Gentirigeoside B from Gentiana rigescens Franch Prolongs Yeast Lifespan via Inhibition of TORC1/Sch9/Rim15/Msn Signaling Pathway and Modification of Oxidative Stress and Autophagy
Abstract
1. Introduction
2. Materials and Methods
2.1. General
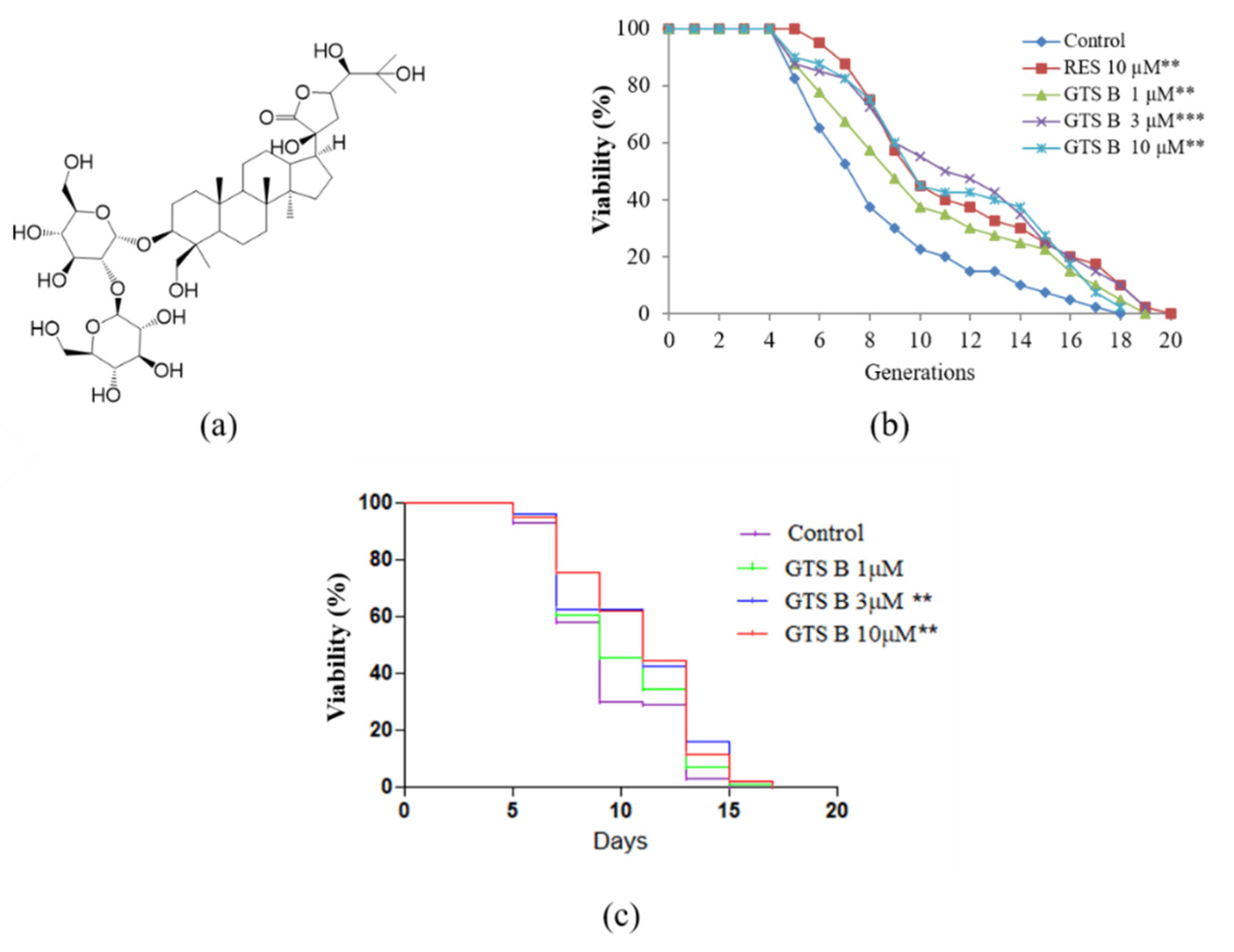
2.2. Preparation of Gentirigeoside B from G. rigescens Franch
2.3. Yeast Strain, Culture Medium and Lifespan Assay
2.4. Real-Time Polymerase Chain Reaction (RT-PCR) Analysis
2.5. Nuclear Translocation of Rim15 and Msn2 Proteins Analysis
2.6. T-Sod, Sod1 and Cat Enzymes Activity Assay
2.7. Yeast Growth Experiment under Oxidative Stress Condition
2.8. Measurement of MDA, ROS Levels of Yeast
2.9. Visualization of Autophagy Induction in Yeast
2.10. Western Blotting Analysis
2.11. Statistical Analysis
3. Results
3.1. GTS B Prolongs Replicative and Chronological Lifespan of Yeast Cells
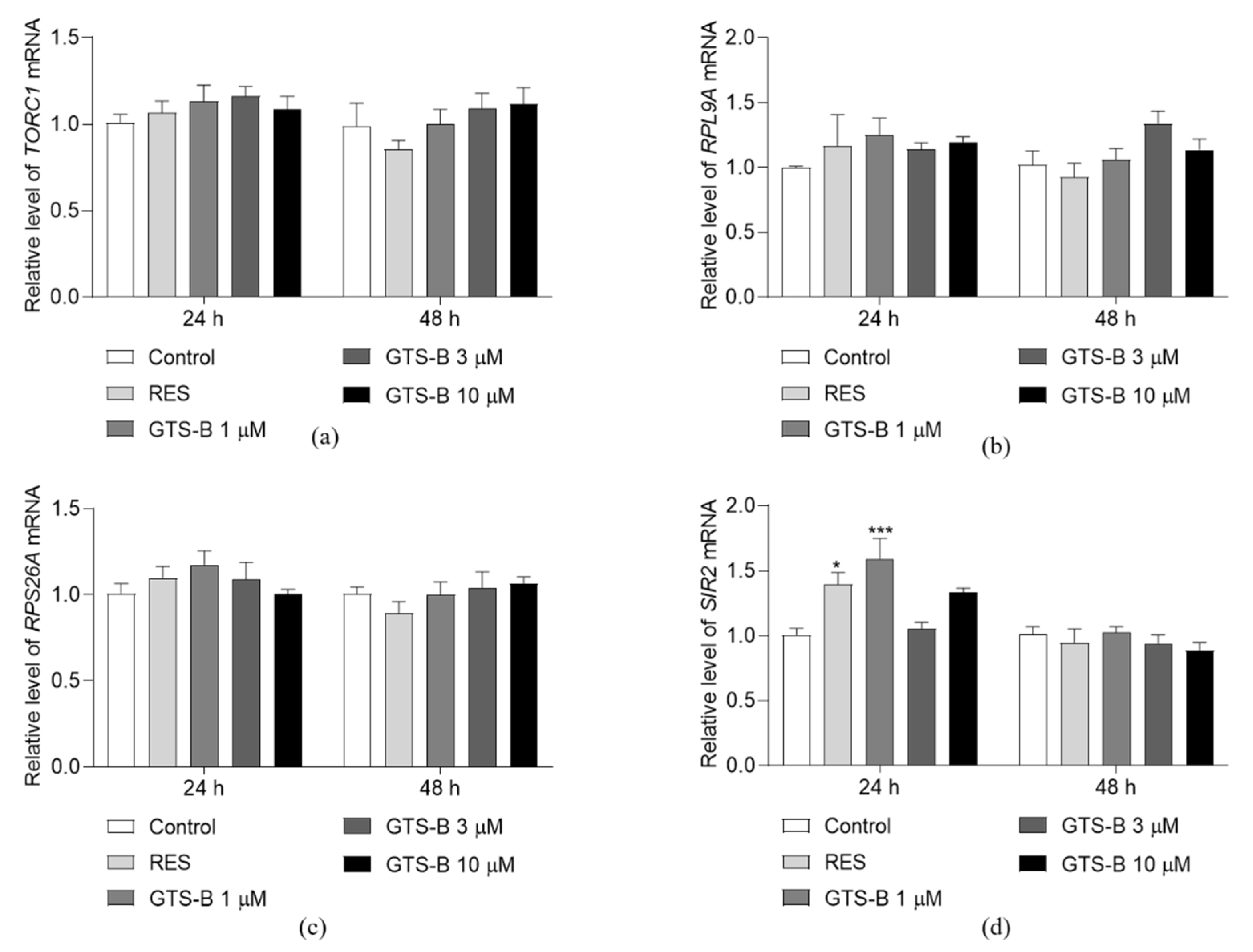
3.2. Effect of GTS B on the Gene Expression of TORC1, RPS26A, RPL9A and SIR2
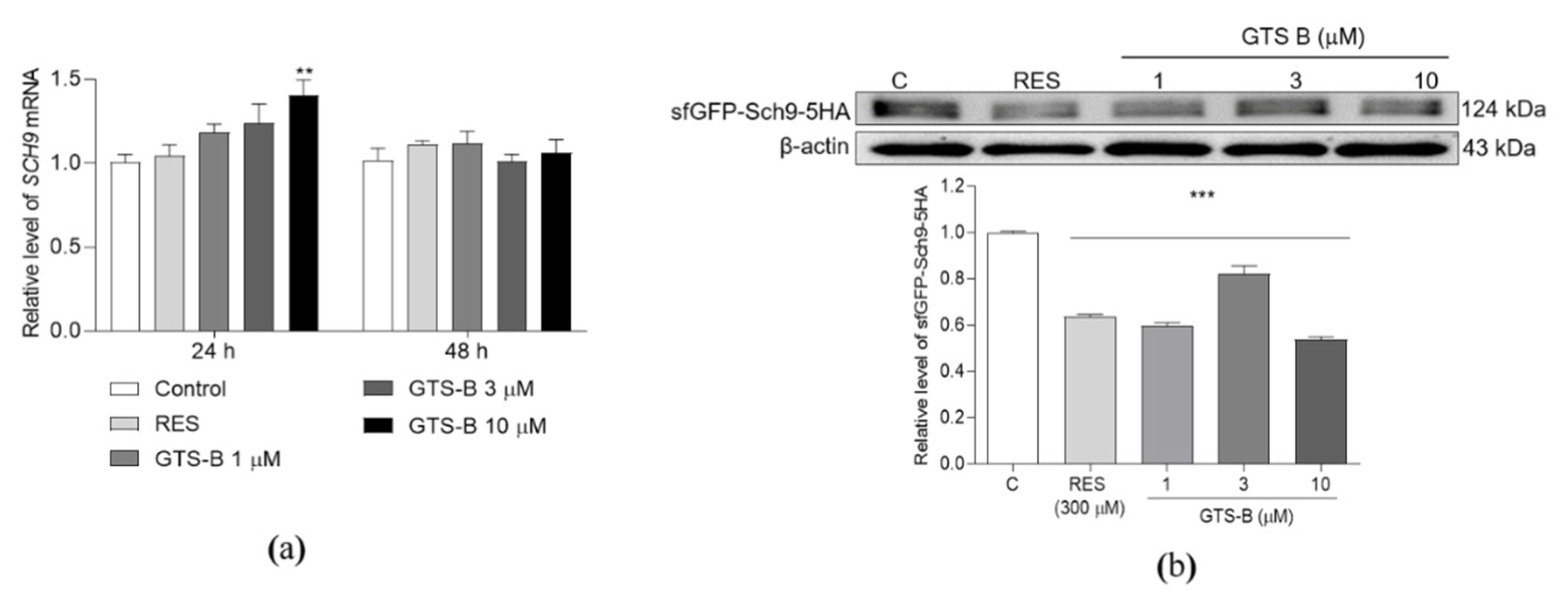
3.3. Effect of GTS B on the Gene Expression and Protein Level of Sch9
3.4. Effect of GTS B on the Gene Expression and Activity of Rim15
3.5. Effect of GTS B on the Gene Expression of MSN2 and MSN4 and Activity of Msn2
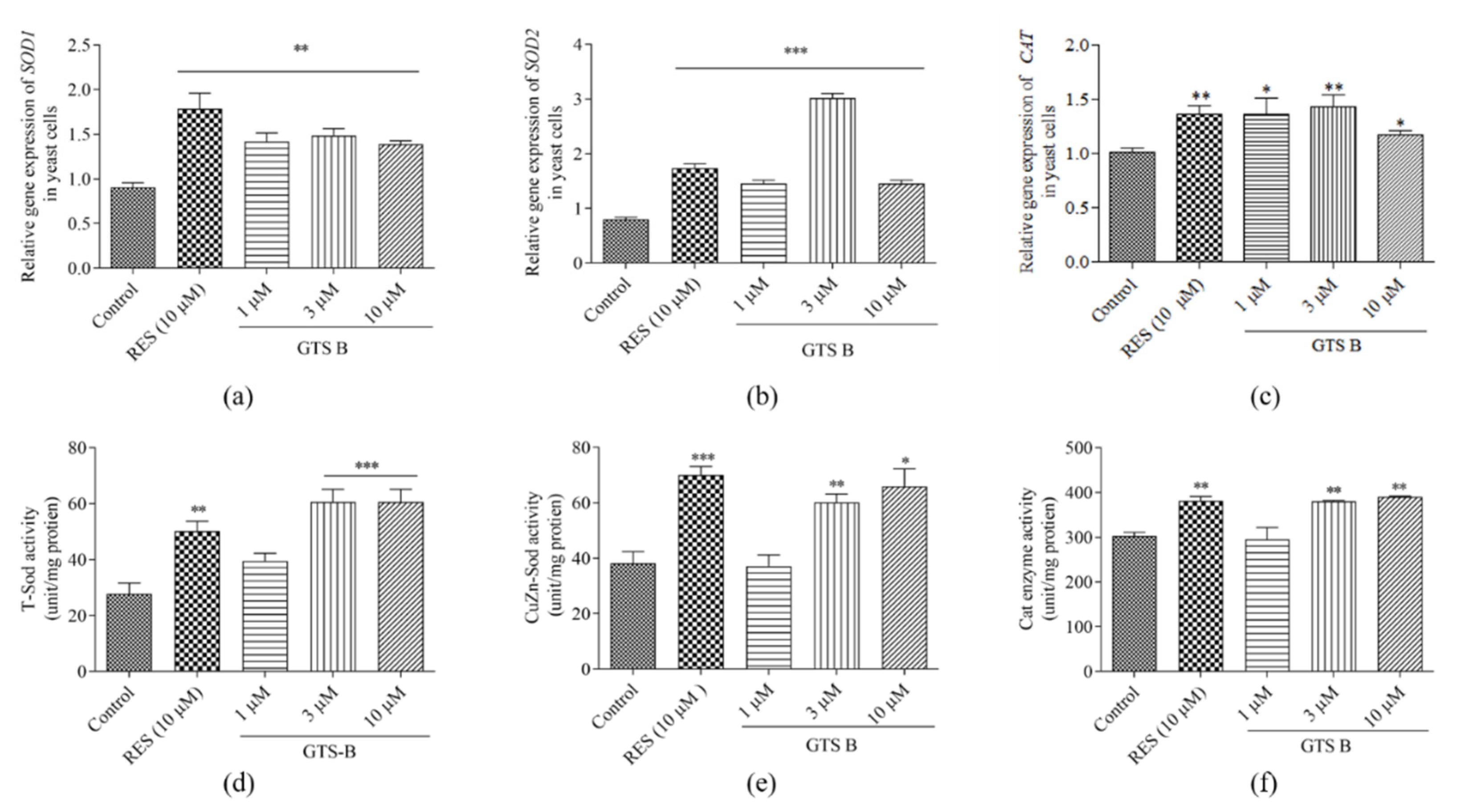
3.6. GTS B Improves Gene Expression of SOD1, SOD2 and CAT
