Capsicum Leaves under Stress: Using Multi-Omics Analysis to Detect Abiotic Stress Network of Secondary Metabolism in Two Species
Abstract
:1. Introduction
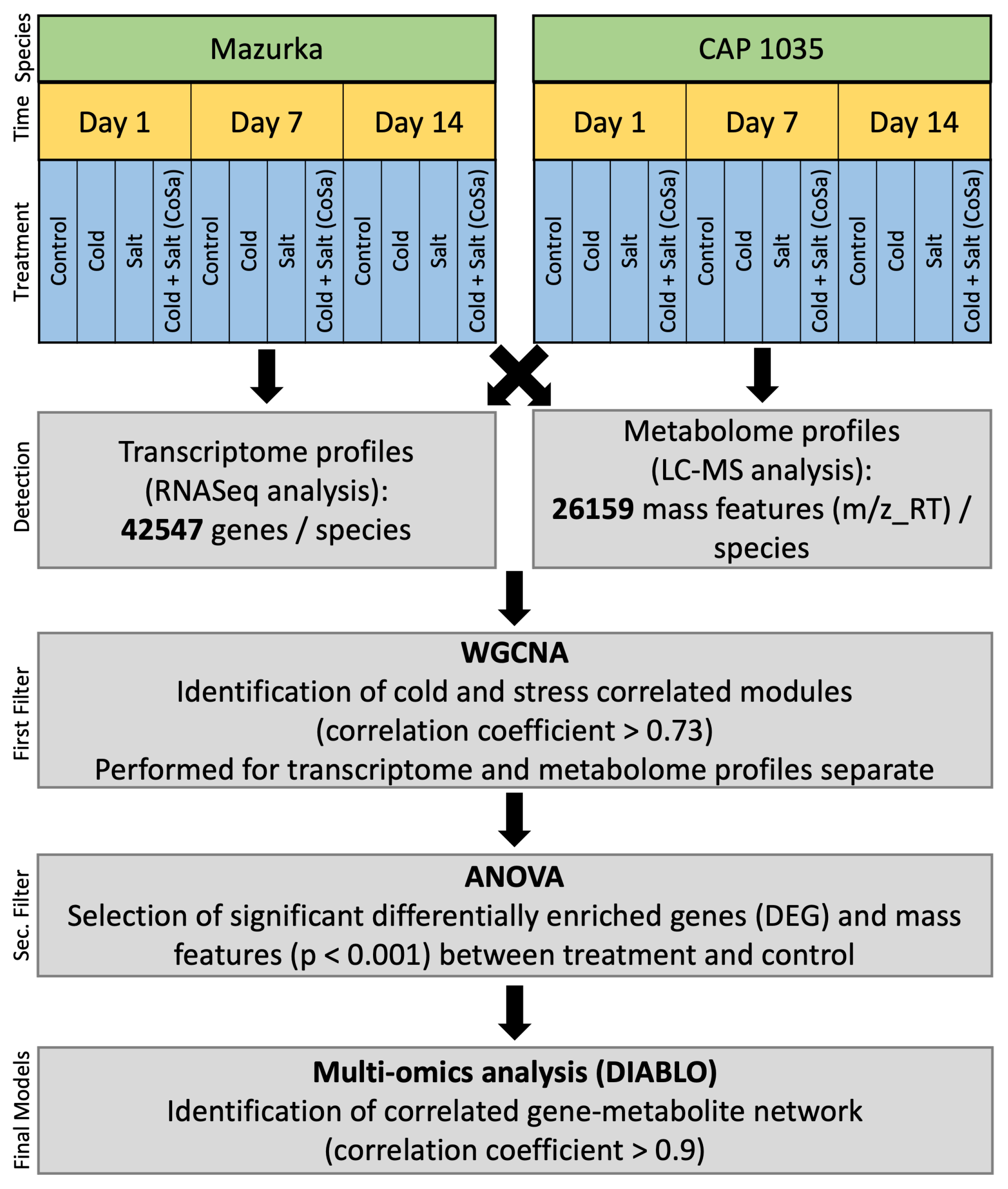
2. Materials and Methods
2.1. Plant Cultivation
2.2. Abiotic Stress Treatments and Harvesting
2.3. Total RNA Extraction and DNase-Digestion
2.4. RNASeq Data Handling and Analysis
2.5. cDNA Synthesis
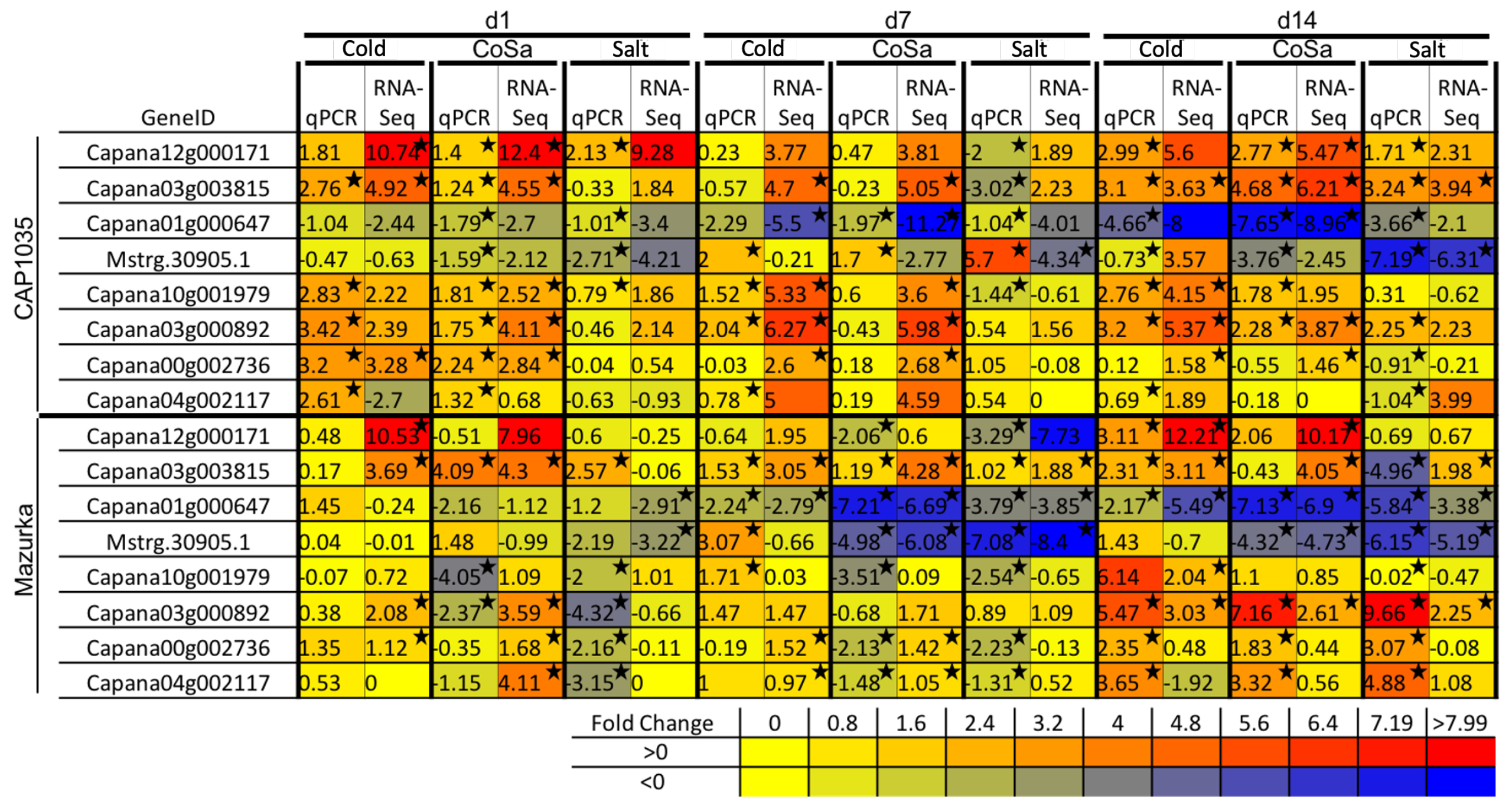
2.6. Quantitative Realtime PCR
2.7. Extraction of Metabolites
2.8. Untargeted Metabolite Screening via QToF MS
2.9. Multi-Omics Analysis
2.10. Identification of Putatively Annotated Metabolites
3. Results
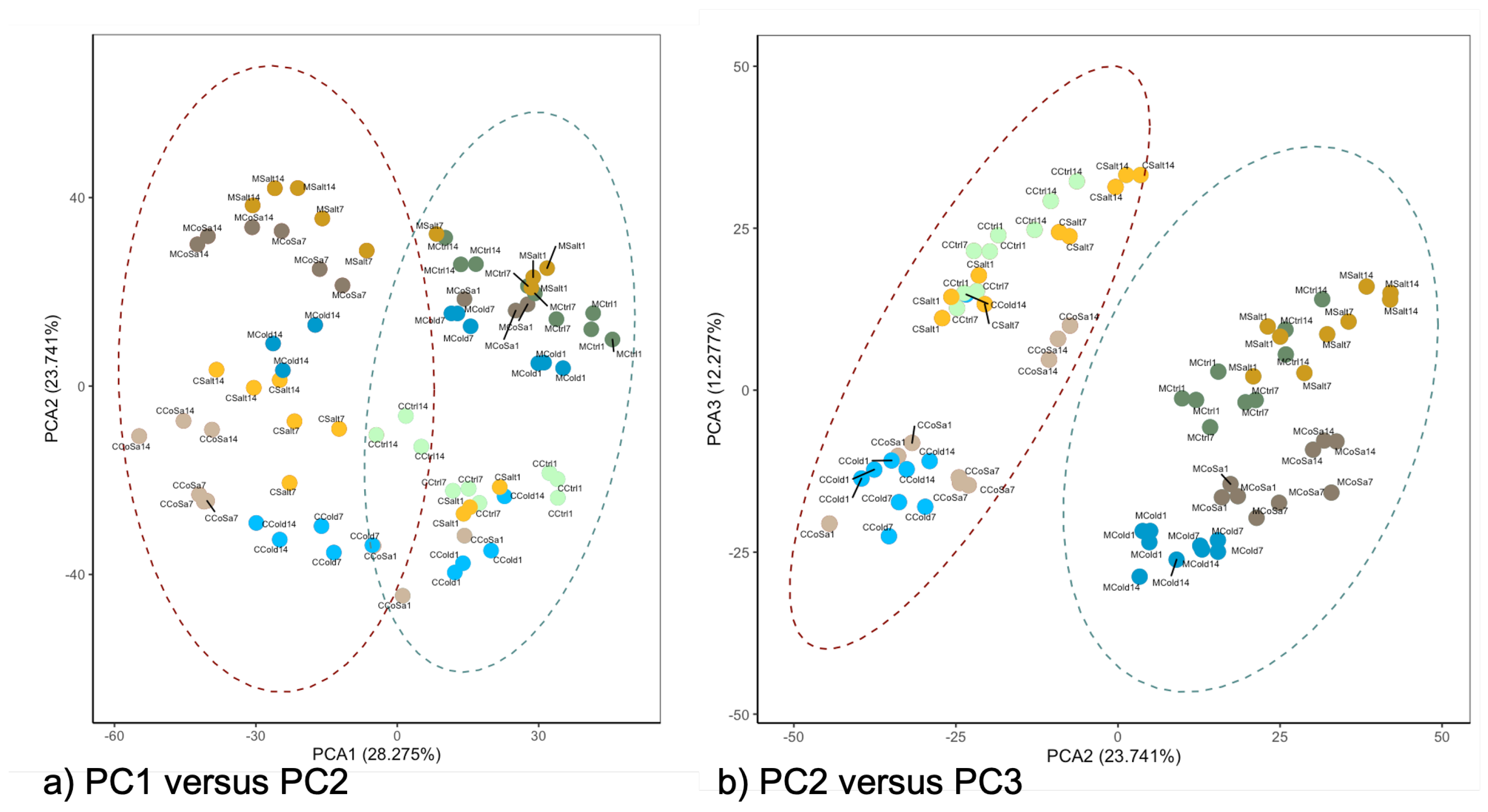
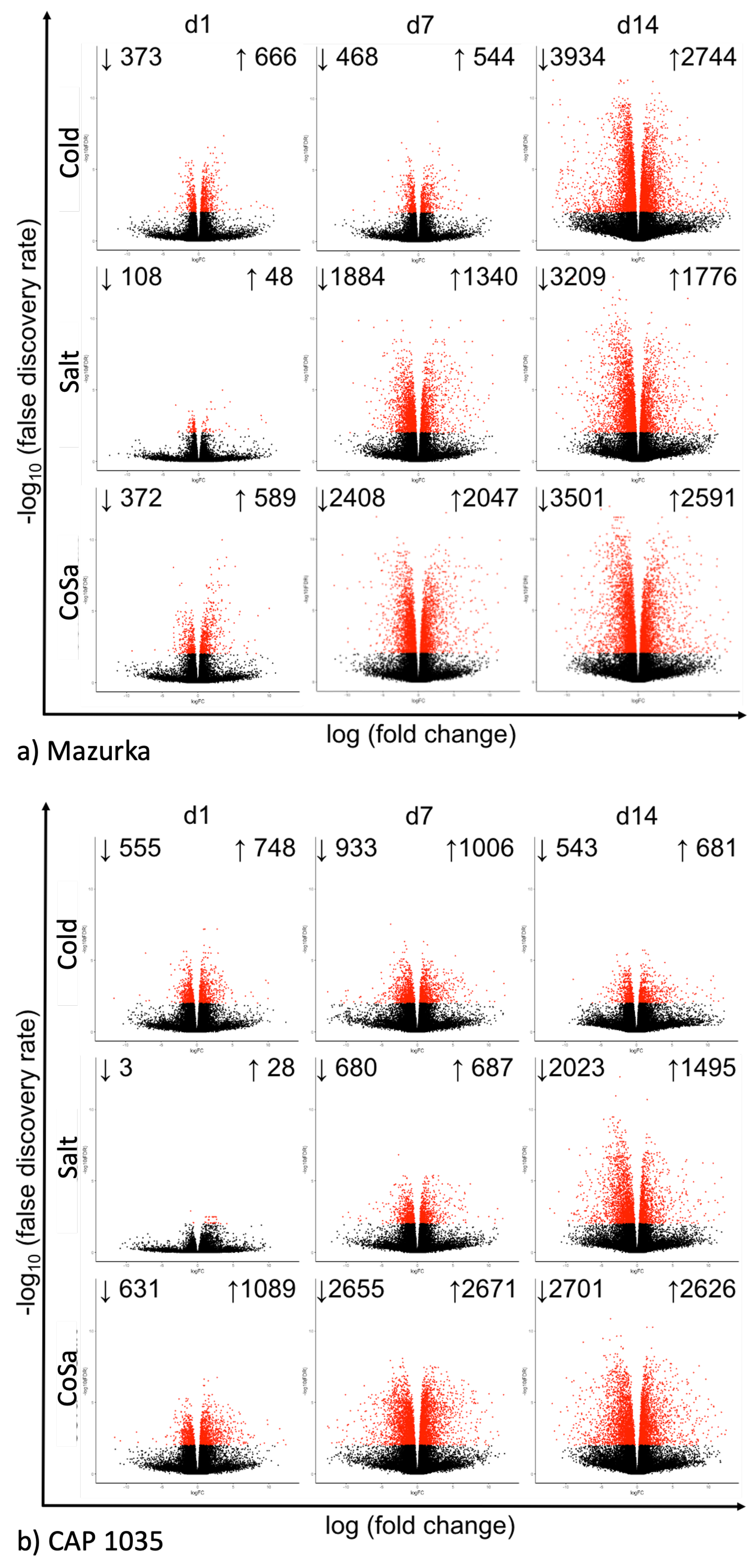
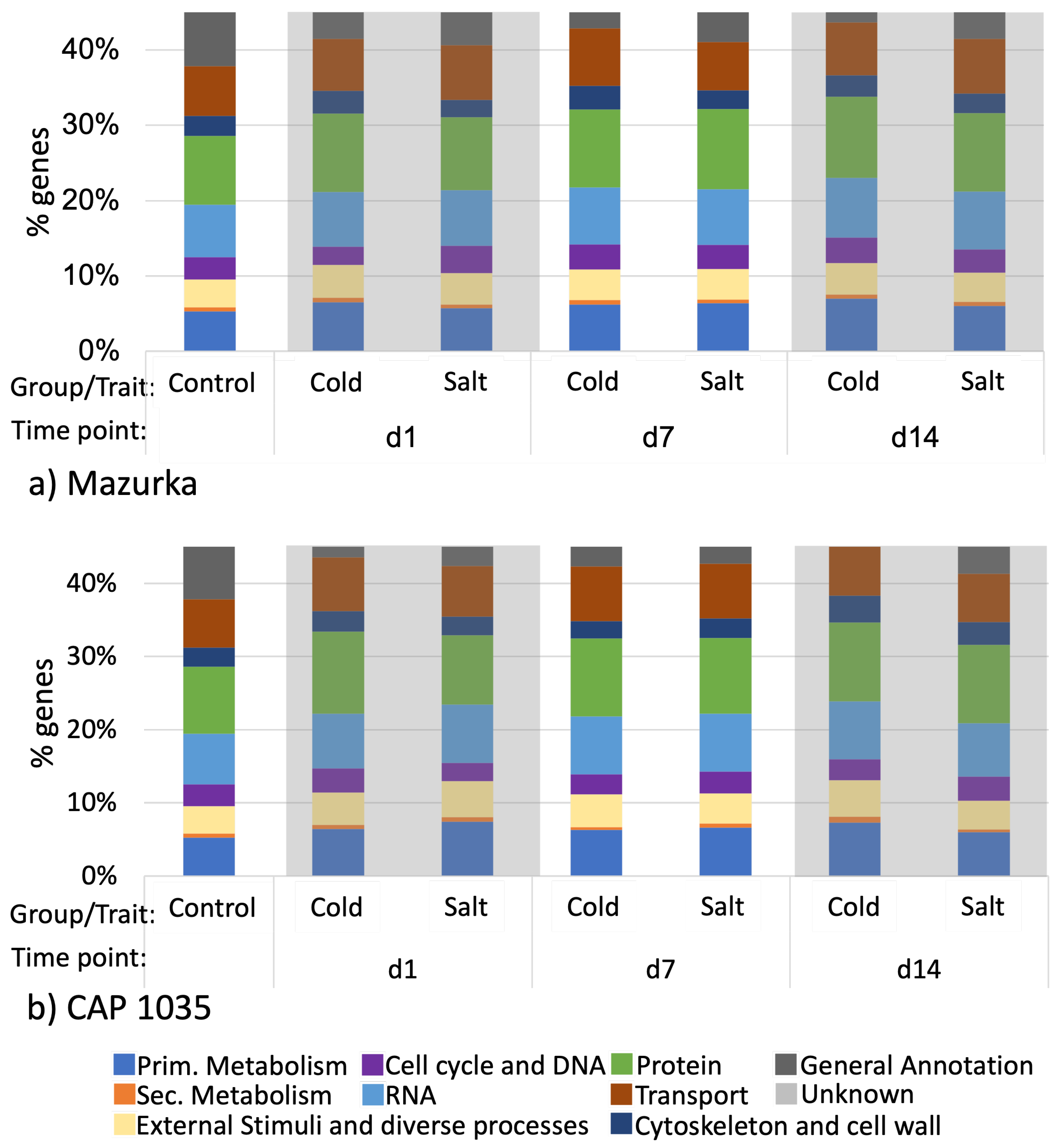
