Hydrogen Peroxide Promotes the Production of Radiation-Derived EVs Containing Mitochondrial Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
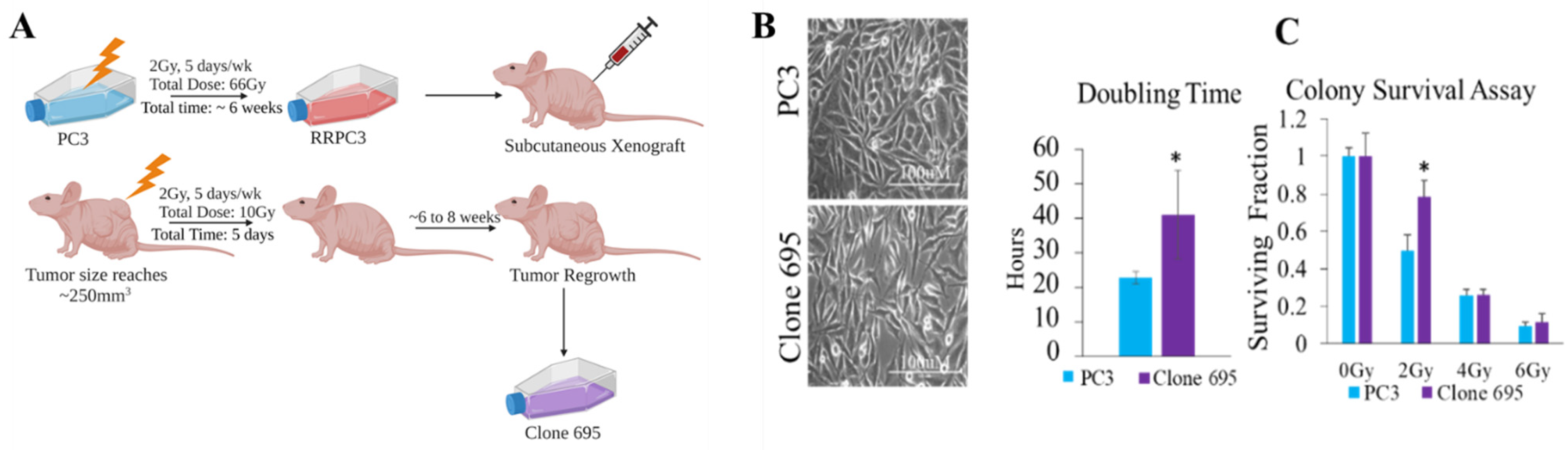
2.2. Radioresistant Prostate Cancer Cell (Clone 695) Development
2.3. Cell Growth and Doubling Time Point
2.4. Colony Survival Assay
2.5. EV Isolation
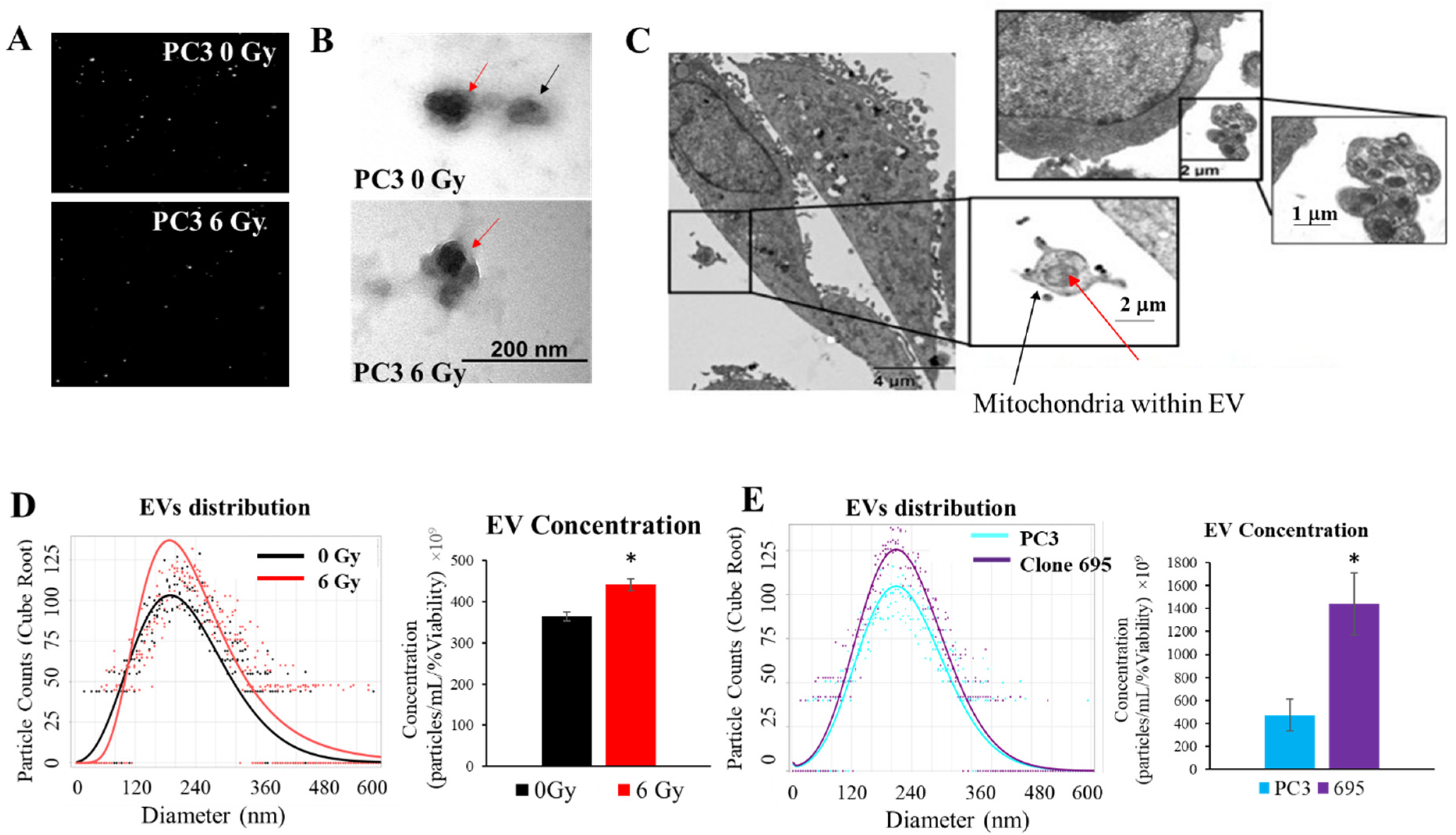
2.6. EV Measurements
2.7. Protein Expression by Capillary Based-Automated Western BLOT JESS
2.8. Metabolic Parameters
2.9. ROS Measurements
2.10. H2O2 Treatment
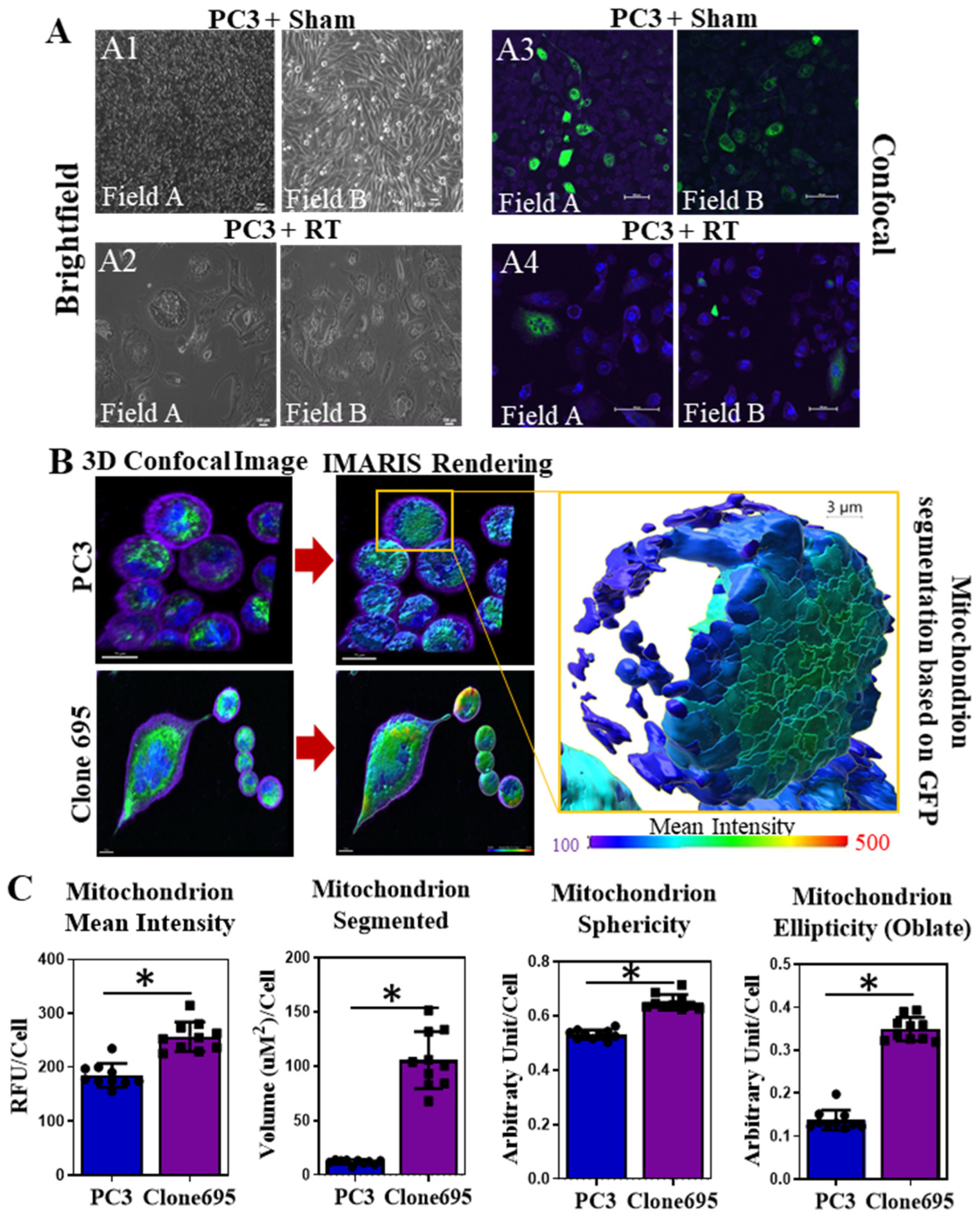
2.11. Mitochondrial Mass Measurements and IMARIS Software
2.12. Electron Microscopy
- EV imaging
- 2.
- Cell Imaging
2.13. Extracellular Vesicle Uptake
2.14. Mitochondrial Treatment
2.15. Statistical Analysis
3. Results
3.1. Radioresistant PCa Cell Line Clone 695 Shows a Significant Difference in Morphology, Growth Rate, and Resistance to Radiation When Compared to Parental PC3 Cells
3.2. Radioresistant PCa Cell Line Clone 695 Shows an Increase in Mitochondrial Mass in Individual Cells
3.3. Radioresistant PCa Cell Line Clone 695 Shows an Increase in H2O2 Production
3.4. Elevation of Mitochondrial Respiratory Activity Correlates with Reprogramming of Mitochondrial Homeostasis in Radioresistant PCa Cell Line Clone 695
3.5. Extracellular Vesicles Are Released upon Radiation Treatment
3.6. Extracellular Vesicles Released during Radiation Treatment Carry Mitochondria as Cargo
3.7. Upon Radiation Treatment, PC3 Cell Line Derived Vesicles Contained H2O2-Responsive Proteins and Mitochondrial Proteins
3.8. Radiation Induces Mitochondrial H2O2 Production in PC3 Cells
3.9. H2O2 Mediates Extracellular Vesicle Release and Impairing Mitochondrial Function