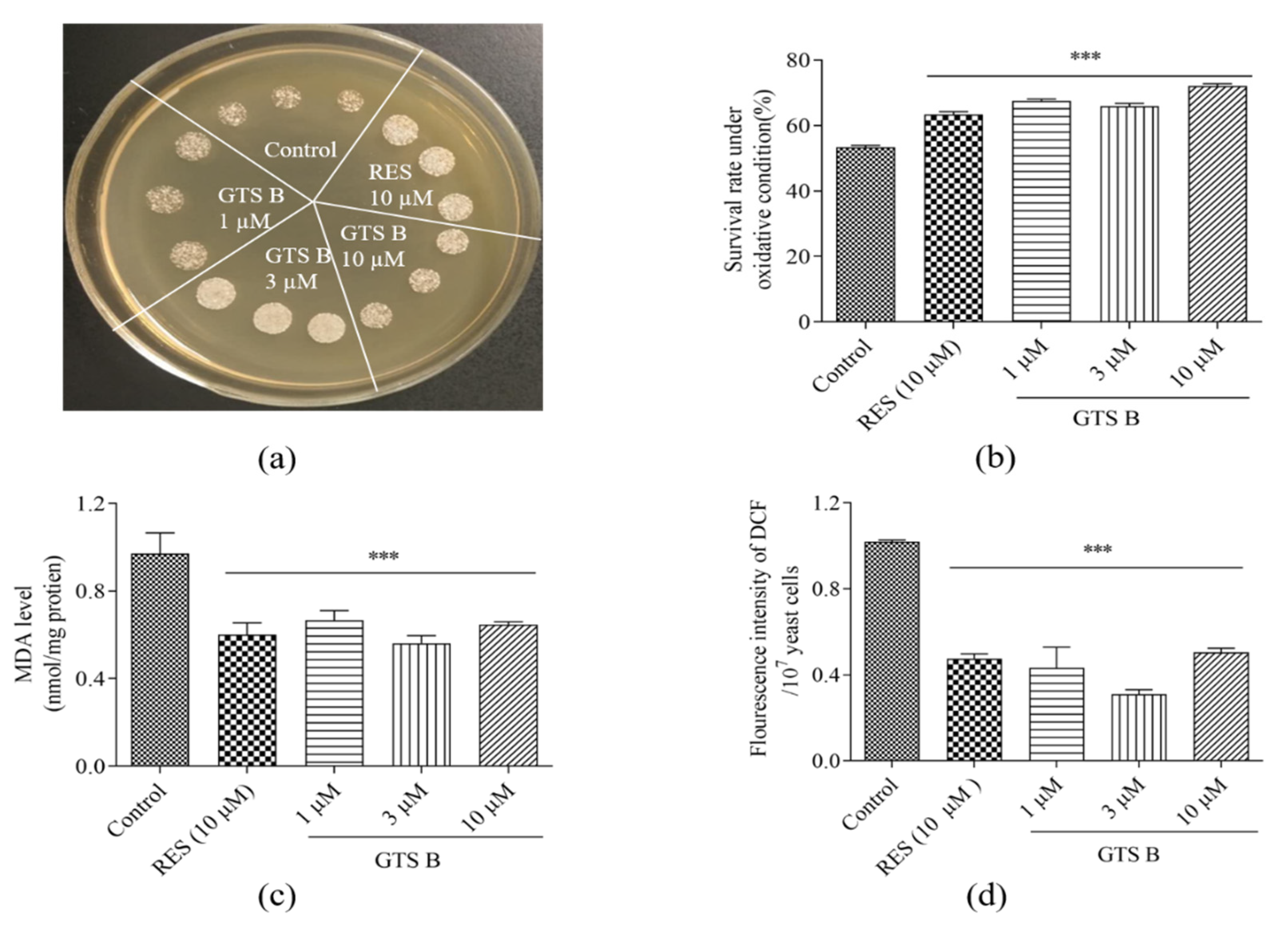
3.7. Effects of GTS B on Oxidative Stress of Yeast
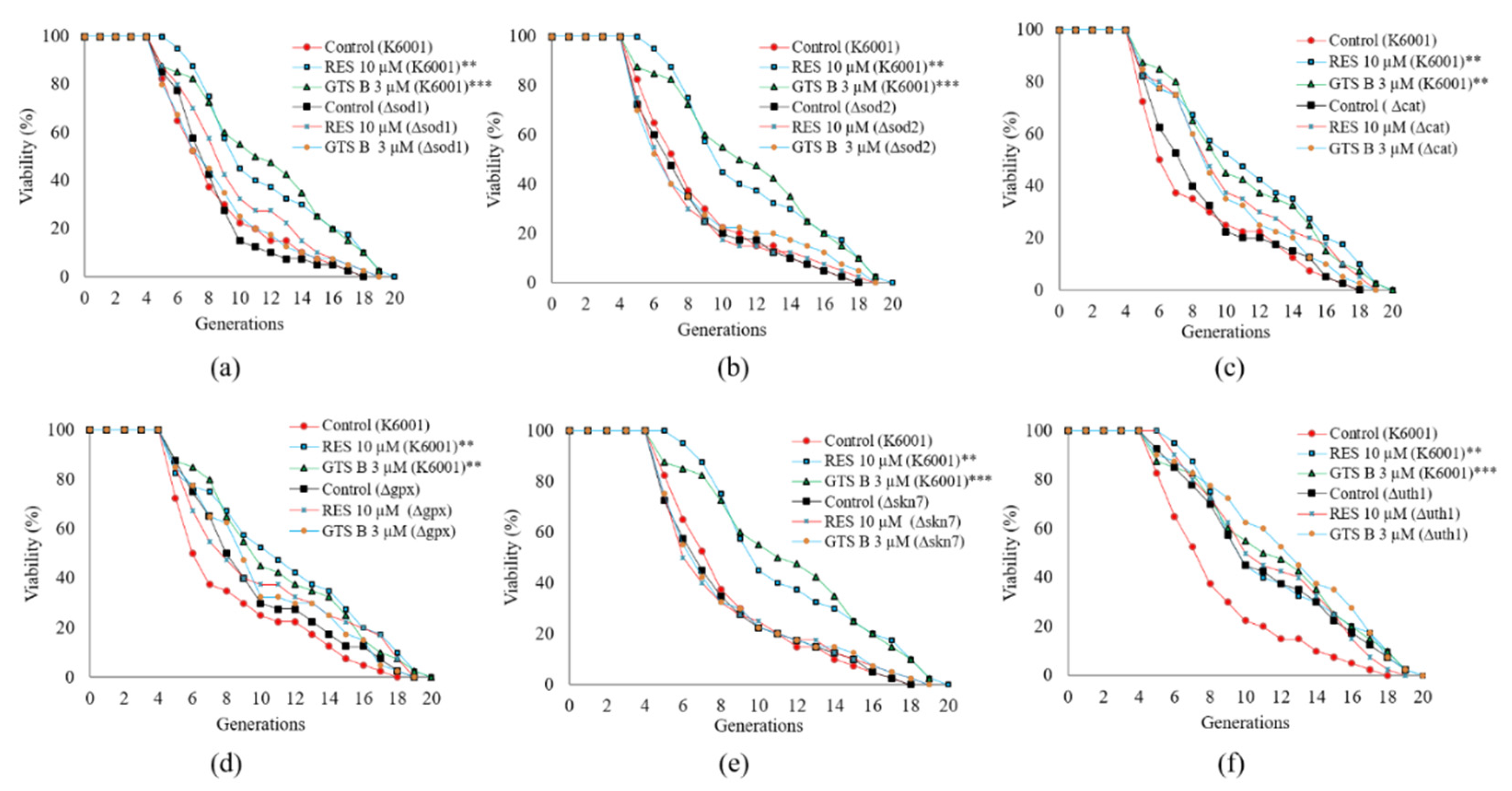
3.8. GTS B Failed to Prolong the Replicative Lifespan of Mutants of K6001 Yeast-Related Oxidative Stress
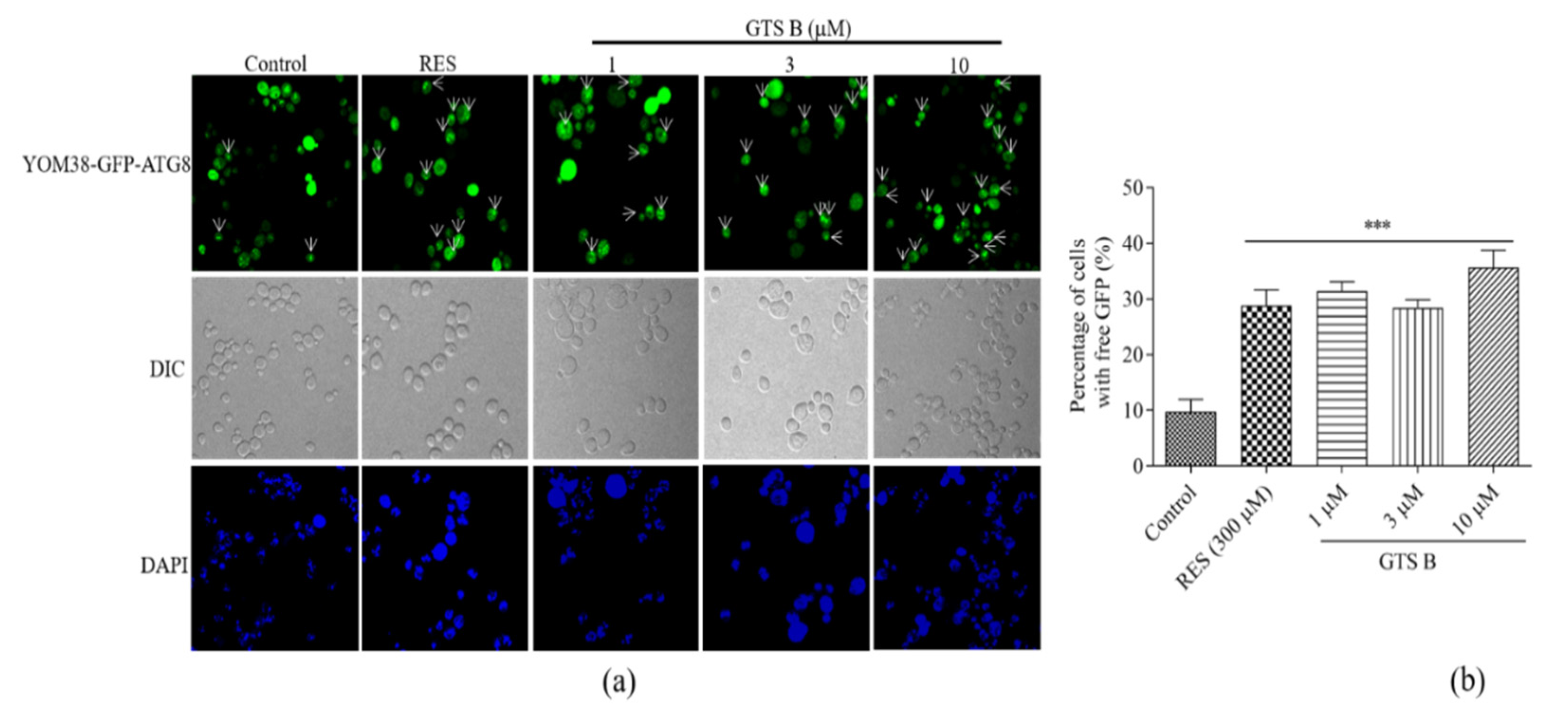
3.9. Effect of GTS B on Autophagic Induction in Yeast
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Eisenberg, T.; Knauer1, H.; Schauer, A.; Buttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Vellai, T.; Taka´cs-Vellai, K.; Sass, M.; Klionsky, D.J. The regulation of aging: Does autophagy underlie longevity? Trends Cell Biol. 2009, 19, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, A.; Hofer, S.; Pendl, T.; Kainz, K.; Madeo, F.; Carmona-Gutierrez, D. Yeast as a tool to identify anti-aging compounds. FEMS Yeast Res. 2018, 18, foy020. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.W.; Huh, W.K. Rapamycin increases rDNA stability by enhancing association of Sir2 with rDNA in Saccharomyces cerevisiae. Nucleic Acids Res. 2011, 39, 1336–1350. [Google Scholar] [CrossRef] [PubMed]
- Devare, M.N.; Kim, Y.H.; Jung, J.; Kang, W.K.; Kwon, K.; Kim, J. TORC1 signaling regulates cytoplasmic pH through Sir2 in yeast. Aging Cell 2020, 19, 13151. [Google Scholar] [CrossRef]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef] [PubMed]
