Arginine-Dependent Nitric Oxide Generation and S-Nitrosation in the Non-Photosynthetic Unicellular Alga Polytomella parva
Abstract
1. Introduction
2. Materials and Methods
2.1. Algal Strain and Cultivation Conditions
2.2. Determination of Arginine Contents
2.3. Determination of Putrescine and Ornithine and Contents
2.4. Measurement of NO
2.5. Confocal Microscopy
2.6. Detection of Protein S-NO Groups
2.7. Protein Extraction and S-Nitrosated Protein Labeling
2.8. Labeled-Protein SDS-PAGE and Western Blotting
2.9. Statistical Analysis
3. Results
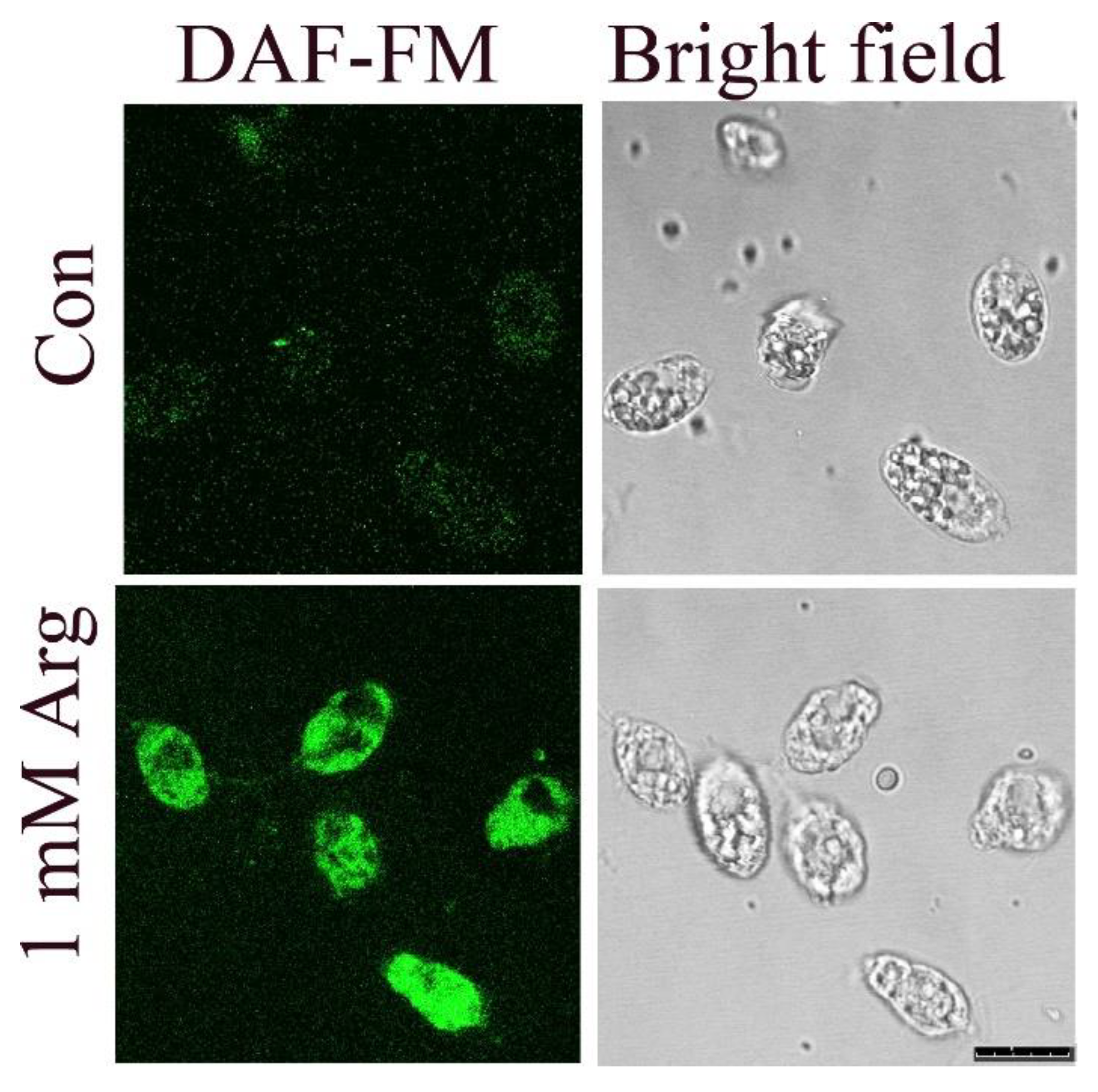
3.1. Effect of L-Arginine Supplementation on NO Production
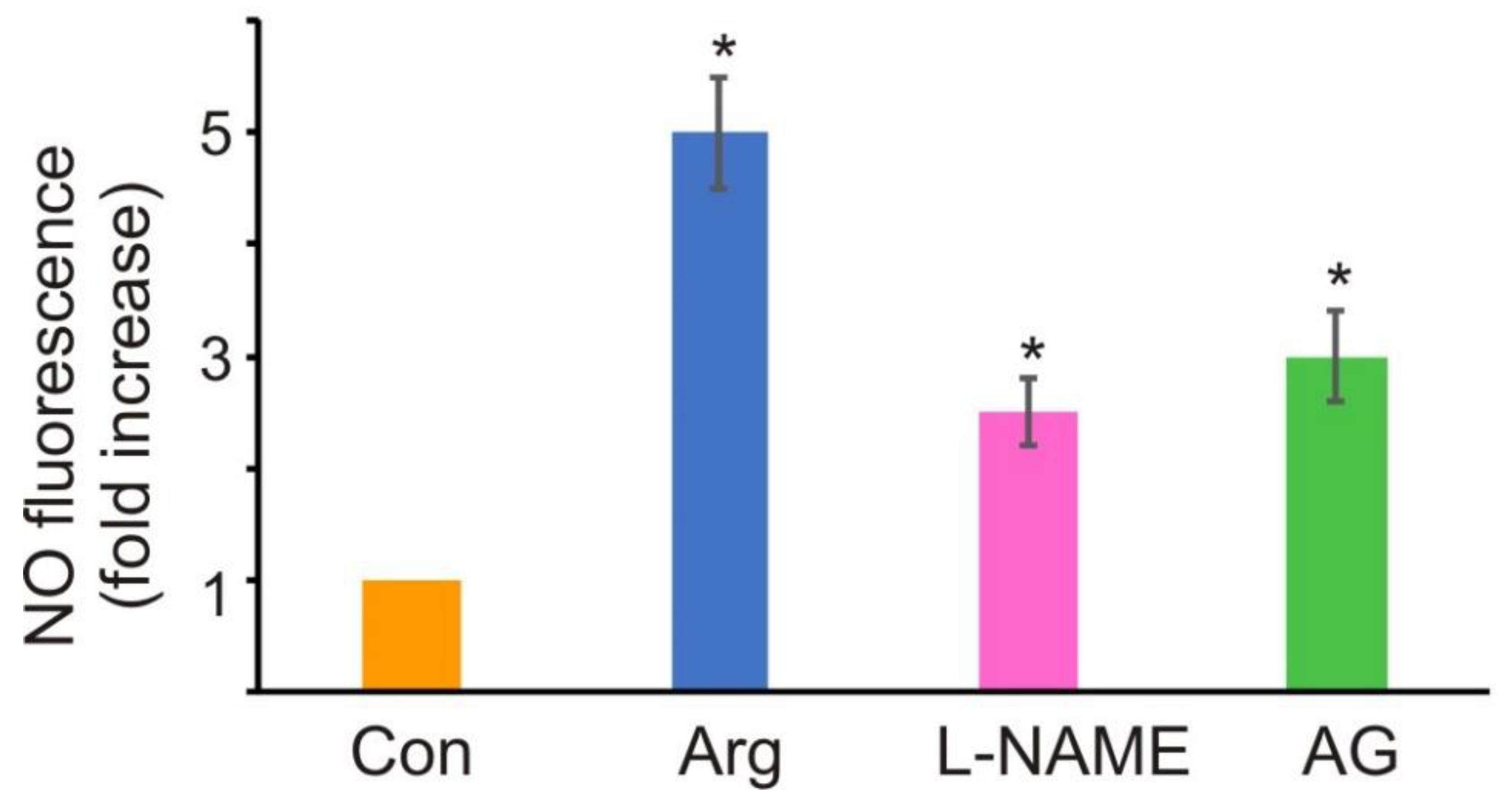
3.2. Source of NO
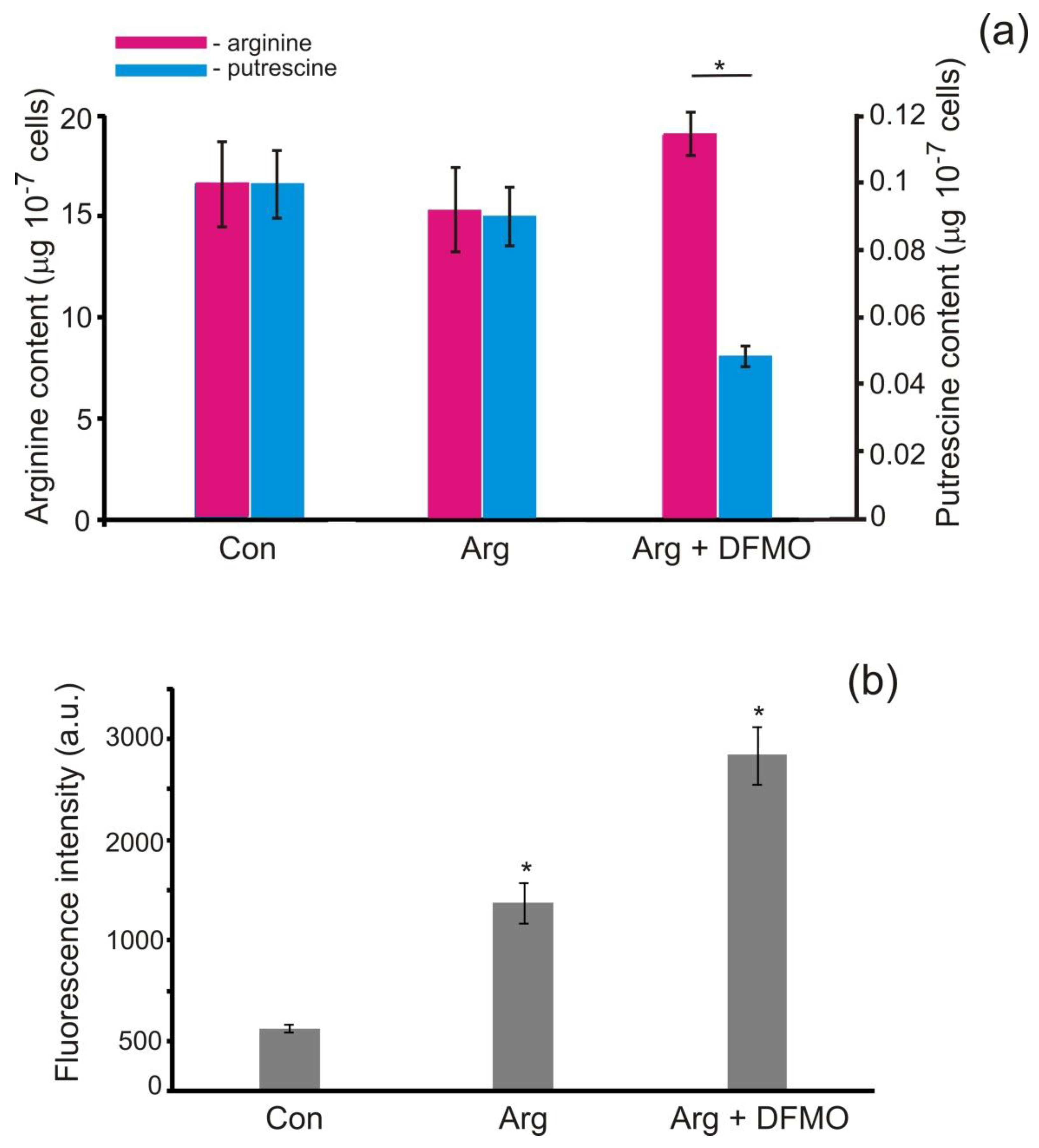
3.3. Putrescine Is Formed in P. parva
3.4. S-Nitrosation of Proteins by NO
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Del Castello, F.; Nejamkin, A.; Cassia, R.; Correa-Aragunde, N.; Fernández, B.; Foresi, N.; Lombardo, C.; Ramirez, L.; Lamattina, L. The era of nitric oxide in plant biology: Twenty years tying up loose ends. Nitric Oxide Biol. Chem. 2019, 85, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Wendehenne, D.; Hancock, J.T. New frontiers in nitric oxide biology in plant. Plant Sci. 2011, 181, 507–508. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Leterrier, M.; Valderrama, R.; Airaki, M.; Chaki, M.; Palma, J.M.; Barroso, J.B. Nitric oxide imbalance provokes a nitrosative response in plants under abiotic stress. Plant Sci. 2011, 181, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.; Prats, E.; Pierre, S.; Hall, M.A.; Hebelstrup, K.H. Integrating nitric oxide into salicylic acid and jasmonic acid/ethylene plant defense pathways. Front. Plant Sci. 2013, 4, 215. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.-E.; Hamdan, M.F.; Pua, T.L.; Saidi, N.B.; Tan, B.C. Plant nitric oxide signaling under drought stress. Plants 2021, 10, 360. [Google Scholar] [CrossRef]