3.10. Uptake of External Mitochondria Correlates with an Increase Mitochondrial Mass and Cancer Survival Post Radiation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Xin, L. Cells of origin for cancer: An updated view from prostate cancer. Oncogene 2013, 32, 3655–3663. [Google Scholar] [CrossRef] [Green Version]
- Society, A.C. Cancer Facts & Figures 2022; American Cancer Society: Atlanta, GA, USA, 2022. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Prostate Cancer Treatment (PDQ(R)). Patient Version. In PDQ Cancer Information Summaries; Prostate Cancer Treatment (PDQ(R)): Bethesda, MD, USA, 2002. [Google Scholar]
- Holley, A.K.; Miao, L.; St Clair, D.K.; St Clair, W.H. Redox-modulated phenomena and radiation therapy: The central role of superoxide dismutases. Antioxid. Redox Signal. 2014, 20, 1567–1589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baskar, R.; Dai, J.; Wenlong, N.; Yeo, R.; Yeoh, K.W. Biological response of cancer cells to radiation treatment. Front. Mol. Biosci. 2014, 1, 24. [Google Scholar] [CrossRef] [Green Version]
- Kim, W.; Lee, S.; Seo, D.; Kim, D.; Kim, K.; Kim, E.; Kang, J.; Seong, K.M.; Youn, H.; Youn, B. Cellular Stress Responses in Radiotherapy. Cells 2019, 8, 1105. [Google Scholar] [CrossRef] [Green Version]
- Chaiswing, L.; Weiss, H.L.; Jayswal, R.D.; Clair, D.K.S.; Kyprianou, N. Profiles of Radioresistance Mechanisms in Prostate Cancer. Crit. Rev. Oncog. 2018, 23, 39–67. [Google Scholar] [CrossRef]
- Kulkarni, R.; Marples, B.; Balasubramaniam, M.; Thomas, R.A.; Tucker, J.D. Mitochondrial gene expression changes in normal and mitochondrial mutant cells after exposure to ionizing radiation. Radiat. Res. 2010, 173, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Cloos, C.R.; Daniels, D.H.; Kalen, A.; Matthews, K.; Du, J.; Goswami, P.C.; Cullen, J.J. Mitochondrial DNA depletion induces radioresistance by suppressing G2 checkpoint activation in human pancreatic cancer cells. Radiat. Res. 2009, 171, 581–587. [Google Scholar] [CrossRef]