- Vellai, T.; Takacs-Vellai, K.; Zhang, Y.; Kovacs, A.L.; Orosz, L.; Müller, F. Genetics-influence of TOR kinase on lifespan in C. elegans. Nature 2003, 426, 620. [Google Scholar] [CrossRef] [PubMed]
- Kapahi, P.; Zid, B.M.; Harper, T.; Koslover, D.; Sapin, V.; Benzer, S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004, 14, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Kaeberlein, M.; Powers, R.W.; Steffen, K.K.; Westman, E.A.; Hu, D.; Dang, N.; Kerr, E.O.; Kirkland, K.T.; Fields, S.; Kennedy, B.K.; et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 2005, 310, 1193–1196. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Partridge, L.; Longo, V.D. Extending healthy life span from yeast to humans. Science 2010, 328, 321–326. [Google Scholar] [CrossRef]
- Wei, M.; Fabrizio, P.; Hu, J.; Ge, H.; Cheng, C.; Li, L.; Longo, V.D. Life span extension by calorie restriction depends on Rim15 and transcription factors downstream of Ras/PKA, Tor, and Sch9. PLoS Genet. 2008, 4, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Medvedik, O.; Lamming, D.W.; Kim, K.D.; Sinclair, D.A. MSN2 and MSN4 Link Calorie Restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisia. PLoS. Biol. 2007, 5, 2330–2340. [Google Scholar] [CrossRef] [PubMed]