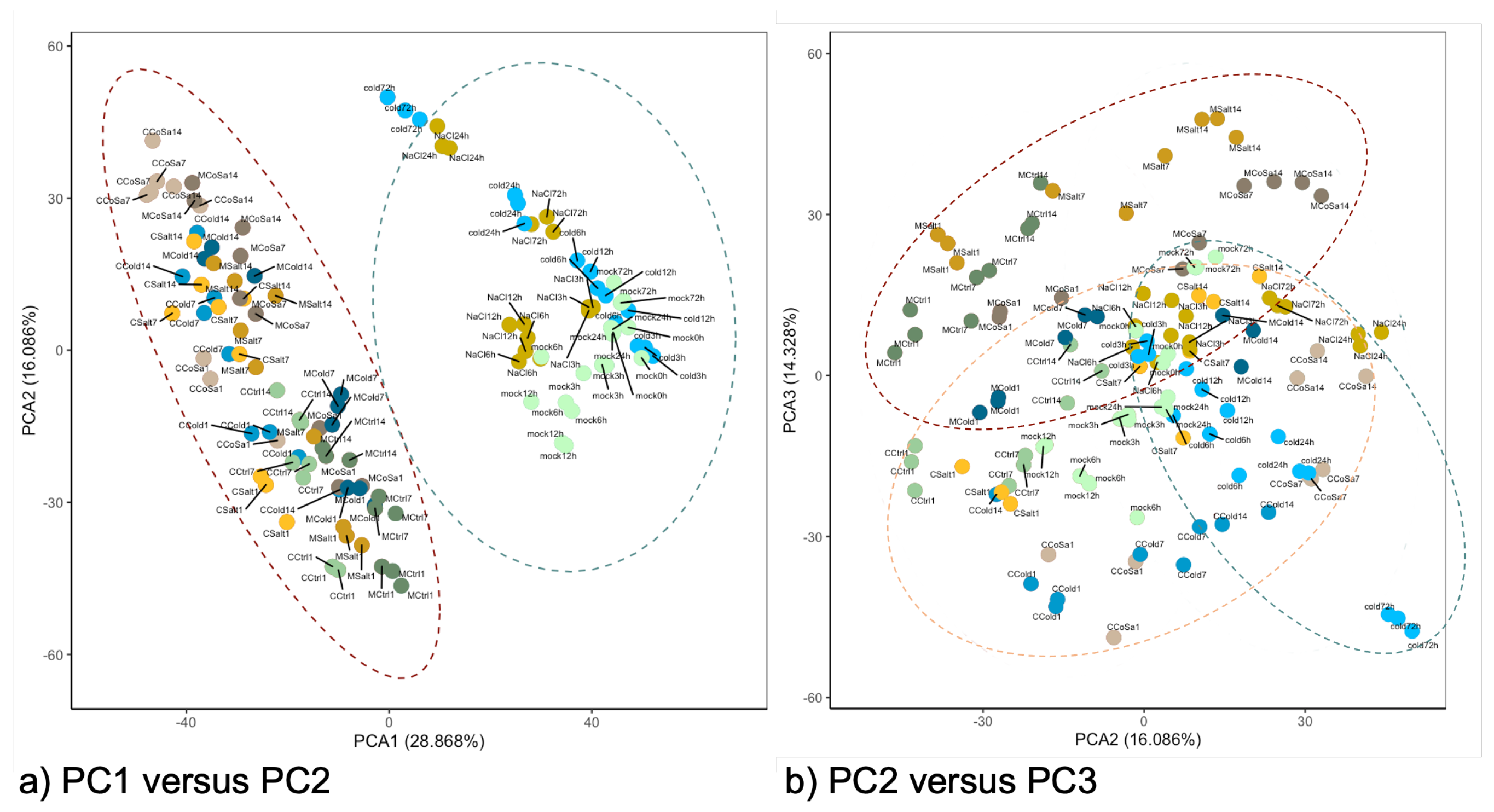
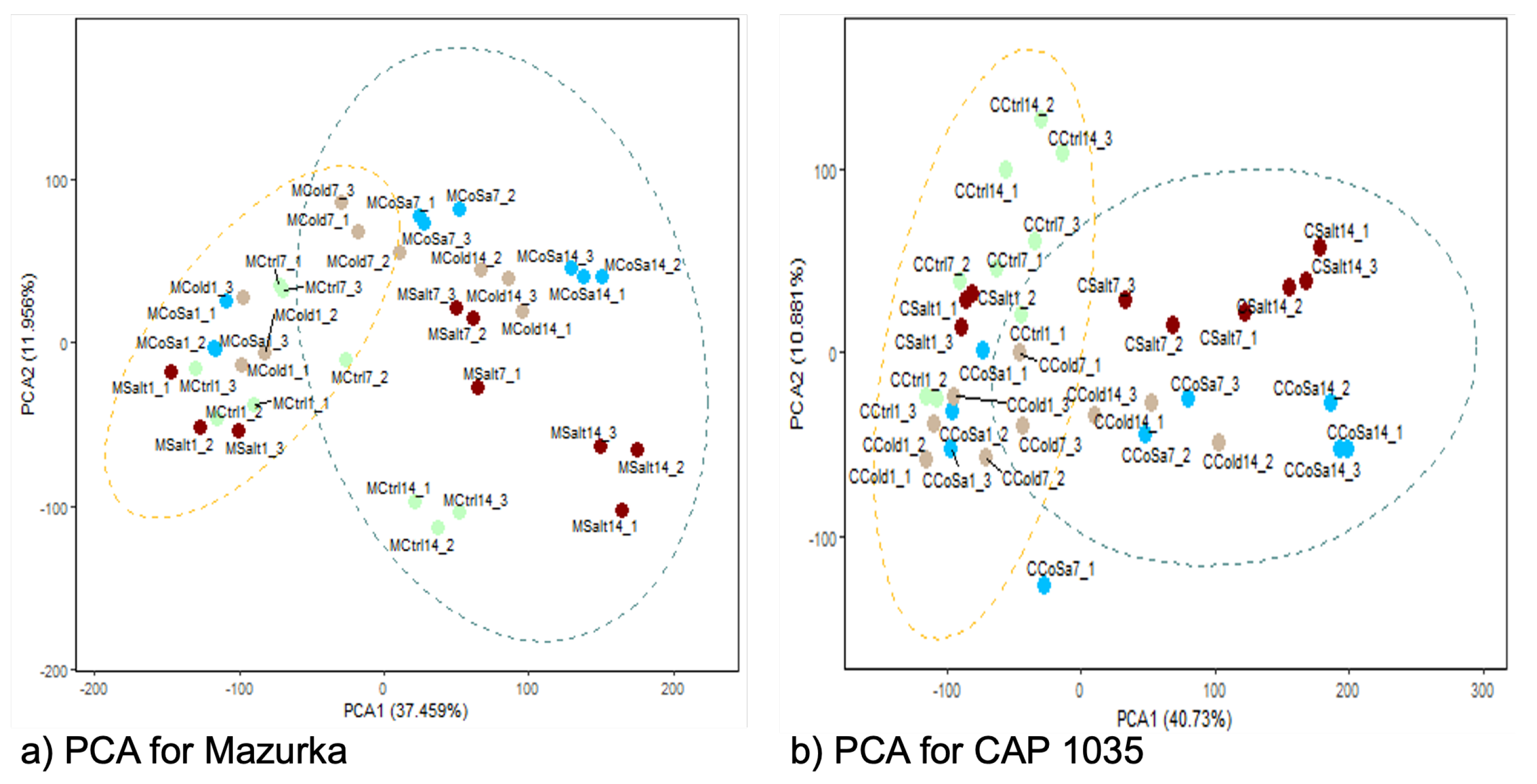
3.1. Global Response of the Transcriptome Is Intensified over a Period of 14 Days
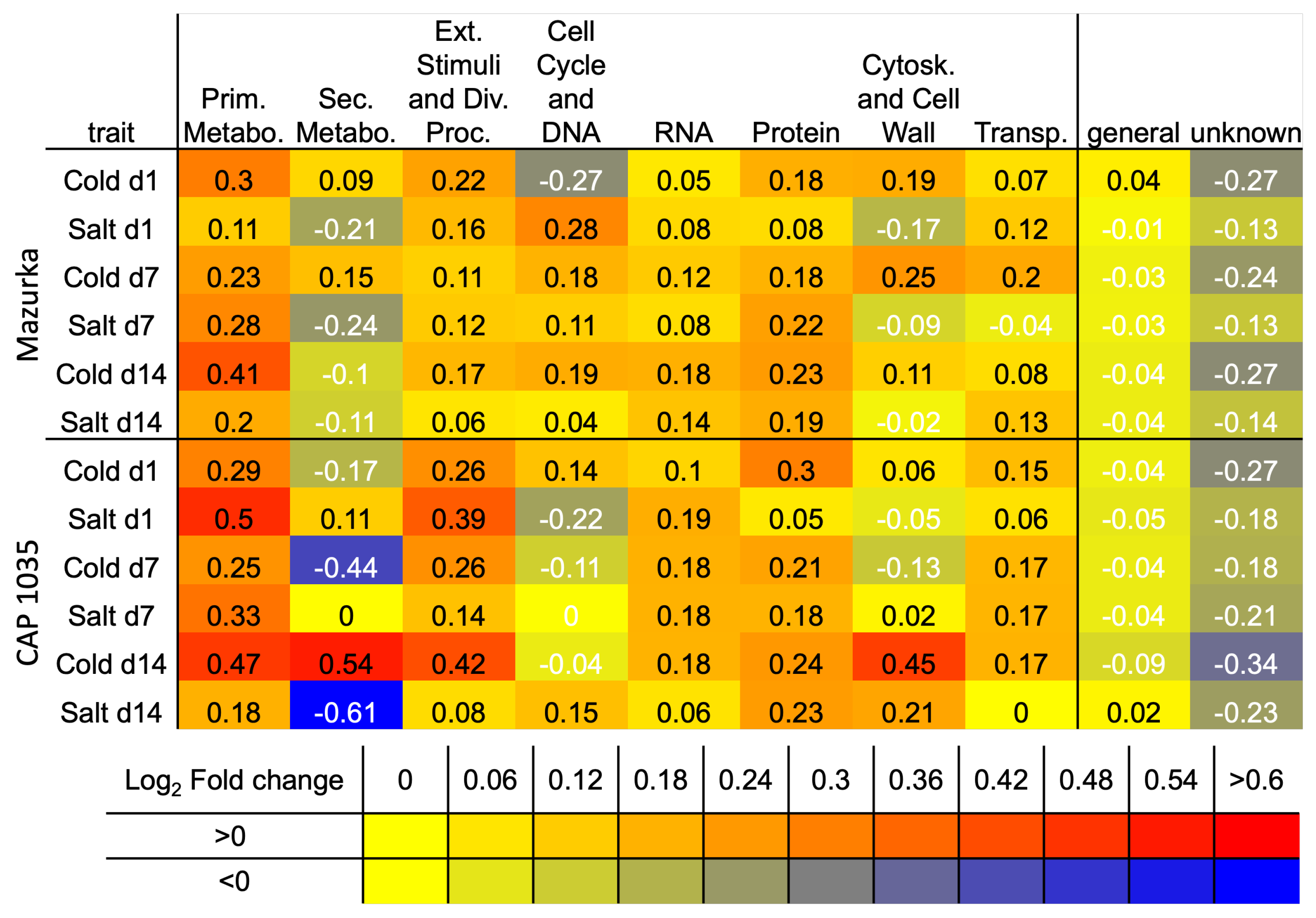
3.2. Differences in the Transcriptional Stress Response of the Two Species
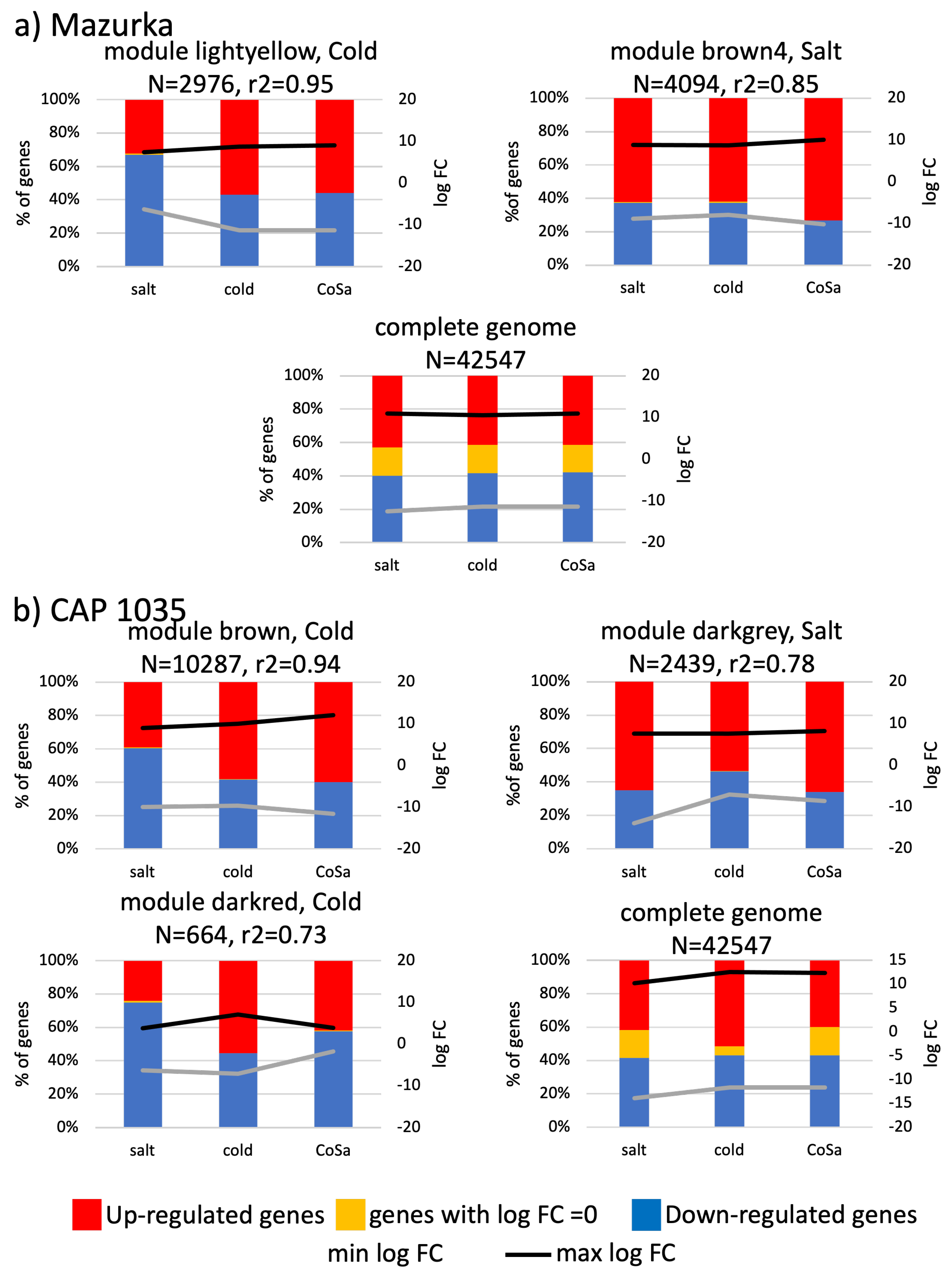
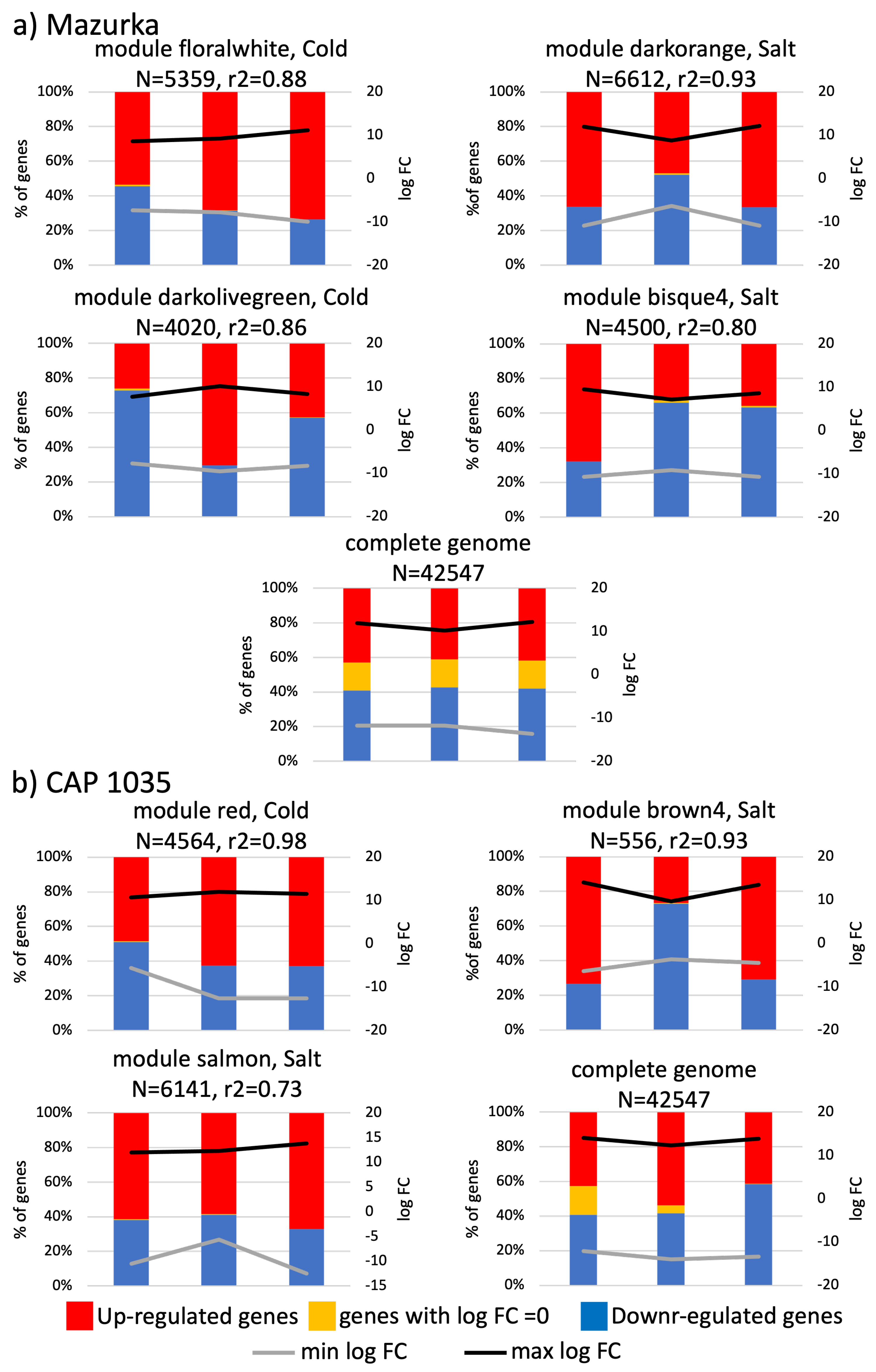
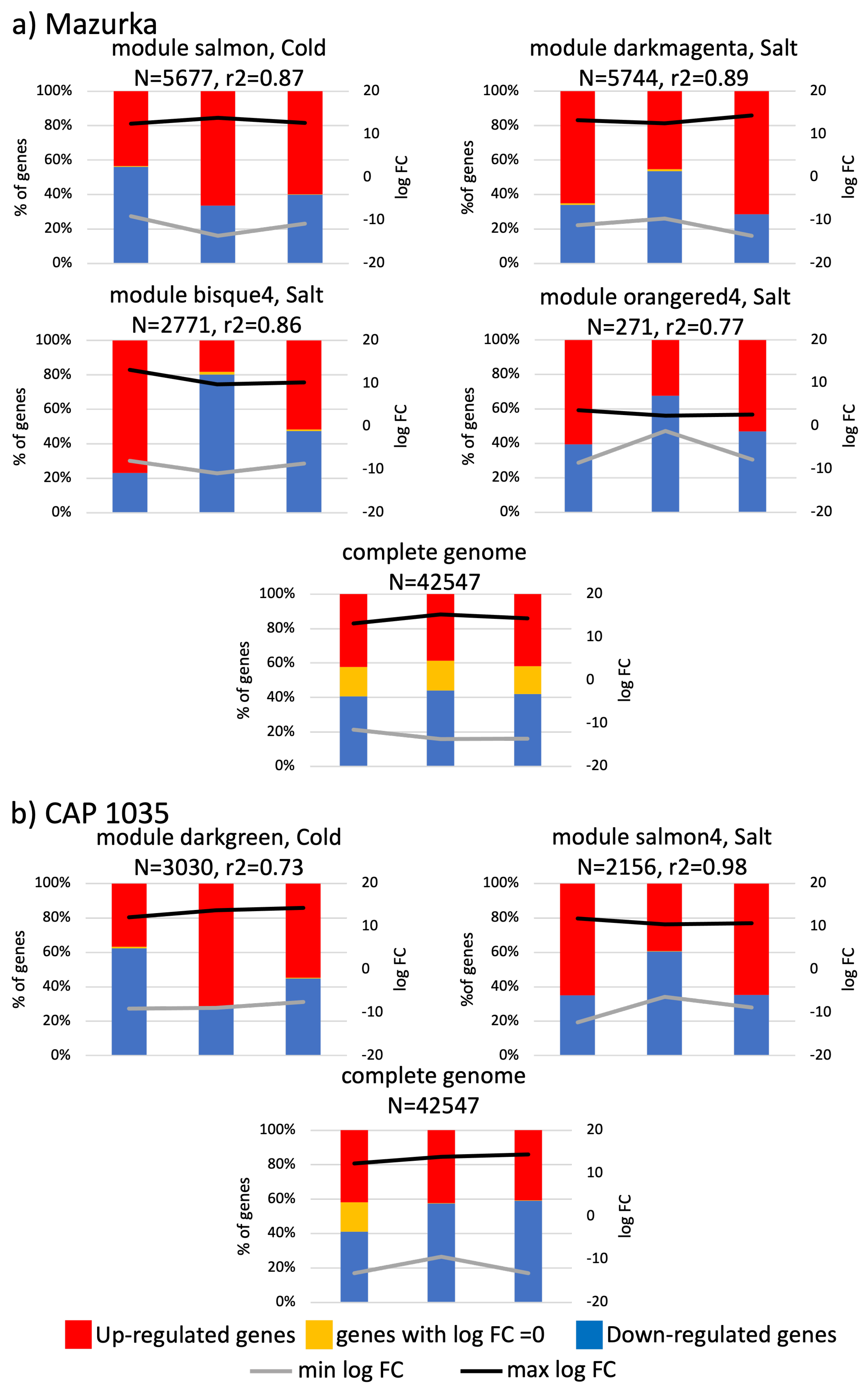
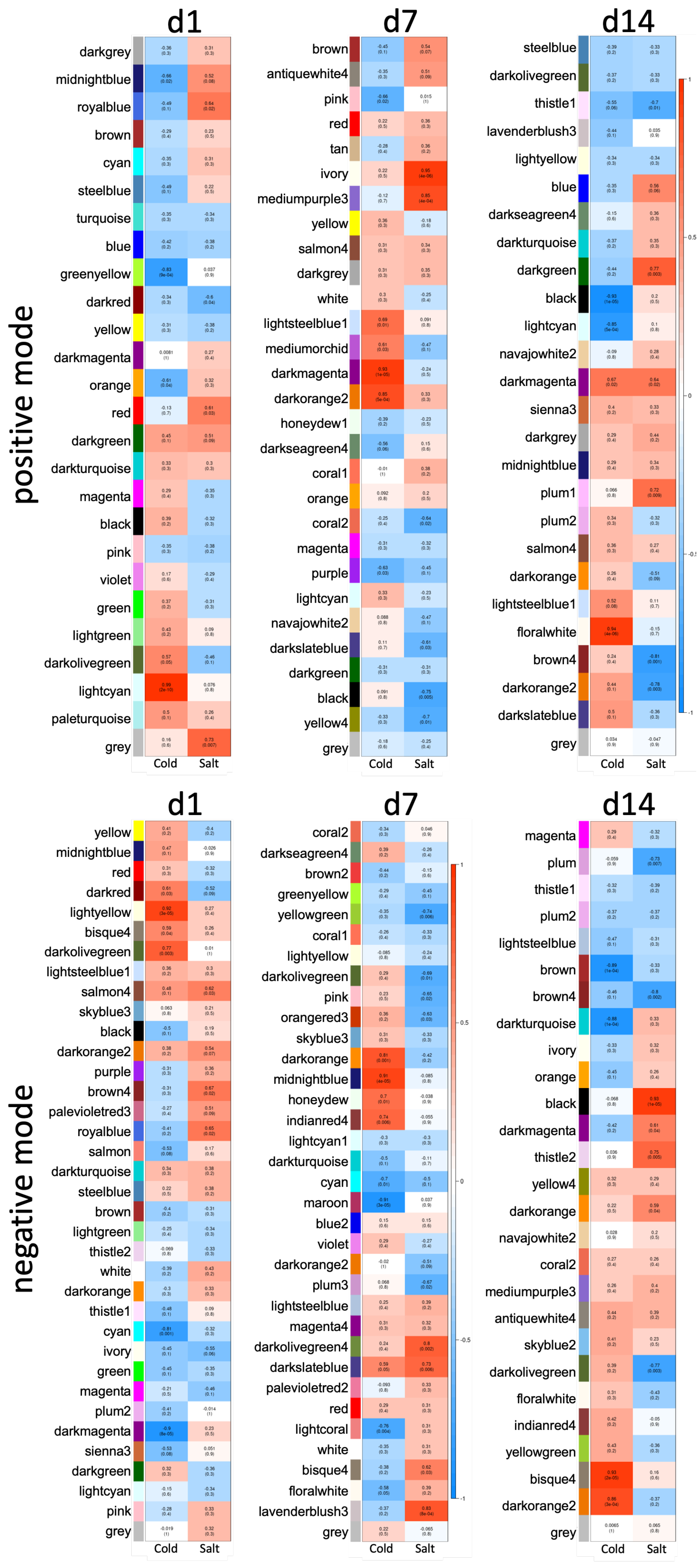
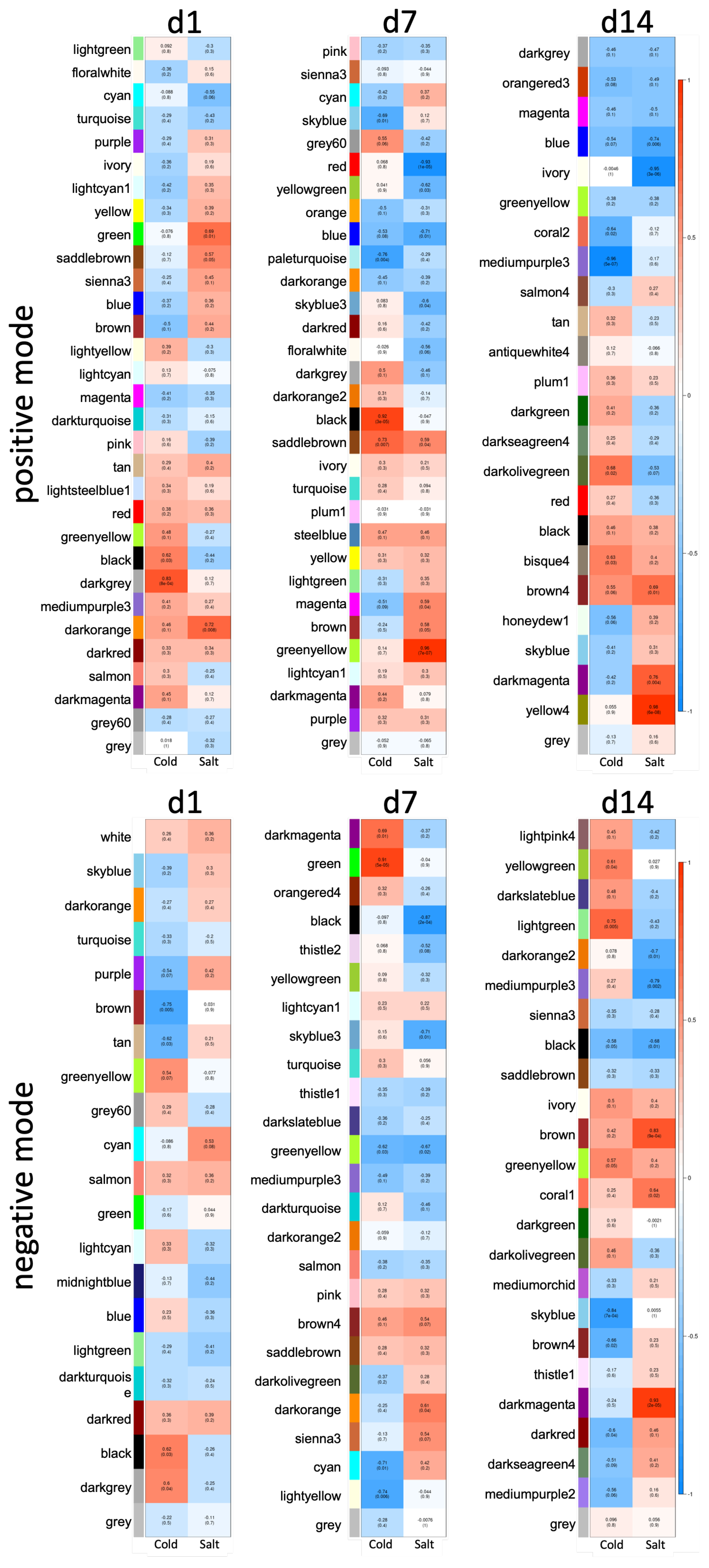
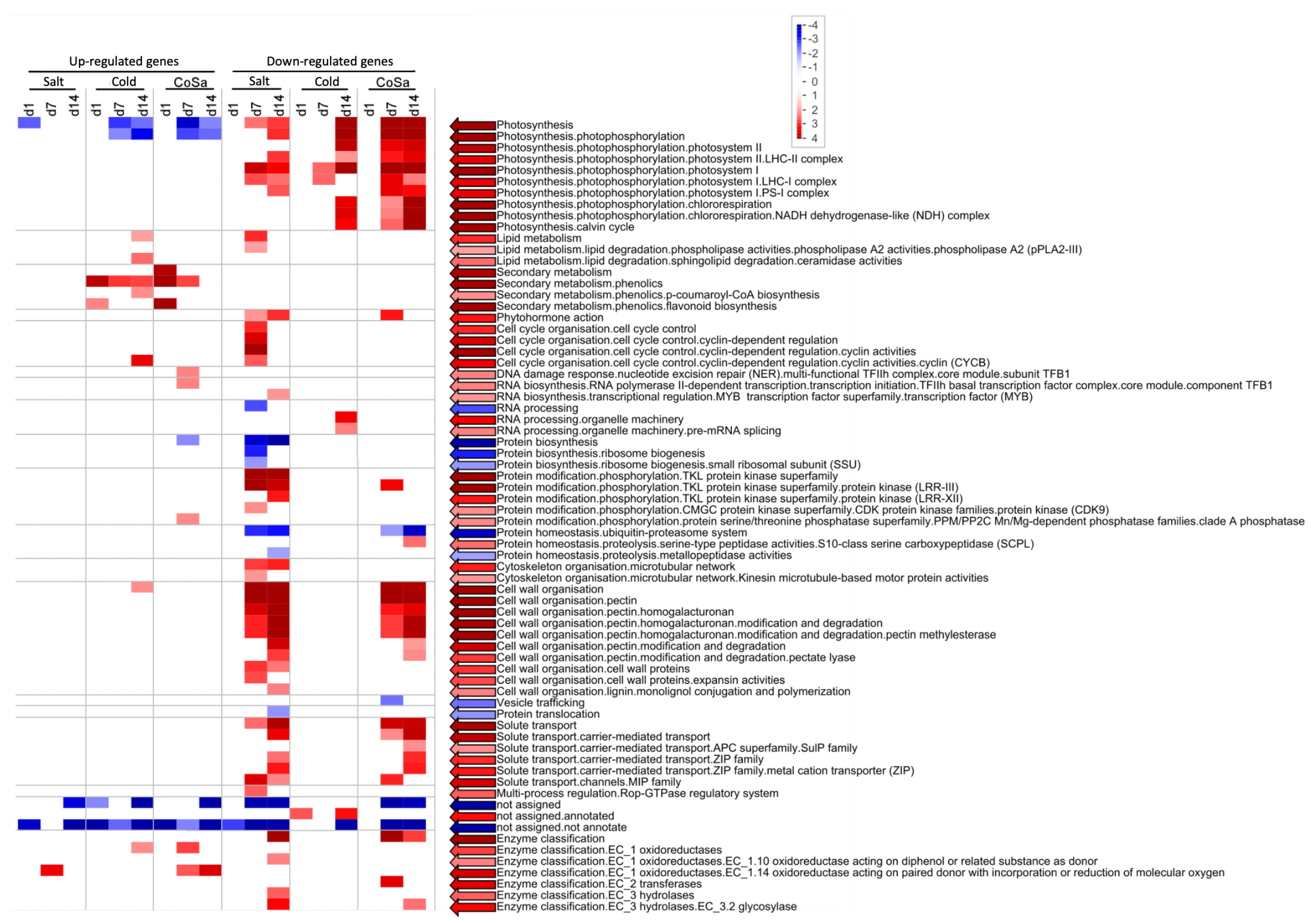
3.3. Differences between Early and Late Response Detected by Co-Expression Networks of the Transcriptome
3.4. Metabolome Analysis
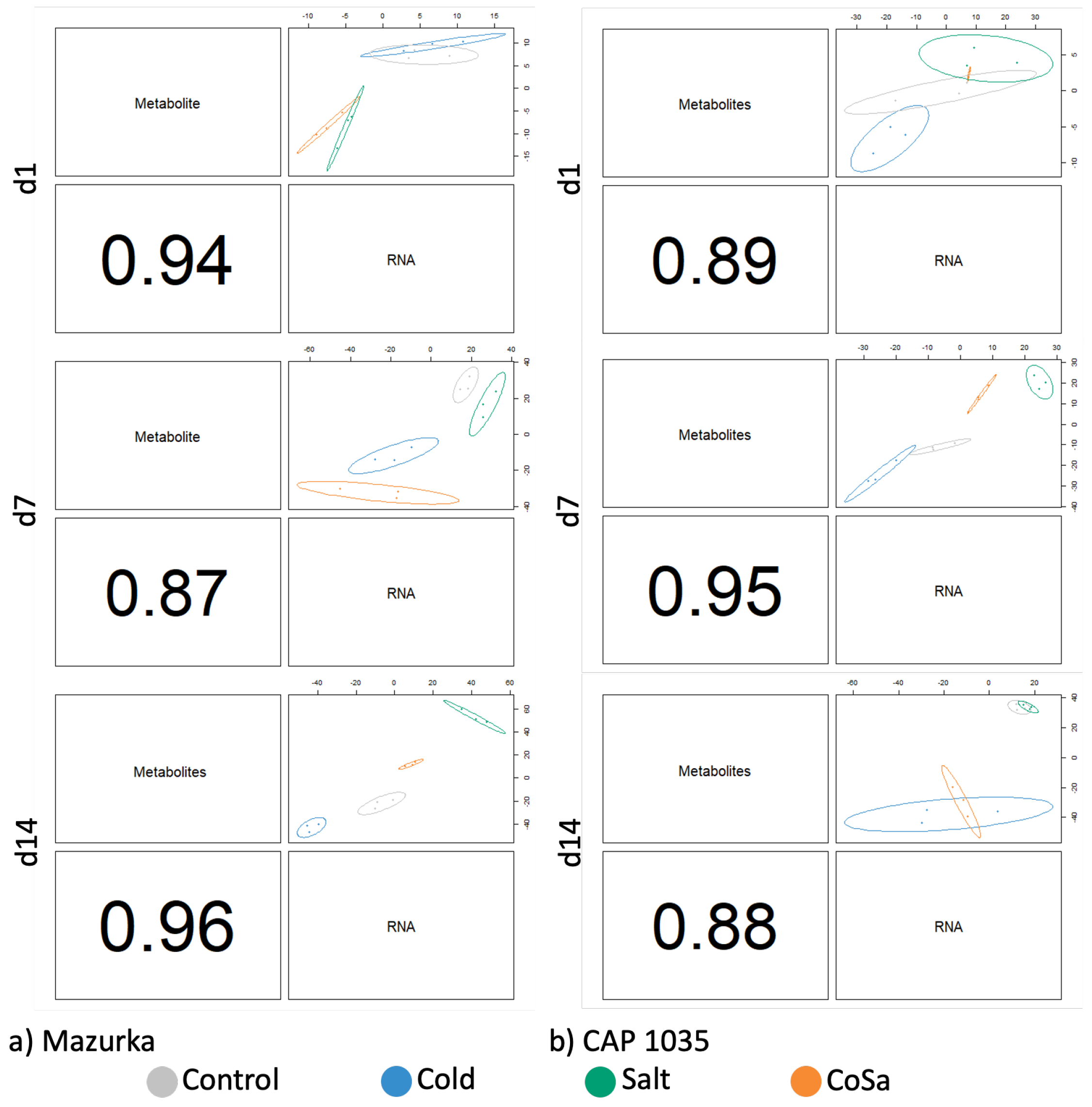
3.5. DIABLO Analysis
4. Discussion