- Astier, J.; Gross, I.; Durner, J. Nitric oxide production in plants: An update. J. Exp. Bot. 2018, 69, 3401–3411. [Google Scholar] [CrossRef]
- Gupta, K.J.; Igamberdiev, A.U.; Kaiser, W.M. New insights into the mitochondrial nitric oxide production pathways. Plant Signal. Behav. 2010, 5, 999–1001. [Google Scholar] [CrossRef][Green Version]
- Chamizo-Ampudia, A.; Sanz-Luque, E.; Llamas, Á.; Ocaña-Calahorro, F.; Mariscal, V.; Carreras, A.; Barroso, J.B.; Galván, A.; Fernández, E. A dual system formed by the ARC and NR molybdoenzymes mediates nitrite-dependent NO production in Chlamydomonas. Plant Cell Environ. 2016, 39, 2097–2107. [Google Scholar] [CrossRef]
- Moncada, S.; Palmer, R.M.; Higgs, E.A. Biosynthesis of nitric oxide from L-arginine. A pathway for the regulation of cell function and communication. Biochem. Pharmacol. 1989, 38, 1709–1715. [Google Scholar] [CrossRef]
- Santolini, J. The molecular mechanism of mammalian NO-synthases: A story of electrons and protons. J. Inorg. Biochem. 2011, 105, 127–141. [Google Scholar] [CrossRef]
- Foresi, N.; Correa-Aragunde, N.; Parisi, G.; Caló, G.; Salerno, G.; Lamattina, L. Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell 2010, 22, 3816–3830. [Google Scholar] [CrossRef] [PubMed]
- Jeandroz, S.; Wipf, D.; Stuehr, D.J.; Lamattina, L.; Melkonian, M.; Tian, Z.; Zhu, Y.; Carpenter, E.J.; Wong, G.K.S.; Wendehenne, D. Occurrence, structure, and evolution of nitric oxide synthase-like proteins in the plant kingdom. Sci. Signal. 2016, 9, re2. [Google Scholar] [CrossRef] [PubMed]
- Tun, N.N.; Santa-Catarina, C.; Begum, T.; Silveira, V.; Handro, W.; Floh, E.I.; Scherer, G.F. Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol. 2006, 47, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Astier, J.; Mounier, A.; Santolini, J.; Jeandroz, S.; Wendehenne, D. The evolution of nitric oxide signalling diverges between animal and green lineages. J. Exp. Bot. 2019, 70, 4355–4364. [Google Scholar] [CrossRef] [PubMed]