- Jarolim, S.; Millen, J.; Heeren, G.; Laun, P.; Goldfarb, D.; Breitenbach, M. A novel assay for replicative lifespan in Saccharomyces cerevisiae. FEMS Yeast Res. 2004, 5, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Cheng, L.H.; Matsuura, A.; Xiang, L.; Qi, J.H. Gentiopicroside, a secoiridoid glycoside from Gentiana rigescens Franch, extends the lifespan of yeast via Inducing mitophagy and antioxidative stress. Oxidative Med. Cell. Longev. 2020, 2020, 9125752. [Google Scholar] [CrossRef]
- Pan, Y.J.; Liu, Y.N.; Rui, F.; Farooq, U.; Cheng, L.H.; Matsuura, A.; Qi, J.H.; Xiang, L. Ehretiquinone from Onosma bracteatum Wall exhibits antiaging effect on yeasts and mammals through antioxidative stress and autophagy induction. Oxidative Med. Cell. Longev. 2021, 2021, 5469849. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.F.; Kotakeyama, Y.; Li, J.; Pan, Y.J.; Matsuura, A.; Ohya, Y.; Yoshida, M.; Xiang, L.; Qi, J.H. Cucurbitacin B exerts antiaging effects in yeast by regulating autophagy and oxidative stress. Oxidative Med. Cell. Longev. 2019, 2019, 4517091. [Google Scholar] [CrossRef]
- Farooq, U.; Pan, Y.J.; Lin, Y.F.; Wang, Y.; Osada, H.; Xiang, L.; Qi, J.H. Structure characterization and action mechanism of an antiaging new compound from Gastrodia elata Blume. Oxidative Med. Cell. Longev. 2019, 2019, 5459862. [Google Scholar] [CrossRef] [PubMed]
