T-helper 1 and 2 and cytokine levels in tumor tissue of mice bearing the 4 T1 mammary carcinoma. Nutrition 2019, 57, 141–147. [Google Scholar] [CrossRef]
- Dawood, M.A.O.; Koshio, S.; Zaineldin, A.I.; Van Doan, H.; Moustafa, E.M.; Abdel-Daim, M.M.; Esteban, M.A.; Hassaan, M.S. Dietary supplementation of selenium nanoparticles modulated systemic and mucosal immune status and stress resistance of red sea bream (Pagrus major). Fish Physiol. Biochem. 2019, 45, 219–230. [Google Scholar] [CrossRef]
- Mahdavi, M.; Mavandadnejad, F.; Yazdi, M.H.; Faghfuri, E.; Hashemi, H.; Homayouni-Oreh, S.; Farhoudi, R.; Shahverdi, A.R. Oral administration of synthetic selenium nanoparticles induced robust Th1 cytokine pattern after HBs antigen vaccination in mouse model. J. Infect. Public Health 2017, 10, 102–109. [Google Scholar] [CrossRef] [Green Version]
- Skalickova, S.; Milosavljevic, V.; Cihalova, K.; Horky, P.; Richtera, L.; Adam, V. Selenium nanoparticles as a nutritional supplement. Nutrition 2017, 33, 83–90. [Google Scholar] [CrossRef]
- Torres, S.K.; Campos, V.L.; Leon, C.G.; Rodriguez-Llamazares, S.M.; Rojas, S.M.; Gonzalez, M.; Smith, C.; Mondaca, M.A. Biosynthesis of selenium nanoparticles by Pantoea agglomerans and their antioxidant activity. J. Nanoparticle Res. 2012, 14, 1236–1245. [Google Scholar] [CrossRef]
- Amin, K.A.; Hashem, K.S.; Alshehri, F.S.; Awad, S.T.; Hassan, M.S. Antioxidant and hepatoprotective efficiency of selenium nanoparticles against acetaminophen-induced hepatic damage. Biol. Trace Elem. Res. 2017, 175, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Puertollano, M.A.; Puertollano, E.; de Cienfuegos, G.A.; de Pablo, M.A. Dietary Antioxidants: Immunity and Host Defense. Curr. Top. Med. Chem. 2011, 11, 1752–1766. [Google Scholar] [CrossRef] [PubMed]
- Pattison, D.J.; Winyard, P.G. Dietary antioxidants in inflammatory arthritis: Do they have any role in etiology or therapy? Nat. Clin. Pract. Rheum. 2008, 4, 590–596. [Google Scholar] [CrossRef]
- Bennike, T.B.; Bellin, M.D.; Xuan, Y.; Stensballe, A.; Moller, F.T.; Beilman, G.J.; Leyy, O.; Cruz-Monserrate, Z.; Andersen, V.; Steen, J.; et al. A cost-effective high-throughput plasma and serum proteomics workflow enables mapping of the molecular impact of total pancreatectomy with islet autotransplantation. J. Proteome Res. 2018, 17, 1983–1992. [Google Scholar] [CrossRef]
- Nayak, S.; Portugal, I.; Zilberg, D. Analyzing complement activity in the serum and body homogenates of different fish species, using rabbit and sheep red blood cells. Vet. Immunol. Immunopathol. 2018, 199, 39–42. [Google Scholar] [CrossRef]
- Kopp, A.; Hebecker, M.; Svobodova, E.; Jozsi, M. Factor H: A complement regulator in health and disease, and a mediator of cellular interactions. Biomolecules 2012, 2, 46–75. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Sun, Q.Q.; Mao, W.L.; Chen, Y. Serum immunoglobulin A (IgA) level is a potential biomarker indicating cirrhosis during chronic hepatitis B infection. Gastroent. Res. Pract. 2016, 2016, 2495073. [Google Scholar] [CrossRef] [Green Version]
- Hajishengallis, G.; Reis, E.S.; Mastellos, D.C.; Ricklin, D.; Lambris, J.D. Novel mechanisms and functions of complement. Nat. Immunol. 2017, 18, 1288–1298. [Google Scholar] [CrossRef]