4.1. Transcriptional Mid- to Long-Term Response to Chilling Temperature and Salinity
4.2. The Transcriptomic Response to a Combination of Abiotic Stresses Differ from Single Stress Responses in Cultivated Bell Pepper
4.3. Metabolic Responses Increases from Mid-to Long-Term Response
4.4. Multi-Omics Network Uncover Gene-Metabolite Relations in Secondary Metabolite Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A




References
- Khoury, C.K.; Carver, D.; Barchenger, D.W.; Barboza, G.E.; van Zonneveld, M.; Jarret, R.; Bohs, L.; Kantar, M.; Uchanski, M.; Mercer, K.; et al. Modelled distributions and conservation status of the wild relatives of chile peppers (Capsicum L.). Divers. Distrib. 2020, 26, 209–225. [Google Scholar] [CrossRef] [Green Version]
- Tripodi, P.; Kumar, S. The Capsicum Crop: An Introduction. In The Capsicum Genome; Springer International Publishing: Cham, Switzerland, 2019; pp. 1–8. [Google Scholar] [CrossRef]
- Olatunji, T.L.; Afolayan, A.J. The suitability of chili pepper (Capsicum annuum L.) for alleviating human micronutrient dietary deficiencies: A review. Food Sci. Nutr. 2018, 6, 2239–2251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahyuni, Y.; Ballester, A.R.; Sudarmonowati, E.; Bino, R.J.; Bovy, A.G. Secondary metabolites of Capsicum species and their importance in the human diet. J. Nat. Prod. 2013, 76, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization (FAO) of the United States, Food and Agriculture Data. 1961–2022. Available online: http://faostat.fao.org/ (accessed on 27 January 2022).