- Disasa, D.; Cheng, L.H.; Manzoor, M.; Liu, Q.; Wang, Y.; Xiang, L.; Qi, J.H. Amarogentin from Gentiana rigescens Franch exhibits antiaging and neuroprotective effects through antioxidative stress. Oxidative Med. Cell. Longev. 2020, 2020, 3184019. [Google Scholar] [CrossRef]
- Xu, M.; Wang, D.; Zhang, Y.; Yang, C. Dammarane triterpenoids from the Roots of Gentiana rigescens. J. Nat. Prod. 2007, 70, 880–883. [Google Scholar] [CrossRef]
- Gao, L.J.; Li, J.Y.; Qi, J.H. Gentisides A and B, two new neuritogenic compounds from the traditional Chinese medicine Gentiana rigescens Franch. Bioorg. Med. Chem. 2010, 18, 2131–2134. [Google Scholar] [CrossRef]
- Zhou, L.B.; Lu, Z.H.; Li, L.; Chen, L.; Qi, J.H.; Chen, L. Pro-neurogenesis and anti-dementia properties of tetradecyl 2,3-dihydroxybenzoate through TrkA receptor-mediated signalling pathways. Int. J. Neuropsychopharmacol. 2014, 17, 1847–1861. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gao, L.J.; Sun, K.Y.; Xiao, D.; Li, W.Y.; Xiang, L.; Qi, J.H. Benzoate fraction from Gentiana rigescens Franch alleviates scopolamine-induced impaired memory in mice model in vivo. J. Ethnopharmacol. 2016, 193, 107–116. [Google Scholar] [CrossRef]
- Huang, Y.; Lu, H.; Yu, X. Dammarane-Type Triterpenoids from Gentianellaazurea. J. Nat. Prod. 2014, 77, 1201–1209. [Google Scholar] [CrossRef]