- Perssonmoschos, M.; Huang, W.; Srikumar, T.S.; Akesson, B.; Lindeberg, S. Selenoprotein-P in serum as a biochemical marker of selenium status. Analyst 1995, 120, 833–836. [Google Scholar] [CrossRef]
- Hubler, M.J.; Kennedy, A.J. Role of lipids in the metabolism and activation of immune cells. J. Nutr. Biochem. 2016, 34, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Rose, A.H.; Hoffmann, P.R. The role of selenium in inflammation and immunity: From molecular mechanisms to therapeutic opportunities. Antioxid. Redox Signal. 2012, 16, 705–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Liang, H.; Yi, J.; Tan, W.; He, S.; Wang, S.; Li, F.; Wu, X.; Ma, J.; Shi, X. Long-term selenium-deficient diet induces liver damage by altering hepatocyte ultrastructure and MMP1/3 and TIMP1/3 expression in growing rats. Biol. Trace Elem. Res. 2017, 175, 396–404. [Google Scholar] [CrossRef]
- Akahoshi, N.; Anan, Y.; Hashimoto, Y.; Tokoro, N.; Mizuno, R.; Hayashi, S.; Yamamoto, S.; Shimada, K.I.; Kamata, S.; Ishii, I. Dietary selenium deficiency or selenomethionine excess drastically alters organ selenium contents without altering the expression of most selenoproteins in mice. J. Nutr. Biochem. 2019, 69, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Li, S.; Zhang, K.; Li, J.; Han, Y.; Zhan, T.; Zhao, Q.; Guo, X.; Zhang, J. Selenium deficiency-induced redox imbalance leads to metabolic reprogramming and inflammation in the liver. Redox Biol. 2020, 36, 101519. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Han, Y.; Zhao, Q.; Zhan, T.; Li, Y.; Sun, W.; Li, S.; Sun, D.; Si, X.; Yu, X. Targeted metabolomics analysis reveals that dietary supranutritional selenium regulates sugar and acylcarnitine metabolism homeostasis in pig liver. J. Nutr. 2020, 150, 704–711. [Google Scholar] [CrossRef] [PubMed]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese Jr, R.V. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlberg, I.; Mannervik, B. Reduction of 2,4,6-trinitrobenzenesulfonate by glutathione reductase and the effect of NADP+ on the electron transfer. J. Biol. Chem. 1986, 261, 1629–1635. [Google Scholar] [CrossRef]
- Babaei, F.; Ramalingam, R.; Tavendale, A.; Liang, Y.; Yan, L.S.; Ajuh, P.; Cheng, S.H.; Lam, Y.W. Novel blood collection method allows plasma proteome analysis from single zebrafish. J. Proteome Res. 2013, 12, 1580–1590. [Google Scholar] [CrossRef]
- Sherrod, S.D.; Myers, M.V.; Li, M.; Myers, J.S.; Carpenter, K.L.; MacLean, B.; MacCoss, M.J.; Liebler, D.C.; Ham, A.J.L. Label-free quantitation of protein modifications by pseudo selected reaction monitoring with internal reference peptides. J. Proteome Res. 2012, 11, 3467–3479. [Google Scholar] [CrossRef]
- Kong, H.K.; Wong, M.H.; Chan, H.M.; Lo, S.C.L. Chronic exposure of adult rats to low doses of methylmercury induced a state of metabolic deficit in the somatosensory cortex. J. Proteome Res. 2013, 12, 5233–5245. [Google Scholar] [CrossRef] [PubMed]