- Gough, D.R.; Cotter, T.G. Hydrogen peroxide: A Jekyll and Hyde signalling molecule. Cell Death Dis. 2011, 2, e213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bienert, G.P.; Schjoerring, J.K.; Jahn, T.P. Membrane transport of hydrogen peroxide. Biochim. Biophys. Acta (BBA)-Biomembr. 2006, 1758, 994–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laporte, A.; Lortz, S.; Schaal, C.; Lenzen, S.; Elsner, M. Hydrogen peroxide permeability of cellular membranes in insulin-producing cells. Biochim. Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183096. [Google Scholar] [CrossRef] [PubMed]
- Rubio, C.; Cerón, J. Spectrophotometric assays for evaluation of Reactive Oxygen Species (ROS) in serum: General concepts and applications in dogs and humans. BMC Vet. Res. 2021, 17, 226. [Google Scholar] [CrossRef] [PubMed]
- Ledo, A.; Fernandes, E.; Salvador, A.; Laranjinha, J.; Barbosa, R.M. In vivo hydrogen peroxide diffusivity in brain tissue supports volume signaling activity. Redox Biol. 2022, 50, 102250. [Google Scholar] [CrossRef]
- Chaiswing, L.; St Clair, W.H.; St Clair, D.K. Redox Paradox: A Novel Approach to Therapeutics-Resistant Cancer. Antioxid. Redox Signal. 2018, 29, 1237–1272. [Google Scholar] [CrossRef]
- Sies, H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: Oxidative eustress. Redox Biol. 2017, 11, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.P.; Sies, H. The Redox Code. Antioxid. Redox Signal. 2015, 23, 734–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lisanti, M.P.; Martinez-Outschoorn, U.E.; Lin, Z.; Pavlides, S.; Whitaker-Menezes, D.; Pestell, R.G.; Howell, A.; Sotgia, F. Hydrogen peroxide fuels aging, inflammation, cancer metabolism and metastasis: The seed and soil also needs “fertilizer”. Cell Cycle 2011, 10, 2440–2449. [Google Scholar] [CrossRef] [Green Version]