- Hernández-Pérez, T.; del Rocío Gómez-García, M.; Valverde, M.E.; Paredes-López, O. Capsicum annuum (hot pepper): An ancient Latin-American crop with outstanding bioactive compounds and nutraceutical potential. A review. Compr. Rev. Food Sci. Food Saf. 2020, 19, 2972–2993. [Google Scholar] [CrossRef]
- Edreva, A. Generation and scavenging of reactive oxygen species in chloroplasts: A submolecular approach. Agric. Ecosyst. Environ. 2005, 106, 119–133. [Google Scholar] [CrossRef]
- Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019, 52, 39. [Google Scholar] [CrossRef] [Green Version]
- Kumar, M.; Prakash, S.; Radha; Kumari, N.; Pundir, A.; Punia, S.; Saurabh, V.; Choudhary, P.; Changan, S.; Dhumal, S.; et al. Beneficial role of antioxidant secondary metabolites from medicinal plants in maintaining oral health. Antioxidants 2021, 10, 1061. [Google Scholar] [CrossRef]
- Odontuya, G.; Hoult, J.R.; Houghton, P.J. Structure-activity relationship for antiinflammatory effect of luteolin and its derived glycosides. Phytother. Res. 2005, 19, 782–786. [Google Scholar] [CrossRef]
- Chiruvella, K.K.; Mohammed, A.; Dampuri, G.; Ghanta, R.G.; Raghavan, S.C. Phytochemical and antimicrobial studies of methyl angolensate and Luteolin-7-O-glucoside isolated from callus cultures of Soymida febrifuga. Int. J. Biomed. Sci. IJBS 2007, 3, 269–278. [Google Scholar]
- Schinor, E.C.; Salvador, M.J.; Ito, I.Y.; Dias, D.A. Evaluation of the antimicrobial activity of crude extracts and isolated constituents from Chresta scapigera. Braz. J. Microbiol. 2007, 38, 145–149. [Google Scholar] [CrossRef] [Green Version]
- Thuphairo, K.; Sornchan, P.; Suttisansanee, U. Bioactive compounds, antioxidant activity and inhibition of key enzymes relevant to Alzheimer’s disease from sweet pepper (Capsicum annuum) extracts. Prev. Nutr. Food Sci. 2019, 24, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Baskar, A.A.; Ignacimuthu, S.; Michael, G.P.; Numair, K.A. Cancer chemopreventive potential of Luteolin-7-O-Glucoside isolated from Ophiorrhiza mungos Linn. Nutr. Cancer 2010, 63, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Thuy, B.T.P.; Nhung, N.T.A.; Duong, T.; Trung, P.V.; Quang, N.M.; Dung, H.T.K.; Tat, P.V. Prediction of anticancer activities of cynaroside and quercetin in leaf of plants Cynara scolymus L and Artocarpus incisa L using structure-activity relationship. Cogent Chem. 2016, 2, 1212452. [Google Scholar] [CrossRef]
- Carvalho Lemos, V.; Reimer, J.J.; Wormit, A. Color for Life: Biosynthesis and Distribution of Phenolic Compounds in Pepper (Capsicum annuum). Agriculture 2019, 9, 81. [Google Scholar] [CrossRef] [Green Version]
- Anaya-Esparza, L.M.; de la Mora, Z.V.; Vázquez-Paulino, O.; Ascencio, F.; Villarruel-López, A. Bell peppers (Capsicum annum L.) losses and wastes: Source for food and pharmaceutical applications. Molecules 2021, 26, 5341. [Google Scholar] [CrossRef] [PubMed]
- Junker-Frohn, L.V.; Lück, M.; Schmittgen, S.; Wensing, J.; Carraresi, L.; Thiele, B.; Groher, T.; Reimer, J.J.; Bröring, S.; Noga, G.; et al. Tomato’s Green Gold: Bioeconomy Potential of Residual Tomato Leaf Biomass as a Novel Source for the Secondary Metabolite Rutin. ACS Omega 2019, 4, 19071–19080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Röhlen-Schmittgen, S.; Ellenberger, J.; Groher, T.; Hunsche, M. Boosting leaf contents of rutin and solanesol in bio-waste of Solanum lycopersicum. Plant Physiol. Biochem. 2020, 155, 888–897. [Google Scholar] [CrossRef]
- Reimer, J.J.; Thiele, B.; Biermann, R.T.; Junker-Frohn, L.V.; Wiese-Klinkenberg, A.; Usadel, B.; Wormit, A. Tomato leaves under stress: A comparison of stress response to mild abiotic stress between a cultivated and a wild tomato species. Plant Mol. Biol. 2021, 107, 177–206. [Google Scholar] [CrossRef]
- Kuwagata, T.; Ishikawa-Sakurai, J.; Hayashi, H.; Nagasuga, K.; Fukushi, K.; Ahamed, A.; Takasugi, K.; Katsuhara, M.; Murai-Hatano, M. Influence of low air humidity and low root temperature on water uptake, growth and aquaporin expression in rice plants. Plant Cell Physiol. 2012, 53, 1418–1431. [Google Scholar] [CrossRef] [Green Version]
- Hatfield, J.L.; Prueger, J.H. Temperature extremes: Effect on plant growth and development. Weather Clim. Extrem. 2015, 10, 4–10. [Google Scholar] [CrossRef] [Green Version]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, F.; Hou, M.; Qiu, Y.; Zhang, T.; Yuan, Y. Salinity stress effects on transpiration and plant growth under different salinity soil levels based on thermal infrared remote (TIR) technique. Geoderma 2020, 357, 113961. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; no, M.F.O.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Cruz, J.L.; Coelho, E.F.; Filho, M.A.C.; dos Santos, A.A. Salinity reduces nutrients absorption and efficiency of their utilization in cassava plants. Cienc. Rural. 2018, 48. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of plant responses to salt stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Raihan, M.R.H.; Masud, A.A.C.; Rahman, K.; Nowroz, F.; Rahman, M.; Nahar, K.; Fujita, M. Regulation of reactive oxygen species and antioxidant defense in plants under salinity. Int. J. Mol. Sci. 2021, 22, 9326. [Google Scholar] [CrossRef]
- Aroca, R.; Vernieri, P.; Irigoyen, J.J.; Sánchez-Díaz, M.; Tognoni, F.; Pardossi, A. Involvement of abscisic acid in leaf and root of maize (Zea mays L.) in avoiding chilling-induced water stress. Plant Sci. 2003, 165, 671–679. [Google Scholar] [CrossRef]
- León-Chan, R.G.; López-Meyer, M.; Osuna-Enciso, T.; nudo Barajas, J.A.S.; Heredia, J.B.; León-Félix, J. Low temperature and ultraviolet-B radiation affect chlorophyll content and induce the accumulation of UV-B-absorbing and antioxidant compounds in bell pepper (Capsicum annuum) plants. Environ. Exp. Bot. 2017, 139, 143–151. [Google Scholar] [CrossRef]
- Zhang, R.X.; Zhu, W.C.; Cheng, G.X.; Yu, Y.N.; Li, Q.H.; ul Haq, S.; Said, F.; Gong, Z.H. A novel gene, CaATHB-12, negatively regulates fruit carotenoid content under cold stress in Capsicum annuum. Food Nutr. Res. 2020, 64, 64. [Google Scholar] [CrossRef]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Pandey, P.; Ramegowda, V.; Senthil-Kumar, M. Shared and unique responses of plants to multiple individual stresses and stress combinations: Physiological and molecular mechanisms. Front. Plant Sci. 2015, 6, 723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, H.A.; Hussain, S.; Khaliq, A.; Ashraf, U.; Anjum, S.A.; Men, S.; Wang, L. Chilling and drought stresses in crop plants: Implications, cross talk, and potential management opportunities. Front. Plant Sci. 2018, 9, 393. [Google Scholar] [CrossRef] [PubMed]
- Jha, U.C.; Bohra, A.; Jha, R. Breeding approaches and genomics technologies to increase crop yield under low-temperature stress. Plant Cell Rep. 2017, 36, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Fita, A.; Rodríguez-Burruezo, A.; Boscaiu, M.; Prohens, J.; Vicente, O. Breeding and domesticating crops adapted to drought and salinity: A new paradigm for increasing food production. Front. Plant Sci. 2015, 6, 978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, L.; Xia, H.; Lu, B.R. Editorial: Crop breeding for drought resistance. Front. Plant Sci. 2019, 10, 314. [Google Scholar] [CrossRef] [PubMed]
- Dempewolf, H.; Baute, G.; Anderson, J.; Kilian, B.; Smith, C.; Guarino, L. Past and future use of wild relatives in crop breeding. Crop Sci. 2017, 57, 1070–1082. [Google Scholar] [CrossRef]
- McCown, B.H.; Zeldin, E.L. ’HyRed’, an Early, High Fruit Color Cranberry Hybrid. HortScience 2003, 38, 304–305. [Google Scholar] [CrossRef] [Green Version]
- Matteo, A.D.; Ruggieri, V.; Sacco, A.; Rigano, M.M.; Carriero, F.; Bolger, A.; Fernie, A.R.; Frusciante, L.; Barone, A. Identification of candidate genes for phenolics accumulation in tomato fruit. Plant Sci. 2013, 205–206, 87–96. [Google Scholar] [CrossRef]
- van Zonneveld, M.; Ramirez, M.; Williams, D.E.; Petz, M.; Meckelmann, S.; Avila, T.; Bejarano, C.; Ríos, L.; na, K.P.; Jäger, M.; et al. Screening genetic resources of Capsicum peppers in their primary center of diversity in Bolivia and Peru. PLoS ONE 2015, 10, e0134663. [Google Scholar] [CrossRef]
- Moscone, E.A.; Scaldaferro, M.A.; Grabiele, M.; Cecchini, N.M.; García, Y.S.; Jarret, R.; na, J.R.D.; Ducasse, D.A.; Barboza, G.E.; Ehrendorfer, F. The evolution of chili peppers (Capsicum -Solanaceae): A cytogenetic perspective. Acta Hortic. 2007, 745, 137–170. [Google Scholar] [CrossRef]
- Yang, H.J.; Chung, K.R.; Kwon, D.Y. DNA sequence analysis tells the truth of the origin, propagation, and evolution of chili (red pepper). J. Ethn. Foods 2017, 4, 154–162. [Google Scholar] [CrossRef]
- e Lima, F.A.; Li, K.; Wen, W.; Yan, J.; Nikoloski, Z.; Willmitzer, L.; Brotman, Y. Unraveling lipid metabolism in maize with time-resolved multi-omics data. Plant J. 2018, 93, 1102–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fondi, M.; Liò, P. Multi-omics and metabolic modelling pipelines: Challenges and tools for systems microbiology. Microbiol. Res. 2015, 171, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Jamil, I.N.; Remali, J.; Azizan, K.A.; Muhammad, N.A.N.; Arita, M.; Goh, H.H.; Aizat, W.M. Systematic Multi-Omics Integration (MOI) Approach in Plant Systems Biology. Front. Plant Sci. 2020, 11, 944. [Google Scholar] [CrossRef]
- Obudulu, O.; Bygdell, J.; Sundberg, B.; Moritz, T.; Hvidsten, T.R.; Trygg, J.; Wingsle, G. Quantitative proteomics reveals protein profiles underlying major transitions in aspen wood development. BMC Genom. 2016, 17, 119. [Google Scholar] [CrossRef] [Green Version]
- Aizat, W.M.; Goh, H.H.; Baharum, S.N. Omics Applications for Systems Biology. Adv. Exp. Med. Biol. 2018. [Google Scholar] [CrossRef]
- Uarrota, V.G.; Fuentealba, C.; Hernández, I.; Defilippi-Bruzzone, B.; Meneses, C.; Campos-Vargas, R.; Lurie, S.; Hertog, M.; Carpentier, S.; Poblete-Echeverría, C.; et al. Integration of proteomics and metabolomics data of early and middle season Hass avocados under heat treatment. Food Chem. 2019, 289, 512–521. [Google Scholar] [CrossRef]
- Singh, A.; Shannon, C.P.; Gautier, B.; Rohart, F.; Vacher, M.; Tebbutt, S.J.; Cao, K.A.L. DIABLO: An integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 2019, 35, 3055–3062. [Google Scholar] [CrossRef]
- Genzel, F.; Dicke, M.D.; Junker-Frohn, L.V.; Neuwohner, A.; Thiele, B.; Putz, A.; Usadel, B.; Wormit, A.; Wiese-Klinkenberg, A. Impact of moderate cold and salt stress on the accumulation of antioxidant flavonoids in the leaves of two Capsicum Cultivars. J. Agric. Food Chem. 2021, 69, 6431–6443. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Kingsford, C. Salmon: Accurate, Versatile and Ultrafast Quantification from RNA-seq Data using Lightweight-Alignment. bioRxiv 2015. [Google Scholar] [CrossRef] [Green Version]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.H.; Sim, Y.M.; Koo, N.; Nam, J.Y.; Lee, J.; Kim, N.; Jang, H.; Kim, Y.M.; Yeom, S.I. Transcriptome profiling of abiotic responses to heat, cold, salt, and osmotic stress of Capsicum annuum L. Sci. Data 2020, 7, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R Foundation for Statistical Computing, R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2019; Available online: https://www.r-project.org/about.html/ (accessed on 4 March 2022).
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2016, 4, 1521. [Google Scholar] [CrossRef] [PubMed]