- Trinh, H.V.; Grossmann, J.; Gehrig, P.; Roschitzki, B.; Schlapbach, R.; Greber, U.F.; Hemmi, S. iTRAQ-based and label-free proteomics approaches for studies of human adenovirus infections. Int. J. Proteom. 2013, 2013, 581862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wong, C.W.; Yan, M.; Li, L.; Liu, T.; Or, P.M.-Y.; Tsui, S.K.-W.; Waye, M.M.-Y.; Chan, A.M.-L. Differential regulation of the pro-inflammatory biomarker, YKL-40/CHI3L1, by PTEN/Phosphoinositide 3-kinase and JAK2/STAT3 pathways in glioblastoma. Cancer Lett. 2018, 429, 54–65. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B Met. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Fang, L.; Zheng, H.K.; Zhang, Y.; Chen, J.; Zhang, Z.J.; Wang, J.; Li, S.T.; Li, R.Q.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Chernick, M.; Ware, M.; Albright, E.; Kwok, K.W.H.; Dong, W.; Zheng, N.; Hinton, D.E. Parental dietary seleno-L-methionine exposure and resultant offspring developmental toxicity. Aquat. Toxicol. 2016, 170, 187–198. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Tan, X.F.; Lim, T.K.; Lin, Q.; Gong, Z. Comprehensive and quantitative proteomic analyses of zebrafish plasma reveals conserved protein profiles between genders and between zebrafish and human. Sci. Rep. 2016, 6, 24329. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.L.; Guo, Y.; Qiao, L.; Ma, L.; Cheng, Y.Y.; Roman, A. Biogenic synthesis of novel functionalized selenium nanoparticles by Lactobacillus casei ATCC 393 and its protective effects on intestinal barrier dysfunction caused by enterotoxigenic Escherichia coli K88. Front. Microbiol. 2018, 9, 1129. [Google Scholar] [CrossRef] [Green Version]
- Ungvari, E.; Monori, I.; Megyeri, A.; Csiki, Z.; Prokisch, J.; Sztrik, A.; Javor, A.; Benko, I. Protective effects of meat from lambs on selenium nanoparticle supplemented diet in a mouse model of polycyclic aromatic hydrocarbon-induced immunotoxicity. Food Chem. Toxicol. 2014, 64, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Cray, C.; Zaias, J.; Altman, N.H. Acute phase response in animals: A review. Comp. Med. 2009, 59, 517–526. [Google Scholar] [PubMed]
- Méplan, C.; Johnson, I.T.; Polley, A.C.; Cockell, S.; Bradburn, D.M.; Commane, D.M.; Arasaradnam, R.P.; Mulholland, F.; Zupanic, A.; Mathers, J.C. Transcriptomics and proteomics show that selenium affects inflammation, cytoskeleton, and cancer pathways in human rectal biopsies. FASEB J. 2016, 30, 2812–2825. [Google Scholar] [CrossRef] [Green Version]
- Pillai, S.S.; Sugathan, J.K.; Indira, M. Selenium downregulates RAGE and NFκB expression in diabetic rats. Biol. Trace Elem. Res. 2012, 149, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Francian, A.; Mann, K.; Kullberg, M. Complement C3-dependent uptake of targeted liposomes into human macrophages, B cells, dendritic cells, neutrophils, and MDSCs. Int. J. Nanomed. 2017, 12, 5149–5161. [Google Scholar] [CrossRef] [Green Version]
- Puchau, B.; Zulet, M.A.; de Echavarri, A.G.; Navarro-Blasco, I.; Martinez, J.A. Selenium intake reduces serum C3, an early marker of metabolic syndrome manifestations, in healthy young adults. Eur. J. Clin. Nutr. 2009, 63, 858–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avery, J.C.; Hoffmann, P.R. Selenium, selenoproteins, and immunity. Nutrients 2018, 10, 1203. [Google Scholar] [CrossRef] [Green Version]