- Besson-Bard, A.; Pugin, A.; Wendehenne, D. New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 2008, 59, 21–39. [Google Scholar] [CrossRef]
- Hancock, J.T. Considerations of the importance of redox state for reactive nitrogen species action. J. Exp. Bot. 2019, 70, 4323–4331. [Google Scholar] [CrossRef]
- Morisse, S.; Zaffagnini, M.; Gao, X.H.; Lemaire, S.D.; Marchand, C.H. Insight into protein S-nitrosylation in Chlamydomonas reinhardtii. Antioxid. Redox Signal. 2014, 21, 1271–1284. [Google Scholar] [CrossRef]
- Berger, H.; De Mia, M.; Morisse, S.; Marchand, C.H.; Lemaire, S.D.; Wobbe, L.; Kruse, O. A light switch based on protein S-nitrosylation fine-tunes photosynthetic light harvesting in Chlamydomonas. Plant Physiol. 2016, 171, 821–832. [Google Scholar] [CrossRef]
- Chen, X.; Tian, D.; Kong, X.; Chen, Q.; Abd_Allah, E.F.; Hu, X.; Jia, A. The role of nitric oxide signalling in response to salt stress in Chlamydomonas reinhardtii. Planta 2016, 244, 651–669. [Google Scholar] [CrossRef]
- Smith, D.R.; Lee, R.W. A plastid without a genome: Evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiol. 2014, 164, 1812–1819. [Google Scholar] [CrossRef]
- Selim, K.; Lapina, T.; Forchhammer, K.; Ermilova, E. Interaction of N-acetyl-l-glutamate kinase with the PII signal transducer in the nonphotosynthetic alga Polytomella parva: Co-evolution towards a hetero-oligomeric enzyme. FEBS J. 2020, 287, 465–482. [Google Scholar] [CrossRef] [PubMed]
- Lapina, T.V.; Kochemasova, L.Y.; Forchhammer, K.; Ermilova, E.V. Effects of arginine on Polytomella parva growth, PII protein levels and lipid body formation. Planta 2019, 250, 1379–1385. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Ramírez, E.O.; Vázquez-Acevedo, M.; Cabrera-Orefice, A.; Guerrero-Castillo, S.; González-Halphen, D. The plastid proteome of the nonphotosynthetic chlorophycean alga Polytomella parva. Microbiol. Res. 2021, 243, 126649. [Google Scholar] [CrossRef] [PubMed]