- Usadel, B.; Nagel, A.; Steinhauser, D.; Gibon, Y.; Bläsing, O.E.; Redestig, H.; Sreenivasulu, N.; Krall, L.; Hannah, M.A.; Poree, F.; et al. PageMan: An interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinform. 2006, 7, 535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. Relative expression software tool (REST(C)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.A.L. mixOmics: An R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A guide to using MapMan to visualize and compare Omics data in plants: A case study in the crop species, Maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- Schwacke, R.; Ponce-Soto, G.Y.; Krause, K.; Bolger, A.M.; Arsova, B.; Hallab, A.; Gruden, K.; Stitt, M.; Bolger, M.E.; Usadel, B. MapMan4: A Refined Protein Classification and Annotation Framework Applicable to Multi-Omics Data Analysis. Mol. Plant 2019, 12, 879–892. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Feiguelman, G.; Fu, Y.; Yalovsky, S. ROP GTPases structure-function and signaling pathways. Plant Physiol. 2018, 176, 57–79. [Google Scholar] [CrossRef] [PubMed]
- Ballard, R.E.; McClure, J.W.; Eshbaugh, W.H.; Wilson, K.G. A chemosystematic study of selected taxa of Capsicum. Am. J. Bot. 1970, 57, 225–233. [Google Scholar] [CrossRef]
- Eshbaugh, W.H. Genetic and biochemical systematic studies of chili peppers (Capsicum -Solanaceae). Bull. Torrey Bot. Club 1975, 102, 396. [Google Scholar] [CrossRef]
- Alves, T.M.A.; Marengo, S.; Machado, C.; Caldeira, R.; Carvalho, O.; dos Santos Isaias, R.M.; ao Renato Stehmann, J.; Zani, C. Morphological, anatomical, macro and micromolecular markers for Solanum cernuum identification. Rev. Bras. Farmacogn. 2007, 17, 542–548. [Google Scholar] [CrossRef]
- Lopez-Lazaro, M. Distribution and biological activities of the flavonoid luteolin. Mini-Rev. Med. Chem. 2009, 9, 31–59. [Google Scholar] [CrossRef] [PubMed]
- Harborne, J.; Corner, J. Plant polyphenols. 4. Hydroxycinnamic acid-sugar derivatives. Biochem. J. 1961, 81, 242–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakimiuk, K.; Wink, M.; Tomczyk, M. Flavonoids of the Caryophyllaceae. Phytochem. Rev. 2021, 21, 179–218. [Google Scholar] [CrossRef]
- El-Sherbeni, S.A.E.H.; Al-Ashmawy, G.M. Myricitrin from Physalis pubescens L. leaves and frankincense decrease resistance of MCF-7 cells and ameliorate efficacy of epirubicin. J. Pharm. Pharmacogn. Res. 2021, 9, 497–508. [Google Scholar]
- Mokhtar, M.; Soukup, J.; Donato, P.; Cacciola, F.; Dugo, P.; Riazi, A.; Jandera, P.; Mondello, L. Determination of the polyphenolic content of a Capsicum annuum L. extract by liquid chromatography coupled to photodiode array and mass spectrometry detection and evaluation of its biological activity. J. Sep. Sci. 2015, 38, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Materska, M. The scavenging effect and flavonoid glycosides content in fractions from fruits of hot pepper Capsicum annuum L. Acta Sci. Pol. Technol. Aliment. 2012, 11, 363–371. [Google Scholar]
- Materska, M.; Konopacka, M.; Rogoliński, J.; Ślosarek, K. Antioxidant activity and protective effects against oxidative damage of human cells induced by X-radiation of phenolic glycosides isolated from pepper fruits Capsicum annuum L. Food Chem. 2015, 168, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, A.R.; Leonardi, B.; Miron, D.; Schapoval, E.; de Oliveira, J.R.; Gosmann, G. Antioxidant and anti-inflammatory properties of Capsicum baccatum: From traditional use to scientific approach. J. Ethnopharmacol. 2012, 139, 228–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadilova, E.; Stintzing, F.C.; Carle, R. Anthocyanins, colour and antioxidant properties of eggplant (Solanum melongena L.) and violet pepper (Capsicum annuum L.) peel extracts. Z. Naturforschung C 2006, 61, 527–535. [Google Scholar] [CrossRef]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef] [PubMed]
- Grace, S.C. Phenolics as Antioxidants. In Antioxidants and Reactive Oxygen Species in Plants; Smirnoff, N., Ed.; Blackwell Publishing Ltd.: Oxford, UK, 2007; pp. 141–168. [Google Scholar] [CrossRef]
- Løvdal, T.; Olsen, K.M.; Slimestad, R.; Verheul, M.; Lillo, C. Synergetic effects of nitrogen depletion, temperature, and light on the content of phenolic compounds and gene expression in leaves of tomato. Phytochemistry 2010, 71, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Selmar, D.; Kleinwächter, M. Stress enhances the synthesis of secondary plant products: The impact of stress-related over-reduction on the accumulation of natural products. Plant Cell Physiol. 2013, 54, 817–826. [Google Scholar] [CrossRef]
- Abewoy Fentik, D. Review on Genetics and Breeding of Tomato (Lycopersicon esculentum Mill). Adv. Crop. Sci. Technol. 2017, 5, 1–6. [Google Scholar] [CrossRef]
- Ksas, B.; Becuwe, N.; Chevalier, A.; Havaux, M. Plant tolerance to excess light energy and photooxidative damage relies on plastoquinone biosynthesis. Sci. Rep. 2015, 5, 10919. [Google Scholar] [CrossRef] [Green Version]
- Chokshi, K.; Pancha, I.; Ghosh, A.; Mishra, S. Nitrogen starvation-induced cellular crosstalk of ROS-scavenging antioxidants and phytohormone enhanced the biofuel potential of green microalga Acutodesmus dimorphus. Biotechnol. Biofuels 2017, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Robles-Rengel, R.; Florencio, F.J.; Muro-Pastor, M.I. Redox interference in nitrogen status via oxidative stress is mediated by 2-oxoglutarate in cyanobacteria. New Phytol. 2019, 224, 216–228. [Google Scholar] [CrossRef]
- Munjal, P.; Munjal, R. Oxidative stress and antioxidant defense in plants under high temperature. In Reactive Oxygen, Nitrogen and Sulfur Species in Plants: Production, Metabolism, Signaling and Defense Mechanisms; Wiley-Blackwell: Hoboken, NJ, USA, 2019. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; Sánchez-calvo, B.; Gómez-rodríguez, M.V.; Chaki, M.; Valderrama, R.; Mata-pérez, C.; López-jaramillo, J.; Corpas, F.J.; Barroso, J.B. Short-term low temperature induces nitro-oxidative stress that deregulates the nadp-malic enzyme function by tyrosine nitration in Arabidopsis thaliana. Antioxidants 2019, 8, 448. [Google Scholar] [CrossRef] [Green Version]
- García-Martí, M.; Piñero, M.C.; García-Sanchez, F.; Mestre, T.C.; López-Delacalle, M.; Martínez, V.; Rivero, R.M. Amelioration of the oxidative stress generated by simple or combined abiotic stress through the K+ and Ca2+ supplementation in tomato plants. Antioxidants 2019, 8, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Wei, W.; Pang, X.; Wang, X.; Zhang, H.; Dong, B.; Xing, Y.; Li, X.; Wang, M. Comparative transcriptome profiling of a desert evergreen shrub, Ammopiptanthus mongolicus, in response to drought and cold stresses. BMC Genom. 2014, 15, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre-Becerra, H.; Vazquez-Hernandez, M.C.; Saenz de la O, D.; Alvarado-Mariana, A.; Guevara-Gonzalez, R.G.; Garcia-Trejo, J.F.; Feregrino-Perez, A.A. Role of stress and defense in plant secondary metabolites production. In Advanced Structured Materials; Springer: Cham, Switzerland, 2021; Volume 140. [Google Scholar] [CrossRef]
- Groher, T.; Schmittgen, S.; Noga, G.; Hunsche, M. Limitation of mineral supply as tool for the induction of secondary metabolites accumulation in tomato leaves. Plant Physiol. Biochem. 2018, 130, 105–111. [Google Scholar] [CrossRef]
- Ellenberger, J.; Siefen, N.; Krefting, P.; Lutum, J.B.S.; Pfarr, D.; Remmel, M.; Schröder, L.; Röhlen-Schmittgen, S. Effect of UV radiation and salt stress on the accumulation of economically relevant secondary metabolites in bell pepper plants. Agronomy 2020, 10, 142. [Google Scholar] [CrossRef] [Green Version]
- Janmohammadi, M.; Zolla, L.; Rinalducci, S. Low temperature tolerance in plants: Changes at the protein level. Phytochemistry 2015, 117, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Wani, S.H.; Roychoudhury, A. Epigenetic control of plant cold responses. Front. Plant Sci. 2017, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T.; Renaut, J. Plant abiotic stress proteomics: The major factors determining alterations in cellular proteome. Front. Plant Sci. 2018, 9, 122. [Google Scholar] [CrossRef] [Green Version]
- Fürtauer, L.; Weiszmann, J.; Weckwerth, W.; Nägele, T. Dynamics of plant metabolism during cold acclimation. Int. J. Mol. Sci. 2019, 20, 5411. [Google Scholar] [CrossRef] [Green Version]
- Friedrich, T.; Faivre, L.; Bäurle, I.; Schubert, D. Chromatin-based mechanisms of temperature memory in plants. Plant Cell Environ. 2019, 42, 762–770. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Chen, S. Physiological and molecular mechanism involved in cold stress tolerance in plants. Plants 2020, 9, 560. [Google Scholar] [CrossRef]
- Pérez-Ilzarbe, J.; Hernández, T.; Estrella, I.; Vendrell, M. Cold storage of apples (cv. Granny Smith) and changes in phenolic compounds. Eur. Food Res. Technol. 1997, 204, 52–55. [Google Scholar] [CrossRef]
- Griffith, M.; Yaish, M.W.F. Antifreeze proteins in overwintering plants: A tale of two activities. Trends Plant Sci. 2004, 9, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Janská, A.; Maršík, P.; Zelenková, S.; Ovesná, J. Cold stress and acclimation—What is important for metabolic adjustment? Plant Biol. 2010, 12, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Minami, A.; Fujiwara, M.; Furuto, A.; Fukao, Y.; Yamashita, T.; Kamo, M.; Kawamura, Y.; Uemura, M. Alterations in detergent-resistant plasma membrane microdomains in Arabidopsis thaliana during cold acclimation. Plant Cell Physiol. 2009, 50, 341–359. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, D.; Imai, H.; Kawamura, Y.; Uemura, M. Lipid profiles of detergent resistant fractions of the plasma membrane in oat and rye in association with cold acclimation and freezing tolerance. Cryobiology 2016, 72, 123–134. [Google Scholar] [CrossRef] [Green Version]
- Huby, E.; Napier, J.A.; Baillieul, F.; Michaelson, L.V.; Dhondt-Cordelier, S. Sphingolipids: Towards an integrated view of metabolism during the plant stress response. New Phytol. 2020, 225, 659–670. [Google Scholar] [CrossRef]
- Spassieva, S.; Hille, J. Plant sphingolipids today - Are they still enigmatic? Plant Biol. 2003, 5, 125–136. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Dai, X.; Xu, Y.; Guo, J.; Liu, Y.; Chen, N.; Xiao, J.; Zhang, D.; Xu, Z.; Zhang, X.; et al. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 2009, 150, 244–256. [Google Scholar] [CrossRef]
- Lin, Y.S.; Medlyn, B.E.; Ellsworth, D.S. Temperature responses of leaf net photosynthesis: The role of component processes. Tree Physiol. 2012, 32, 219–231. [Google Scholar] [CrossRef] [Green Version]
- Nievola, C.C.; Carvalho, C.P.; Carvalho, V.; Rodrigues, E. Rapid responses of plants to temperature changes. Temperature 2017, 4, 371–405. [Google Scholar] [CrossRef]