- Gan, L.; Liu, Q.O.; Xu, H.B.; Zhu, Y.S.; Yang, X.L. Effects of selenium overexposure on glutathione peroxidase and thioredoxin reductase gene expressions and activities. Biol. Trace Elem. Res. 2002, 89, 165–175. [Google Scholar] [CrossRef]
- Wu, G.Y.; Fang, Y.Z.; Yang, S.; Lupton, J.R.; Turner, N.D. Glutathione metabolism and its implications for health. J. Nutr. 2004, 134, 489–492. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Ren, Z.; Zhang, J.; Chuang, C.C.; Kandaswamy, E.; Zhou, T.; Zuo, L. Role of ROS and nutritional antioxidants in human diseases. Front. Physiol. 2018, 9, 477. [Google Scholar] [CrossRef] [Green Version]
- Azadmanesh, J.; Borgstahl, G.E.O. A review of the catalytic mechanism of human manganese superoxide dismutase. Antioxidants 2018, 7, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosnedlova, B.; Kepinska, M.; Skalickova, S.; Fernandez, C.; Ruttkay-Nedecky, B.; Peng, Q.M.; Baron, M.; Melcova, M.; Opatrilova, R.; Zidkova, J.; et al. Nano-selenium and its nanomedicine applications: A critical review. Int. J. Nanomed. 2018, 13, 2107–2128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Meo, S.; Napolitano, G.; Venditti, P. Physiological and pathological role of ROS: Benefits and limitations of antioxidant treatment. Int. J. Mol. Sci. 2019, 20, 4810. [Google Scholar] [CrossRef] [Green Version]
- Ristow, M. Unraveling the truth about antioxidants: Mitohormesis explains ROS-induced health benefits. Nat. Med. 2014, 20, 709–711. [Google Scholar] [CrossRef]
- Wowk, P.F.; Zardo, M.L.; Miot, H.T.; Goldenberg, S.; Carvalho, P.C.; Morking, P.A. Proteomic profiling of extracellular vesicles secreted from Toxoplasma gondii. Proteomics 2017, 17, 15–16. [Google Scholar] [CrossRef]
- Li, Y.H.; Lin, Z.F.; Guo, M.; Xia, Y.; Zhao, M.Q.; Wang, C.B.; Xu, T.T.; Chen, T.F.; Zhu, B. Inhibitory activity of selenium nanoparticles functionalized with oseltamivir on H1N1 influenza virus. Int. J. Nanomed. 2017, 12, 5733–5743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferro, C.; Florindo, H.F.; Santos, H.A. Selenium Nanoparticles for Biomedical Applications: From Development and Characterization to Therapeutics. Adv. Healthc. Mater. 2021, 10, 2100598. [Google Scholar] [CrossRef]
- Foroozandeh, P.; Aziz, A.A. Insight into cellular uptake and intracellular trafficking of nanoparticles. Nanoscale Res. Lett. 2018, 13, 339. [Google Scholar] [CrossRef]
- de Salamanca, A.E.; Diebold, Y.; Calonge, M.; Garcia-Vazquez, C.; Callejo, S.; Vila, A.; Alonso, M.J. Chitosan nanoparticles as a potential drug delivery system for the ocular surface: Toxicity, uptake mechanism and in vivo tolerance. Investig. Ophthalmol. Vis. Sci. 2006, 47, 1416–1425. [Google Scholar] [CrossRef]

| Analysis | ID | Molecules in Network | Score | Focus Molecules | Top Diseases and Functions |
|---|---|---|---|---|---|
| Healthy (SeNP diet vs. Control diet) | 1 | A2M, adhesion molecule, AKR1B1, APOA1, Cg, Cpla2, F10, GGCT, GLRX, glutathione peroxidase, GPX1, GPX3, GPX4, Growth hormone, GST, GSTM3, GSTP1, Hif, Ldh (complex), LDHA, LDHB, LDL, MTPN, NFkβ (complex), peroxidase (miscellaneous), PGAM2, PRDX1, PRDX2, PRDX5, PRDX6, PROC, SHBG, Sod, SOD1, SYK/ZAP | 45 | 22 | Free Radical Scavenging, Small Molecule Biochemistry, Drug Metabolism |