- Atteia, A.; van Lis, R.; Ramírez, J.; González-Halphen, D. Polytomella spp. growth on ethanol. Extracellular pH affects the accumulation of mitochondrial cytochrome c550. FEBS J. 2000, 267, 2850–2858. [Google Scholar]
- Baker, C.J.; Mock, N.M. An improved method for monitoring cell death in cell suspension and leaf disc assays using Evans blue. Plant Cell Tissue Organ Cult. 1994, 39, 7–12. [Google Scholar] [CrossRef]
- Sakaguchi, S. A new method for the colorimetric determination of arginine. J. Biochem. 1950, 37, 231–236. [Google Scholar] [CrossRef]
- Zalutskaya, Z.; Derkach, V.; Puzanskiy, R.; Ermilova, E. Impact of nitric oxide on proline and putrescine biosynthesis in Chlamydomonas via transcriptional regulation. Biol. Plant. 2020, 64, 653–659. [Google Scholar] [CrossRef]
- Puzanskiy, R.; Tarakhovskaya, E.; Shavarda, A.; Shishova, M. Metabolomic and physiological changes of Chlamydomonas reinhardtii (Chlorophyceae, Chlorophyta) during batch culture development. J. Appl. Phycol. 2017, 30, 803–818. [Google Scholar] [CrossRef]
- Johnsen, L.G.; Skou, P.B.; Khakimov, B.; Bro, R. Gas chromatography—Mass spectrometry data processing made easy. J. Chromatogr. A 2017, 1503, 57–64. [Google Scholar] [CrossRef]
- Ma, Z.; Marsolais, F.; Bykova, N.V.; Igamberdiev, A.U. Nitric oxide and reactive oxygen species mediate metabolic changes in barley seed embryo during germination. Front. Plant Sci. 2016, 7, 138. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Weisslocker-Schaetzel, M.; André, F.; Touazi, N.; Foresi, N.; Lembrouk, M.; Dorlet, P.; Frelet-Barrand, A.; Lamattina, L.; Santolini, J. The NOS-like protein from the microalgae Ostreococcus tauri is a genuine and ultrafast NO-producing enzyme. Plant Sci. 2017, 265, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Palma, J.M.; Del Río, L.A.; Barroso, J.B. Evidence supporting the existence of L-arginine-dependent nitric oxide synthase activity in plants. New Phytol. 2009, 184, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Slocum, R.D. Tissue and subcellular localization of polyamines and enzymes of polyamine metabolism. In Biochemistry and Physiology of Polyamines in Plants; Slocum, R.D., Flores, H.E., Eds.; CRC Press: Boca Raton, FL, USA, 1991; pp. 23–40. [Google Scholar]
- Astier, J.; Rossi, J.; Chatelain, P.; Klinguer, A.; Besson-Bard, A.; Rosnoblet, C.; Jeandroz, S.; Nicolas-Francès, V.; Wendehenne, D. Nitric oxide production and signalling in algae. J. Exp. Bot. 2021, 72, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Docimo, T.; Reichelt, M.; Schneider, B.; Al, E. The first step in the biosynthesis of cocaine in Erythroxylum coca: The characterization of arginine and ornithine decarboxylases. Plant Mol. Biol. 2012, 78, 599–615. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine function in plants: Metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 2019, 9, 1945. [Google Scholar] [CrossRef] [PubMed]
- Voigt, J.; Deinert, B.; Bohley, P. Subcellular localization and light-dark control of ornithine decarboxylase in the unicellular green alga Chlamydomonas reinhardtii. Physiol. Plant. 2003, 108, 353–360. [Google Scholar] [CrossRef]
- Yamamoto, A.; Shim, I.S.; Fujihara, S. Inhibition of putrescine biosynthesis enhanced salt stress sensitivity and decreased spermidine content in rice seedlings. Biol. Plant. 2016, 61, 385–388. [Google Scholar] [CrossRef]
- Grinko, A.; Alqoubaili, R.; Lapina, T.; Ermilova, E. Truncated hemoglobin 2 modulates phosphorus deficiency response by controlling of gene expression in nitric oxide-dependent pathway in Chlamydomonas reinhardtii. Planta 2021, 254, 39. [Google Scholar] [CrossRef]