- Calixto, C.P.; Guo, W.; James, A.B.; Tzioutziou, N.A.; Entizne, J.C.; Panter, P.E.; Knight, H.; Nimmo, H.G.; Zhang, R.; Brown, J.W. Rapid and dynamic alternative splicing impacts the arabidopsis cold response transcriptome. Plant Cell 2018, 30, 1424–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichersky, E.; Raguso, R.A. Why do plants produce so many terpenoid compounds? New Phytol. 2018, 220, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.; Wu, X.N.; Gilbert, M.; Schulze, W.X. Classification and interactions of LRR receptors and co-receptors within the Arabidopsis Plasma Membrane: An overview. Front. Plant Sci. 2019, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Dievart, A.; Gottin, C.; Périn, C.; Ranwez, V.; Chantret, N. Origin and diversity of plant receptor-like kinases. Annu. Rev. Plant Biol. 2020, 71, 131–156. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Xun, Q.; Guo, Y.; Zhang, J.; Cheng, K.; Shi, T.; He, K.; Hou, S.; Gou, X.; Li, J. Genome-wide expression pattern analyses of the Arabidopsis leucine-rich repeat receptor-like kinases. Mol. Plant 2016, 9, 289–300. [Google Scholar] [CrossRef] [Green Version]
- Shiu, S.H.; Bleecker, A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 2001, 98, 10763–10768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lehti-Shiu, M.D.; Shiu, S.H. Diversity, classification and function of the plant protein kinase superfamily. Philos. Trans. R. Soc. Biol. Sci. 2012, 367, 2619–2639. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.S.; Choi, J.H.; Heinz, J.; Chetty, C.S. Domain-specific positive selection contributes to the evolution of Arabidopsis leucine-rich repeat receptor-like kinase (LRR RLK) genes. J. Mol. Evol. 2006, 63, 612–621. [Google Scholar] [CrossRef]
- Afzal, A.J.; Wood, A.J.; Lightfoot, D.A. Plant receptor-like serine threonine kinases: Roles in signaling and plant defense. Mol.-Plant-Microbe Interact. 2008, 21, 507–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jose, J.; Ghantasala, S.; Choudhury, S.R. Arabidopsis transmembrane Receptor-Like Kinases (RLKs): A bridge between extracellular signal and intracellular regulatory machinery. Int. J. Mol. Sci. 2020, 21, 4000. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Yang, J.; Peng, M.; Liu, X.; He, H. Genome-wide characterization, evolution, and expression analysis of the leucine-rich repeat receptor-like protein kinase (LRR-RLK) gene family in Medicago truncatula. Life 2020, 10, 176. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Ma, X.; Wan, P.; Liu, L. Plant salt-tolerance mechanism: A review. Biochem. Biophys. Res. Commun. 2018, 495, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; Gómez, M.; Hernández, J.A.; del Río, L.A. Metabolism of activated oxygen in peroxisomes from two Pisum sativum L. cultivars with different sensitivity to sodium chloride. J. Plant Physiol. 1993, 141, 160–165. [Google Scholar] [CrossRef]
- Mittova, V.; Tal, M.; Volokita, M.; Guy, M. Up-regulation of the leaf mitochondrial and peroxisomal antioxidative systems in response to salt-induced oxidative stress in the wild salt-tolerant tomato species (Lycopersicon pennellii). Plant Cell Environ. 2003, 26, 845–856. [Google Scholar] [CrossRef] [PubMed]
- Mittova, V.; Guy, M.; Tal, M.; Volokita, M. Salinity up-regulates the antioxidative system in root mitochondria and peroxisomes of the wild salt-tolerant tomato species Lycopersicon pennellii. J. Exp. Bot. 2004, 55, 1105–1113. [Google Scholar] [CrossRef]
- Yang, Z.; Li, J.L.; Liu, L.N.; Xie, Q.; Sui, N. Photosynthetic regulation under salt stress and salt-tolerance mechanism of sweet Sorghum. Front. Plant Sci. 2020, 10, 10. [Google Scholar] [CrossRef]
- Hnilickova, H.; Kraus, K.; Vachova, P.; Hnilicka, F. Salinity stress affects photosynthesis, malondialdehyde formation, and proline content in Portulaca oleracea L. Plants 2021, 10, 845. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef]
- Endler, A.; Kesten, C.; Schneider, R.; Zhang, Y.; Ivakov, A.; Froehlich, A.; Funke, N.; Persson, S. A mechanism for sustained cellulose synthesis during salt stress. Cell 2015, 162, 1353–1364. [Google Scholar] [CrossRef] [Green Version]
- Kesten, C.; Wallmann, A.; Schneider, R.; McFarlane, H.E.; Diehl, A.; Khan, G.A.; van Rossum, B.J.; Lampugnani, E.R.; Szymanski, W.G.; Cremer, N.; et al. The companion of cellulose synthase 1 confers salt tolerance through a Tau-like mechanism in plants. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, W.; Kita, D.; Peaucelle, A.; Cartwright, H.N.; Doan, V.; Duan, Q.; Liu, M.C.; Maman, J.; Steinhorst, L.; Schmitz-Thom, I.; et al. The FERONIA Receptor Kinase Maintains Cell-Wall Integrity during Salt Stress through Ca2+ Signaling. Curr. Biol. 2018, 28, 666–675.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chun, H.J.; Baek, D.; Cho, H.M.; Lee, S.H.; Jin, B.J.; Yun, D.J.; Hong, Y.S.; Kim, M.C. Lignin biosynthesis genes play critical roles in the adaptation of Arabidopsis plants to high-salt stress. Plant Signal. Behav. 2019, 14, 1625697. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Kurepa, J.; Hashimoto, T.; Smalle, J.A. Salt stress-induced disassembly of Arabidopsis cortical microtubule arrays involves 26S proteasome-dependent degradation of SPIRAL1. Plant Cell 2011, 23, 3412–3427. [Google Scholar] [CrossRef] [Green Version]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Schluttenhofer, C.; Yuan, L. Regulation of specialized metabolism by WRKY transcription factors. Plant Physiol. 2015, 167, 295–306. [Google Scholar] [CrossRef] [Green Version]
- Kato, N.; Dubouzet, E.; Kokabu, Y.; Yoshida, S.; Taniguchi, Y.; Dubouzet, J.G.; Yazaki, K.; Sato, F. Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol. 2007, 48, 8–18. [Google Scholar] [CrossRef] [Green Version]
- Ma, D.; Pu, G.; Lei, C.; Ma, L.; Wang, H.; Guo, Y.; Chen, J.; Du, Z.; Wang, H.; Li, G.; et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 2009, 50, 2146–2161. [Google Scholar] [CrossRef] [Green Version]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar] [CrossRef] [Green Version]
- Barco, B.; Clay, N.K. Hierarchical and dynamic regulation of defense-responsive specialized metabolism by WRKY and MYB transcription factors. Front. Plant Sci. 2020, 10, 1775. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Qiao, Z.; Muchero, W.; Chen, J.G. Lectin Receptor-Like Kinases: The sensor and mediator at the plant cell surface. Front. Plant Sci. 2020, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Passricha, N.; Saifi, S.K.; Kharb, P.; Tuteja, N. Rice lectin receptor-like kinase provides salinity tolerance by ion homeostasis. Biotechnol. Bioeng. 2020, 117, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Wang, G.; Zhao, J.L.; Zhang, L.Q.; Ai, L.F.; Han, Y.F.; Sun, D.Y.; Zhang, S.W.; Sun, Y. The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 2014, 26, 2538–2553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.L.; Zhang, L.Q.; Liu, N.; Xu, S.L.; Yue, Z.L.; Zhang, L.L.; Deng, Z.P.; Burlingame, A.L.; Sun, D.Y.; Wang, Z.Y.; et al. Mutual regulation of receptor-like kinase SIT1 and B’K-PP2A shapes the early response of rice to salt stress. Plant Cell 2019, 31, 2131–2151. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, S.; Barah, P.; Suarez-Rodriguez, M.C.; Bressendorff, S.; Friis, P.; Costantino, P.; Bones, A.M.; Nielsen, H.B.; Mundy, J. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiol. 2013, 161, 1783–1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rejeb, I.; Pastor, V.; Mauch-Mani, B. Plant responses to simultaneous biotic and abiotic stress: Molecular mechanisms. Plants 2014, 3, 458–475. [Google Scholar] [CrossRef]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Nishimura, N.; Kitahata, N.; Kuromori, T.; Ito, T.; Asami, T.; Shinozaki, K.; Hirayama, T. ABA-hypersensitive germination 3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 2006, 140, 115–126. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Wei, Q.; Shi, J.; Jin, X.; He, Y.; Zhang, Y.; Luo, Q.; Wang, Y.; Chang, J.; Yang, G.; et al. Brachypodium distachyon BdPP2CA6 interacts with BdPYLs and BdSnRK2 and positively regulates salt tolerance in transgenic Arabidopsis. Front. Plant Sci. 2017, 8, 264. [Google Scholar] [CrossRef] [Green Version]
- Lu, T.; Zhang, G.; Wang, Y.; He, S.; Sun, L.; Hao, F. Genome-wide characterization and expression analysis of PP2CA family members in response to ABA and osmotic stress in Gossypium. PeerJ 2019, 7, e7105. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Wu, J.; Sun, X.; Dai, M. The maize clade A PP2C phosphatases play critical roles in multiple abiotic stress responses. Int. J. Mol. Sci. 2019, 20, 3573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruelland, E.; Vaultier, M.N.; Zachowski, A.; Hurry, V. Chapter 2: Cold signalling and cold acclimation in plants. Adv. Bot. Res. 2009, 49, 35–150. [Google Scholar] [CrossRef]
- Hussain, S.; Khan, F.; Cao, W.; Wu, L.; Geng, M. Seed priming alters the production and detoxification of reactive oxygen intermediates in rice seedlings grown under sub-optimal temperature and nutrient supply. Front. Plant Sci. 2016, 7, 439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Wang, P.; Hou, H.; Zhang, H.; Wang, Y.; Yan, S.; Huang, Y.; Li, H.; Tan, J.; Hu, A.; et al. Transcriptional regulation of cell cycle genes in response to abiotic stresses correlates with dynamic changes in histone modifications in maize. PLoS ONE 2014, 9, e106070. [Google Scholar] [CrossRef]
- Qi, F.; Zhang, F. Cell cycle regulation in the plant response to stress. Front. Plant Sci. 2020, 10, 1765. [Google Scholar] [CrossRef] [Green Version]
- Rymen, B.; Fiorani, F.; Kartal, F.; Vandepoele, K.; Inzé, D.; Beemster, G.T. Cold nights impair leaf growth and cell cycle progression in maize through transcriptional changes of cell cycle genes. Plant Physiol. 2007, 143, 1429–1438. [Google Scholar] [CrossRef] [Green Version]
- Vallarino, J.G.; de Abreu e Lima, F.; Soria, C.; Tong, H.; Pott, D.M.; Willmitzer, L.; Fernie, A.R.; Nikoloski, Z.; Osorio, S. Genetic diversity of strawberry germplasm using metabolomic biomarkers. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Pott, D.M.; Osorio, S.; Vallarino, J.G. From Central to specialized metabolism: An overview of some secondary compounds derived from the primary metabolism for their role in conferring nutritional and organoleptic characteristics to fruit. Front. Plant Sci. 2019, 10, 835. [Google Scholar] [CrossRef] [Green Version]
- Bhat, M.A.; Ahmad, S.; Aslam, J.; Mujib, A.; Mahmooduzzfar. Salinity stress enhances production of solasodine in Solanum nigrum L. Chem. Pharm. Bull. 2008, 56, 17–21. [Google Scholar] [CrossRef] [Green Version]
- Rabara, R.C.; Tripathi, P.; Reese, R.N.; Rushton, D.L.; Alexander, D.; Timko, M.P.; Shen, Q.J.; Rushton, P.J. Tobacco drought stress responses reveal new targets for Solanaceae crop improvement. BMC Genom. 2015, 16, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Rivero, R.M.; Mestre, T.C.; Mittler, R.; Rubio, F.; Garcia-Sanchez, F.; Martinez, V. The combined effect of salinity and heat reveals a specific physiological, biochemical and molecular response in tomato plants. Plant Cell Environ. 2014, 37, 1059–1073. [Google Scholar] [CrossRef] [PubMed]
- Ou, L.J.; Wei, G.; Zhang, Z.Q.; Dai, X.Z.; Zou, X.X. Effects of low temperature and low irradiance on the physiological characteristics and related gene expression of different pepper species. Photosynthetica 2015, 53, 85–94. [Google Scholar] [CrossRef]
- Yanagawa, Y.; Komatsu, S. Ubiquitin/proteasome-mediated proteolysis is involved in the response to flooding stress in soybean roots, independent of oxygen limitation. Plant Sci. 2012, 185–186, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Liang, L.; Xie, Y.; Zhao, Z.; Su, L.; Tang, Y.; Sun, B.; Lai, Y.; Li, H. Transcriptome and metabolome analyses reveal molecular responses of two pepper (Capsicum annuum L.) cultivars to cold stress. Front. Plant Sci. 2022, 13, 574. [Google Scholar] [CrossRef]