- Kaeberlein, M.; Burtner, C.R.; Kennedy, B.K. Recent Developments in Yeast Aging. PLoS Genet. 2007, 3, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Matsui, A.; Kamada, Y.; Matsuura, A. The role of autophagy in genome stability through suppression of abnormal mitosis under starvation. PLoS Genet. 2013, 9, e1003245. [Google Scholar] [CrossRef] [PubMed]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alexandria J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef]
- Wang, X.K.; Michaelis, E.K. Selective neuronal vulnerability to oxidative stress in the brain. Front. Aging Neurosci. 2010, 2, 12. [Google Scholar] [CrossRef] [PubMed]
- Mates, J.M.; Perez-Gomez, C.; De Castro, I.N. Antioxidant enzymes and human diseases. Clin. Biochem. 1999, 32, 595–603. [Google Scholar] [CrossRef]
- Oroojan, A.A.; Chenani, N.; An’aam, M. Antioxidant effects of eugenol on oxidative stress induced by hydrogen peroxide in islets of langerhans isolated from male mouse. Int. J. Hepatol. 2021, 2020, 5890378. [Google Scholar] [CrossRef]
- Sharma, D.; Trivedi, S.S.; Bhattacharjee, J. Oxidative stress and eNOS (Glu298Asp) gene polymorphism in preeclampsia in Indian population. Mol. Cell. Biochem. 2011, 353, 189–193. [Google Scholar] [CrossRef]
- Longo, V.D.; Gralla, E.B.; Valentine, J.S. Superoxide dismutase activity Is essential for stationary phase survival in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 12275–12280. [Google Scholar] [CrossRef] [PubMed]