- Hemschemeier, A.; Düner, M.; Casero, D.; Merchant, S.S.; Winkler, M.; Happe, T. Hypoxic survival requires a 2-on-2 hemoglobin in a process involving nitric oxide. Proc. Natl. Acad. Sci. USA 2013, 110, 10854–10859. [Google Scholar] [CrossRef]
- Galatro, A.; Ramos-Artuso, F.; Luquet, M.; Buet, A.; Simontacchi, M. An update on nitric oxide production and role under phosphorus scarcity in plants. Front. Plant Sci. 2020, 11, 413. [Google Scholar] [CrossRef] [PubMed]
- Wimalasekera, R.; Tebartz, F.; Scherer, G.F. Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci. 2011, 181, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Zhang, Y.; Liu, L.; Liu, X.; Li, B.; Jin, C.; Lin, X. Molecular functions of nitric oxide and its potential applications in horticultural crops. Hortic. Res. 2021, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Chatelain, P.; Astier, J.; Wendehenne, D.; Rosnoblet, C.; Jeandroz, S. Identification of partner proteins of the algae Klebsormidium nitens NO synthases: Toward a better understanding of NO signaling in eukaryotic photosynthetic organisms. Front. Plant Sci. 2021, 12, 797451. [Google Scholar] [CrossRef]
- De Mia, M.; Lemaire, S.D.; Choquet, Y.; Wollman, F.A. Nitric oxide remodels the photosynthetic apparatus upon S-starvation in Chlamydomonas reinhardtii. Plant Physiol. 2019, 179, 718–731. [Google Scholar] [CrossRef]
- Kumar, A.; Castellano, I.; Patti, F.P.; Palumbo, A.; Buia, M.C. Nitric oxide in marine photosynthetic organisms. Nitric Oxide 2015, 47, 34–39. [Google Scholar] [CrossRef]
- Fröhlich, A.; Durner, J. The hunt for plant nitric oxide synthase (NOS): Is one really needed? Plant Sci. 2011, 181, 401–404. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B. Nitric oxide synthase-like activity in higher plants. Nitric Oxide 2017, 68, 5–6. [Google Scholar] [CrossRef]