| 2 | AHSG, Alpha 1 antitrypsin, APOA4, APOB, BHMT, Ces, CES1, CETP, CFH, CP, creatine kinase, ERK1/2, ESD, Ferritin, FGA, GC, HDL, HDL-cholesterol, hemoglobin, HPX, ITIH4, ITLN1, MHC Class II (complex), Nos, Nr1h, PRKAA, RBP4, SERPINA1, SERPINA9, SERPINC1, SERPINF2, SH3BGRL, TF, VLDL, VLDL-cholesterol | 43 | 21 | Lipid Metabolism, Small Molecule Biochemistry, Vitamin and Mineral Metabolism | |
| 3 | Akt, C3, C5, C6, C7, C8, C1q, C3-Cfb, C5-C6-C7, C5-C6-C7-C8, C5-C6-C7-C8-C9, C8A, C8B, C8G, CFI, Collagen Alpha1, Collagen type IV, Collagen(s), Complement, Complement component 1, CSTB, elastase, Fibrin, Fibrinogen, HABP2, HBE1, HBZ, Kallikrein, Laminin (complex), LRG1, MAC, NME3, Pdi, PLG, SERPING1 | 30 | 16 | Immunological Disease, Developmental Disorder, Hereditary Disorder | |
| Disease (SeNP diet vs. Control diet) | 1 | 20s proteasome, ACLY, AK1, ALDOA, AMPK, Dynamin, EGLN, ENO3, ERK, GAPDH, GAPDHS, GOT, GOT1, Hif1, Ikb, Immunoproteasome Pa28/20s, LDHA, LDHB, LRG1, NDPK, NME1, NME2, NME3, PGK1, PGM1, PRDX2, PRDX5, Proteasome PA700/20s, PSMA, PSMA4, PSMA5, PSMA8, PSMB3, Rsk, SOD1 | 44 | 22 | Carbohydrate Metabolism, Nucleic Acid Metabolism, Small Molecule Biochemistry |
| 2 | AHSG, Alpha 1 antitrypsin, AMBP, APOA4, APOB, CES1, CETP, CFH, CP, creatine kinase, ERK1/2, Ferritin, FGA, GC, HBZ, HDL, HDL-cholesterol, hemoglobin, HPX, Iti, ITIH3, ITIH4, ITLN1, MHC Class II (complex), Nos, Nr1h, PRKAA, RBP4, SERPINA1, SERPINA9, SERPINC1, SERPINF2, TF, VLDL, VLDL-cholesterol | 41 | 21 | Lipid Metabolism, Small Molecule Biochemistry, Vitamin and Mineral Metabolism | |
| 3 | A2M, C6, C7, C3-Cfb, C5-C6-C7, C5-C6-C7-C8, C5-C6-C7-C8-C9, C8A, C8B, CAPNS1, CFB, CFD, CFI, CFP, chymotrypsin, coagulation factor, CORO1A, Ecm, elastase, F9, F10, glutathione peroxidase, Hif, Kallikrein, Ldh (complex), MASP2, NFkβ (complex), PAPSS2, PLG, PROC, PRSS2, Serine Protease, SERPING1, SOD3, trypsin | 38 | 20 | Immunological Disease, Developmental Disorder, Hereditary Disorder |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, I.F.; Kong, H.-K.; Wu, M.M.H.; Lu, Y.; Wong, K.-H.; Kwok, K.W.H. Selenium Nanoparticles (SeNPs) Immunomodulation Is More Than Redox Improvement: Serum Proteomics and Transcriptomic Analyses. Antioxidants 2022, 11, 964. https://doi.org/10.3390/antiox11050964
Xia IF, Kong H-K, Wu MMH, Lu Y, Wong K-H, Kwok KWH. Selenium Nanoparticles (SeNPs) Immunomodulation Is More Than Redox Improvement: Serum Proteomics and Transcriptomic Analyses. Antioxidants. 2022; 11(5):964. https://doi.org/10.3390/antiox11050964
Chicago/Turabian StyleXia, Ivan Fan, Hang-Kin Kong, Margaret M. H. Wu, Yishan Lu, Ka-Hing Wong, and Kevin W. H. Kwok. 2022. "Selenium Nanoparticles (SeNPs) Immunomodulation Is More Than Redox Improvement: Serum Proteomics and Transcriptomic Analyses" Antioxidants 11, no. 5: 964. https://doi.org/10.3390/antiox11050964
APA StyleXia, I. F., Kong, H.-K., Wu, M. M. H., Lu, Y., Wong, K.-H., & Kwok, K. W. H. (2022). Selenium Nanoparticles (SeNPs) Immunomodulation Is More Than Redox Improvement: Serum Proteomics and Transcriptomic Analyses. Antioxidants, 11(5), 964. https://doi.org/10.3390/antiox11050964