- Bandara, P.D.S.; Flattery-O’Brien, J.A.; Grant, C.M.; Dawes, I.W. Involvement of the Saccharomyces cerevisiae UTH1 gene in the oxidative-stress response. Curr. Genet. 1998, 34, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2022, 10, 457–468. [Google Scholar]
- Wullschleger, S.; Loewith, R.; Hal, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef]
- Wang, X.; Proud, C.G. Nutrient control of TORC1, a cell-cycle regulator. Trends Cell Biol. 2009, 19, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Sheen, J. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 2014, 164, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Takahara, T.; Maeda, T. Evolutionrily conserved regulation of TOR signalling. J. Biochem. 2013, 154, 1–10. [Google Scholar] [CrossRef]
- Hu, G.; McQuiston, T.; Bernard, A.; Park, Y.-D.; Qiu, J.; Vural, A.; Zhang, N.N.; Waterman, S.R.; Blewett, N.H.; Myers, T.G.; et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat. Cell Biol. 2015, 17, 930–942. [Google Scholar] [CrossRef]
- Hu, G.; McQuiston, T.; Bernard, A.; Park, Y.-D.; Qiu, J.; Vural, A.; Zhang, N.N.; Waterman, S.R.; Blewett, N.H.; Myers, T.G.; et al. TOR-dependent post-transcriptional regulation of autophagy. Autophagy 2015, 11, 2390–2392. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiang, L.; Disasa, D.; Liu, Y.; Fujii, R.; Yang, M.; Wu, E.; Matsuura, A.; Qi, J. Gentirigeoside B from Gentiana rigescens Franch Prolongs Yeast Lifespan via Inhibition of TORC1/Sch9/Rim15/Msn Signaling Pathway and Modification of Oxidative Stress and Autophagy. Antioxidants 2022, 11, 2373. https://doi.org/10.3390/antiox11122373
Xiang L, Disasa D, Liu Y, Fujii R, Yang M, Wu E, Matsuura A, Qi J. Gentirigeoside B from Gentiana rigescens Franch Prolongs Yeast Lifespan via Inhibition of TORC1/Sch9/Rim15/Msn Signaling Pathway and Modification of Oxidative Stress and Autophagy. Antioxidants. 2022; 11(12):2373. https://doi.org/10.3390/antiox11122373
Chicago/Turabian StyleXiang, Lan, Dejene Disasa, Yanan Liu, Rui Fujii, Mengya Yang, Enchan Wu, Akira Matsuura, and Jianhua Qi. 2022. "Gentirigeoside B from Gentiana rigescens Franch Prolongs Yeast Lifespan via Inhibition of TORC1/Sch9/Rim15/Msn Signaling Pathway and Modification of Oxidative Stress and Autophagy" Antioxidants 11, no. 12: 2373. https://doi.org/10.3390/antiox11122373
APA StyleXiang, L., Disasa, D., Liu, Y., Fujii, R., Yang, M., Wu, E., Matsuura, A., & Qi, J. (2022). Gentirigeoside B from Gentiana rigescens Franch Prolongs Yeast Lifespan via Inhibition of TORC1/Sch9/Rim15/Msn Signaling Pathway and Modification of Oxidative Stress and Autophagy. Antioxidants, 11(12), 2373. https://doi.org/10.3390/antiox11122373