- Zaffagnini, M.; De Mia, M.; Morisse, S.; Di Giacinto, N.; Marchand, C.H.; Maes, A.; Lemaire, S.D.; Trost, P. Protein S-nitrosylation in photosynthetic organisms: A comprehensive overview with future perspectives. Biochim. Biophys. Acta 2016, 1864, 952–966. [Google Scholar] [CrossRef]
- Astier, J.; Kulik, A.; Koen, E.; Besson-Bard, A.; Bourque, S.; Jeandroz, S.; Lamotte, O.; Wendehenne, D. Protein S-nitrosylation: What’s going on in plants? Free Radic. Biol. Med. 2012, 53, 1101–1110. [Google Scholar] [CrossRef]
- De Pinto, M.C.; Locato, V.; Sgobba, A.; Romero-Puertas, M.D.; Gadaleta, C.; Delledonne, M.; De Gara, L. S-Nitrosylation of ascorbate peroxidase is part of programmed cell death signaling in tobacco bright yellow-2 cells. Plant Physiol. 2013, 163, 1766–1775. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lapina, T.; Statinov, V.; Puzanskiy, R.; Ermilova, E. Arginine-Dependent Nitric Oxide Generation and S-Nitrosation in the Non-Photosynthetic Unicellular Alga Polytomella parva. Antioxidants 2022, 11, 949. https://doi.org/10.3390/antiox11050949
Lapina T, Statinov V, Puzanskiy R, Ermilova E. Arginine-Dependent Nitric Oxide Generation and S-Nitrosation in the Non-Photosynthetic Unicellular Alga Polytomella parva. Antioxidants. 2022; 11(5):949. https://doi.org/10.3390/antiox11050949
Chicago/Turabian StyleLapina, Tatiana, Vladislav Statinov, Roman Puzanskiy, and Elena Ermilova. 2022. "Arginine-Dependent Nitric Oxide Generation and S-Nitrosation in the Non-Photosynthetic Unicellular Alga Polytomella parva" Antioxidants 11, no. 5: 949. https://doi.org/10.3390/antiox11050949
APA StyleLapina, T., Statinov, V., Puzanskiy, R., & Ermilova, E. (2022). Arginine-Dependent Nitric Oxide Generation and S-Nitrosation in the Non-Photosynthetic Unicellular Alga Polytomella parva. Antioxidants, 11(5), 949. https://doi.org/10.3390/antiox11050949