- Zhao, Q.; Brkljacic, J.; Meier, I. Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 2008, 20, 1639–1651. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Groves, N.R.; Meier, I. Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1. Nucleus 2015, 6, 144–153. [Google Scholar] [CrossRef] [Green Version]
- Soto, G.; Stritzler, M.; Lisi, C.; Alleva, K.; Pagano, M.E.; Ardila, F.; Mozzicafreddo, M.; Cuccioloni, M.; Angeletti, M.; Ayub, N.D. Acetoacetyl-CoA thiolase regulates the mevalonate pathway during abiotic stress adaptation. J. Exp. Bot. 2011, 62, 5699–5711. [Google Scholar] [CrossRef]
- Zhao, C.; Lang, Z.; Zhu, J.K. Cold responsive gene transcription becomes more complex. Trends Plant Sci. 2015, 20, 466–468. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Silva, J.A.D.; Niu, M.; Li, M.; He, C.; Zhao, J.; Zeng, S.; Duan, J.; Ma, G. Physiological and transcriptomic analyses reveal a response mechanism to cold stress in Santalum album L. leaves. Sci. Rep. 2017, 7, 42165. [Google Scholar] [CrossRef] [Green Version]
- Mageney, V.; Baldermann, S.; Albach, D.C. Intraspecific variation in carotenoids of Brassica oleracea var. sabellica. J. Agric. Food Chem. 2016, 64, 3251–3257. [Google Scholar] [CrossRef] [PubMed]
- Mathesius, U. Flavonoid functions in plants and their interactions with other organisms. Plants 2018, 7, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yonekura-Sakakibara, K.; Higashi, Y.; Nakabayashi, R. The origin and evolution of plant flavonoid metabolism. Front. Plant Sci. 2019, 10, 943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tohge, T.; Alseekh, S.; Fernie, A.R. On the regulation and function of secondary metabolism during fruit development and ripening. J. Exp. Bot. 2013, 65, 4599–4611. [Google Scholar] [CrossRef] [Green Version]
- Antonio, A.S.; Wiedemann, L.S.M.; Junior, V.F.V. The genus Capsicum: A phytochemical review of bioactive secondary metabolites. RSC Adv. 2018, 8, 25767–25784. [Google Scholar] [CrossRef] [Green Version]
- Tohge, T.; de Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef]
- Knekt, P.; Kumpulainen, J.; Järvinen, R.; Rissanen, H.; Heliövaara, M.; Reunanen, A.; Hakulinen, T.; Aromaa, A. Flavonoid intake and risk of chronic diseases. Am. J. Clin. Nutr. 2002, 76, 560–568. [Google Scholar] [CrossRef] [Green Version]
- Tohge, T.; Fernie, A.R. An overview of compounds derived from the shikimate and phenylpropanoid pathways and their medicinal importance. Mini-Rev. Med. Chem. 2017, 17, 1013–1027. [Google Scholar] [CrossRef]
- Sun, Q.; Lu, N.N.; Feng, L. Apigetrin inhibits gastric cancer progression through inducing apoptosis and regulating ROS-modulated STAT3/JAK2 pathway. Biochem. Biophys. Res. Commun. 2018, 498, 164–170. [Google Scholar] [CrossRef]
- Kim, W.R.; Kim, E.O.; Kang, K.; Oidovsambuu, S.; Jung, S.H.; Kim, B.S.; Nho, C.W.; Um, B.H. Antioxidant activity of phenolics in leaves of three red pepper (Capsicum annuum) cultivars. J. Agric. Food Chem. 2014, 62, 850–859. [Google Scholar] [CrossRef]
- Zietz, M.; Weckmüller, A.; Schmidt, S.; Rohn, S.; Schreiner, M.; Krumbein, A.; Kroh, L.W. Genotypic and climatic influence on the antioxidant activity of flavonoids in kale (Brassica oleracea var. sabellica). J. Agric. Food Chem. 2010, 58, 2123–2130. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Shi, R.; Wang, X.; Shen, H.M. Luteolin, a flavonoid with potential for cancer prevention and therapy. Curr. Cancer Drug Targets 2008, 8, 634–646. [Google Scholar] [CrossRef] [PubMed]
- Jan, R.; Kim, N.; Lee, S.H.; Khan, M.A.; Asaf, S.; Lubna; Park, J.R.; Asif, S.; Lee, I.J.; Kim, K.M. Enhanced flavonoid accumulation reduces combined salt and heat stress through regulation of transcriptional and hormonal mechanisms. Front. Plant Sci. 2021, 12, 796956. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Morishita, T.; Kim, S.J.; Park, S.U.; Woo, S.H.; Noda, T.; Takigawa, S. Physiological roles of rutin in the buckwheat plant. Jpn. Agric. Res. Q. JARQ 2015, 49, 37–43. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, T.; Honda, Y.; Mukasa, Y. Effects of UV-B radiation, cold and desiccation stress on rutin concentration and rutin glucosidase activity in tartary buckwheat (Fagopyrum tataricum) leaves. Plant Sci. 2005, 168, 1303–1307. [Google Scholar] [CrossRef]
- Acquaviva, R.; Russo, A.; Galvano, F.; Galvano, G.; Barcellona, M.L.; Volti, G.L.; Vanella, A. Cyanidin and cyanidin 3-O-β-D-glucoside as DNA cleavage protectors and antioxidants. Cell Biol. Toxicol. 2003, 19, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Serraino, I.; Dugo, L.; Dugo, P.; Mondello, L.; Mazzon, E.; Dugo, G.; Caputi, A.P.; Cuzzocrea, S. Protective effects of cyanidin-3-O-glucoside from blackberry extract against peroxynitrite-induced endothelial dysfunction and vascular failure. Life Sci. 2003, 73, 1097–1114. [Google Scholar] [CrossRef]
- Pace, E.; Jiang, Y.; Clemens, A.; Crossman, T.; Rupasinghe, H.P.V. Impact of thermal degradation of cyanidin-3-O-glucoside of haskap berry on cytotoxicity of hepatocellular carcinoma HepG2 and breast cancer MDA-MB-231 cells. Antioxidants 2018, 7, 24. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Liang, H.; Guo, Y.; Yang, D. Cyanidin 3-O-galactoside: A Natural Compound with multiple health benefits. Int. J. Mol. Sci. 2021, 22, 2261. [Google Scholar] [CrossRef]
- Schulz, E.; Tohge, T.; Zuther, E.; Fernie, A.R.; Hincha, D.K. Natural variation in flavonol and anthocyanin metabolism during cold acclimation in Arabidopsis thaliana accessions. Plant Cell Environ. 2015, 38, 1658–1672. [Google Scholar] [CrossRef]
- Ahmed, N.U.; Park, J.I.; Jung, H.J.; Hur, Y.; Nou, I.S. Anthocyanin biosynthesis for cold and freezing stress tolerance and desirable color in Brassica rapa. Funct. Integr. Genom. 2015, 15, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhai, J.; Shao, L.; Lin, W.; Peng, C. Accumulation of anthocyanins: An adaptation strategy of Mikania micrantha to low temperature in winter. Front. Plant Sci. 2019, 10, 1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Q.; Ren, Y.; Zhao, W.; Li, R.; Zhang, L. Low temperature promotes anthocyanin biosynthesis and related gene expression in the seedlings of purple head chinese cabbage (Brassica rapa L.). Genes 2020, 11, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
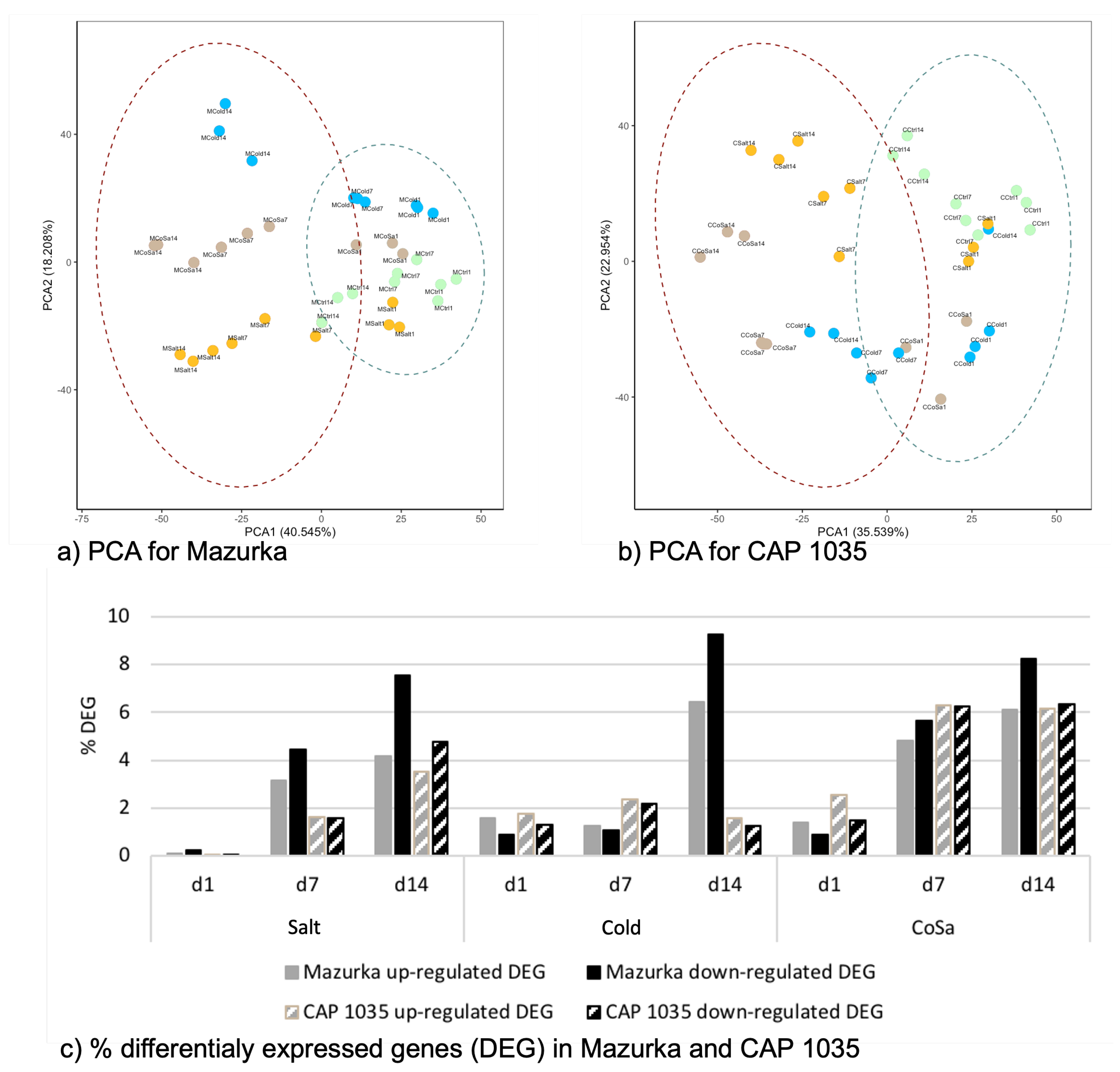


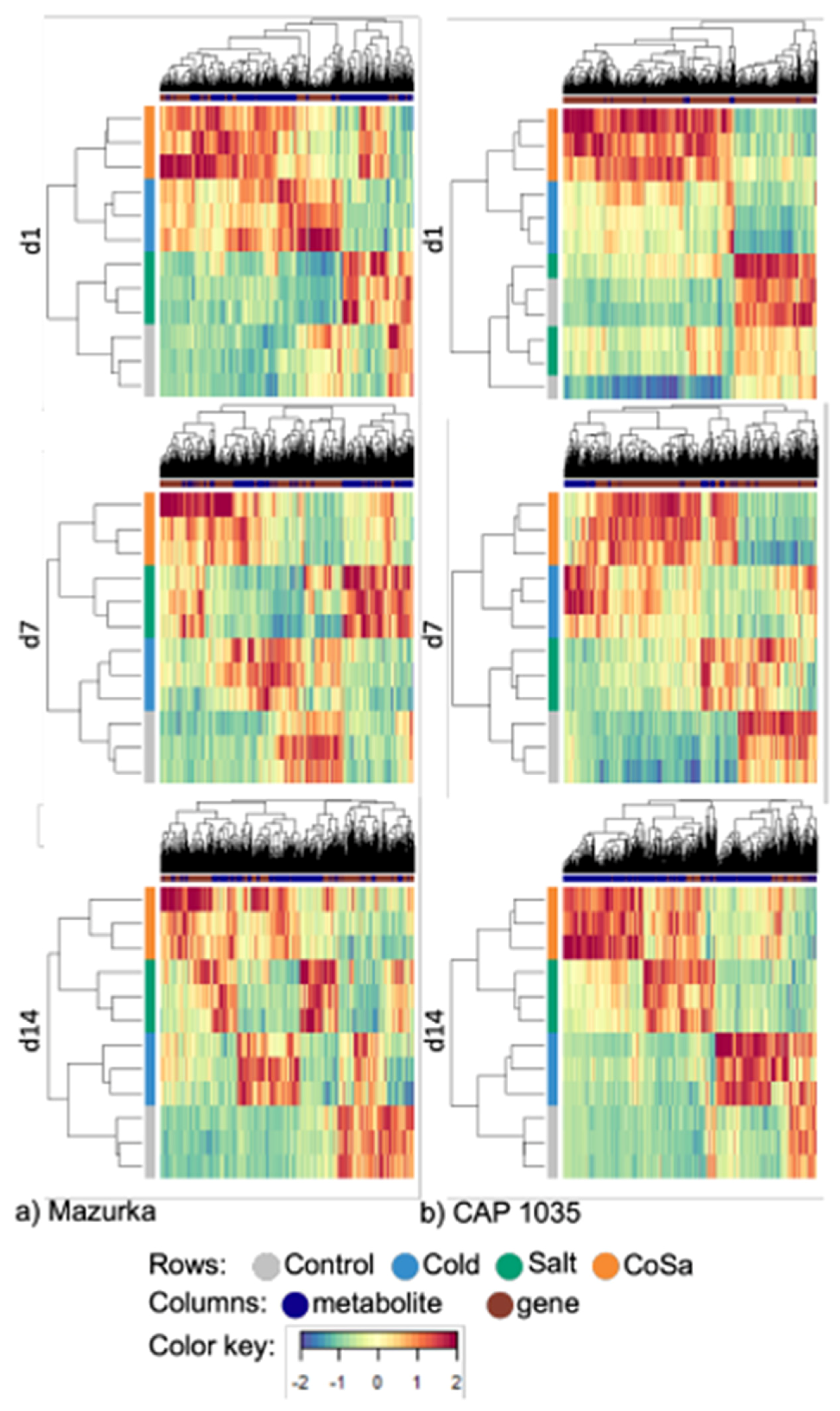
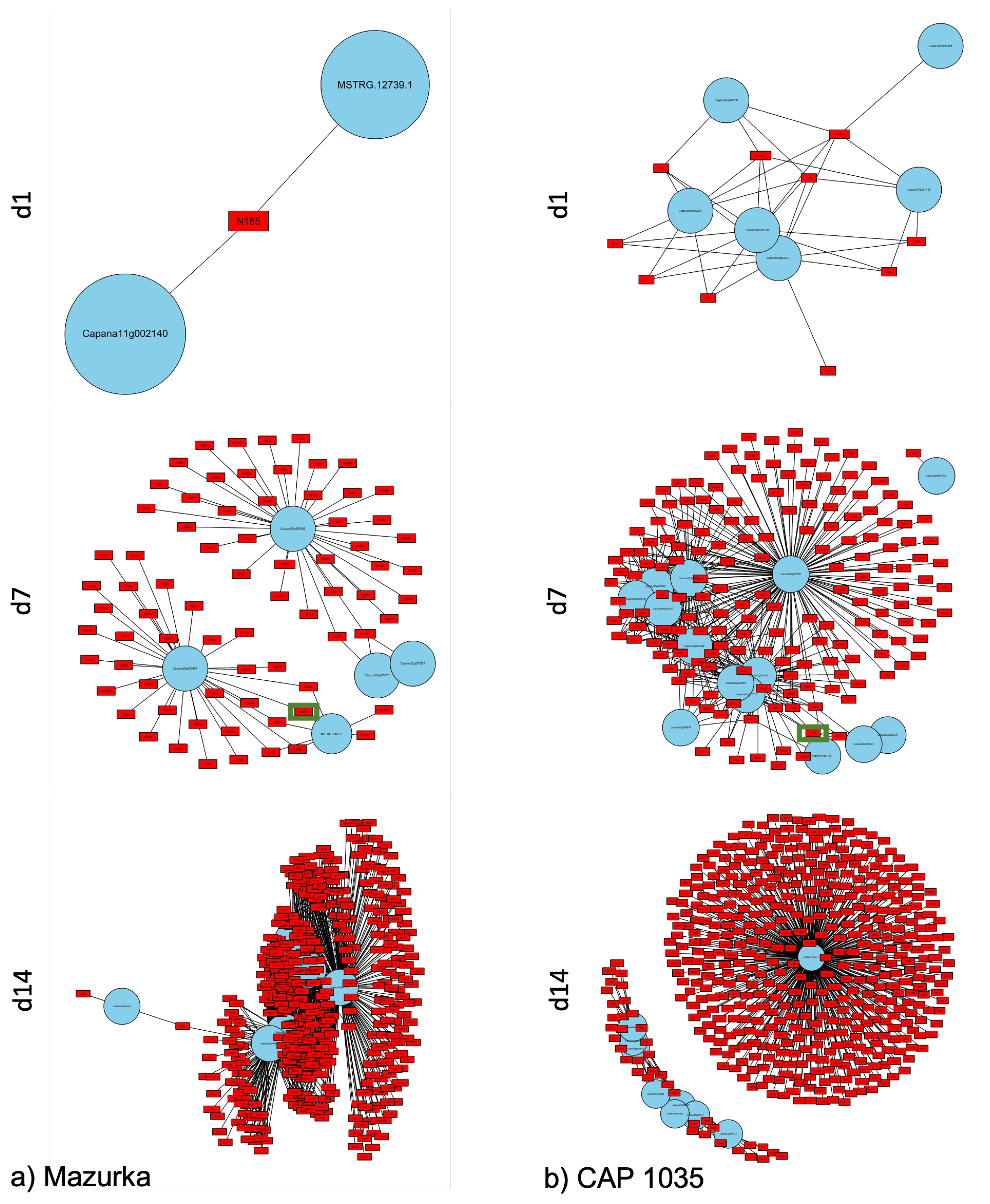
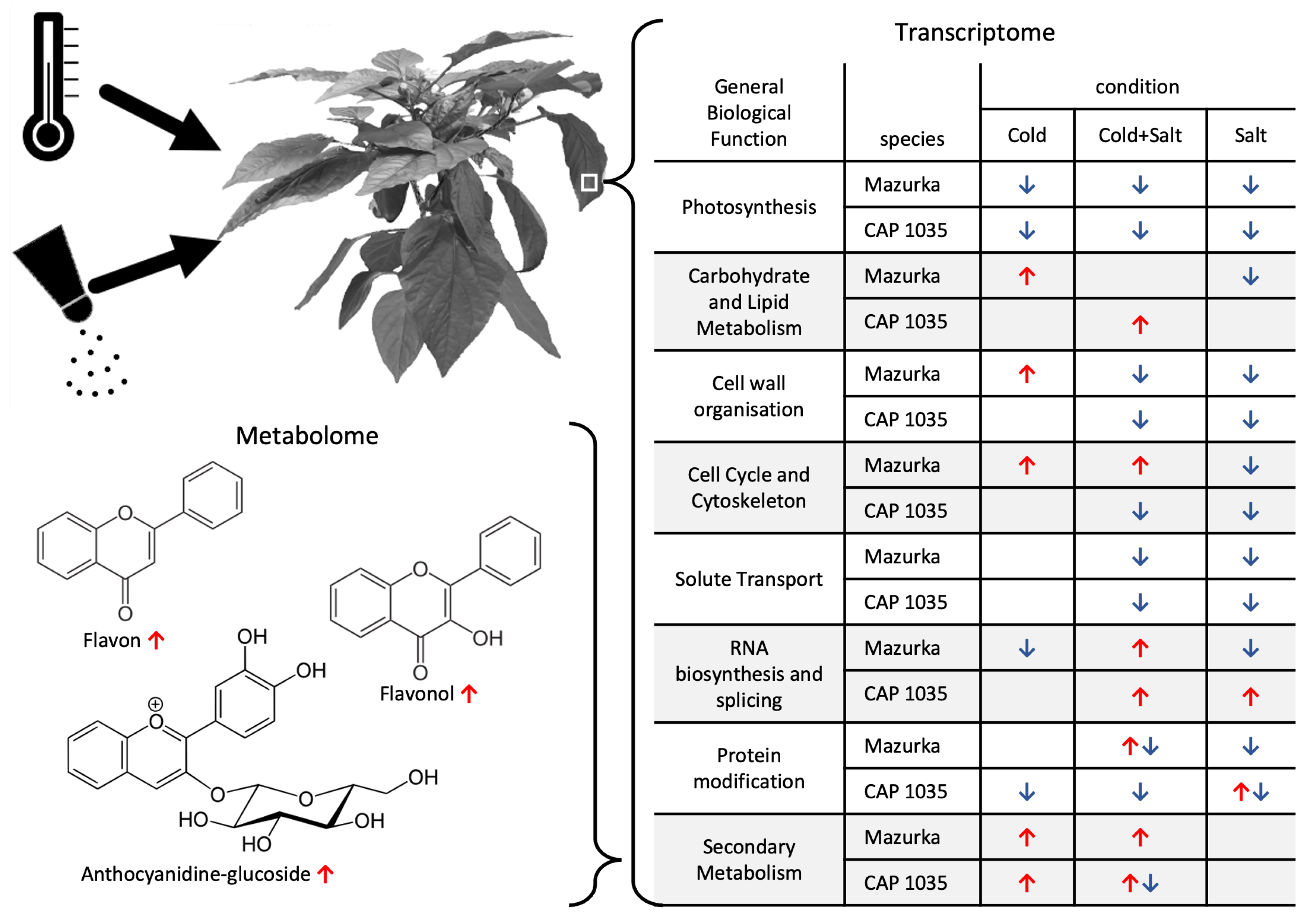
| Species | Harvest Time Point | No. of Sig. DEG | No. of Sig. Mass Features |
|---|---|---|---|
| Day 1 | 437 | 352 | |
| Mazurka | Day 7 | 2320 | 2040 |
| Day 14 | 5623 | 3385 | |
| Day 1 | 1489 | 70 | |
| CAP 1035 | Day 7 | 3413 | 956 |
| Day 14 | 1818 | 4691 |
| Condition | ||||||||
|---|---|---|---|---|---|---|---|---|
| Timepoint | Total No. of | Cold | Cold+Salt | Salt | ||||
| Affect. Mass Features | Increase | Decrease | Increase | Decrease | Increase | Decrease | ||
| Day 1 | 352 | 256 | 93 | 279 | 71 | 264 | 83 | |
| M | Day 7 | 2040 | 1314 | 686 | 1587 | 439 | 1530 | 500 |
| Day 14 | 3385 | 2262 | 1099 | 2652 | 726 | 2536 | 789 | |
| Day 1 | 70 | 46 | 23 | 54 | 15 | 41 | 28 | |
| C | Day 7 | 956 | 766 | 159 | 754 | 194 | 559 | 366 |
| Day 14 | 4691 | 3690 | 729 | 3933 | 741 | 3578 | 1041 | |
| Gene ID | Gene Name | Put. | Reg. | d1 | d7 | d14 |
|---|---|---|---|---|---|---|
| Capana06g000272 | trans-cinnamate 4-monooxygenase | up | C | C | C | |
| Capana09g002190 | phenylalanine ammonia-lyase-like | up | C | C | ||
| Capana09g002200 | phenylalanine ammonia-lyase-like | up | C | C | ||
| Capana06g000273 | trans-cinnamate 4-monooxygenase-like | up | C | C, M | ||
| Capana00g003499 | phenylalanine ammonia-lyase | x | up | C | M | |
| Capana07g001146 | acetyl-CoA acetyltransferase | x | up | C | ||
| Capana11g002140 | WPP domain-interacting tail-anchored | x | up | M | ||
| protein 2 | ||||||
| Capana06g001083 | 4-coumarate-CoA ligase 2 | x | up | C | ||
| MSTRG.44643.1 | chalcone synthase J-like | x | up | C | ||
| Capana02g002404 | 3-hydroxy-3-methylglutaryl-coenzyme A reductase | x | up | C | M | |
| reductase | ||||||
| Capana05g002274 | chalcone synthase 2 | up | M | |||
| Capana01g000948 | carotenoid cleavage dioxygenase 4 | x | down | C | C, M | |
| Capana05g002107 | chalcone-flavonone isomerase 3 | x | up | C | C, M | |
| Capana03g000578 | 4-coumarate_coenzyme A ligase (4CL) | up | C, M | C, M | ||
| Capana08g001733 | 2-C-methyl-D-erythritol 4-phosphate | x | down | C | ||
| cytidylyltransferase | ||||||
| Capana09g000404 | beta-amyrin synthase-like | x | up | C | ||
| Capana04g002519 | bifunctional 15-cis-phytoene synthase | up | C | |||
| Capana00g002736 | chalcone-flavonone isomerase 3 | x | up | C | ||
| Capana06g002959 | cytochrome P450 | x | down | C | ||
| Capana03g000892 | flavonoid 3’-monooxygenase | x | up | C | ||
| Capana06g000463 | licodione synthase-like | x | up | C | ||
| Capana02g000638 | select squalene epoxidase 3-like | x | up | C | ||
| Capana05g000154 | acetyl-CoA acetyltransferase, cytosolic 2 | x | up | M | ||
| MSTRG.13875.1 | hydroxymethylglutaryl-CoA synthase-like | x | up | M | ||
| Capana12g001287 | acetyl-CoA carboxylase 1 | x | up | C, M |
| Group | Compound | Stress | Putatively Annotated Metabolites | Reference |
|---|---|---|---|---|
| M d7 | N2109 | Salt | Isovitexin | [74,75] |
| Vitexin | [75] | |||
| Apigetrin | [52] | |||
| Afzelin | [76] | |||
| C d7 | N2744 | Salt | Kaempferol-3-O-rutinoside | [74] |
| Luteolin 7-beta-neohesperidoside | [77] | |||
| M d14 | C3609 | CoSa | p-Coumaroyl quinic acid | [78] |
| N6446 | CoSa | Sophoraflavonoloside | [79] | |
| N3794 | CoSa | Myricitrin | [80] | |
| N6025 | CoSa | Kaempferol | [81] | |
| Luteolin | [81] | |||
| N3036, N3037, N4704 | CoSa | Apiin | [52] | |
| Quercetin 3-O-rhamnoside 7-O-glucoside | [82,83] | |||
| Rutin | [84] | |||
| Cyanidin 3,7-di-O-beta-D-glucoside | [85] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reimer, J.J.; Shaaban, B.; Drummen, N.; Sanjeev Ambady, S.; Genzel, F.; Poschet, G.; Wiese-Klinkenberg, A.; Usadel, B.; Wormit, A. Capsicum Leaves under Stress: Using Multi-Omics Analysis to Detect Abiotic Stress Network of Secondary Metabolism in Two Species. Antioxidants 2022, 11, 671. https://doi.org/10.3390/antiox11040671
Reimer JJ, Shaaban B, Drummen N, Sanjeev Ambady S, Genzel F, Poschet G, Wiese-Klinkenberg A, Usadel B, Wormit A. Capsicum Leaves under Stress: Using Multi-Omics Analysis to Detect Abiotic Stress Network of Secondary Metabolism in Two Species. Antioxidants. 2022; 11(4):671. https://doi.org/10.3390/antiox11040671
Chicago/Turabian StyleReimer, Julia Jessica, Basel Shaaban, Noud Drummen, Sruthy Sanjeev Ambady, Franziska Genzel, Gernot Poschet, Anika Wiese-Klinkenberg, Björn Usadel, and Alexandra Wormit. 2022. "Capsicum Leaves under Stress: Using Multi-Omics Analysis to Detect Abiotic Stress Network of Secondary Metabolism in Two Species" Antioxidants 11, no. 4: 671. https://doi.org/10.3390/antiox11040671
APA StyleReimer, J. J., Shaaban, B., Drummen, N., Sanjeev Ambady, S., Genzel, F., Poschet, G., Wiese-Klinkenberg, A., Usadel, B., & Wormit, A. (2022). Capsicum Leaves under Stress: Using Multi-Omics Analysis to Detect Abiotic Stress Network of Secondary Metabolism in Two Species. Antioxidants, 11(4), 671. https://doi.org/10.3390/antiox11040671