- Averbeck, D.; Rodriguez-Lafrasse, C. Role of Mitochondria in Radiation Responses: Epigenetic, Metabolic, and Signaling Impacts. Int. J. Mol. Sci. 2021, 22, 11047. [Google Scholar] [CrossRef]
- Rossmann, M.P.; Dubois, S.M.; Agarwal, S.; Zon, L.I. Mitochondrial function in development and disease. Dis. Model. Mech. 2021, 14, dmm048912. [Google Scholar] [CrossRef]
- Brand, M.D. Mitochondrial generation of superoxide and hydrogen peroxide as the source of mitochondrial redox signaling. Free Radic. Biol. Med. 2016, 100, 14–31. [Google Scholar] [CrossRef]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.; Sun, L.; Chen, X.; Zhang, D. Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regen. Res. 2013, 8, 2003–2014. [Google Scholar] [CrossRef]
- Jornayvaz, F.R.; Shulman, G.I. Regulation of mitochondrial biogenesis. Essays Biochem. 2010, 47, 69–84. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Gao, Y.; Liu, J.; Huang, Y.; Yin, J.; Feng, Y.; Shi, L.; Meloni, B.P.; Zhang, C.; Zheng, M.; et al. Intercellular mitochondrial transfer as a means of tissue revitalization. Signal Transduct. Target. Ther. 2021, 6, 65. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, Y.; Qi, Z.; Cao, L.; Ding, S. Mitochondrial transfer/transplantation: An emerging therapeutic approach for multiple diseases. Cell Biosci. 2022, 12, 66. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Borges, F.T.; Reis, L.A.; Schor, N. Extracellular vesicles: Structure, function, and potential clinical uses in renal diseases. Braz. J. Med. Biol. Res. 2013, 46, 824–830. [Google Scholar] [CrossRef] [Green Version]
- Bapst, A.M.; Knöpfel, T.; Nolan, K.A.; Imeri, F.; Schuh, C.D.; Hall, A.M.; Guo, J.; Katschinski, D.M.; Wenger, R.H. Neurogenic and pericytic plasticity of conditionally immortalized cells derived from renal erythropoietin-producing cells. J. Cell. Physiol. 2022, 237, 2420–2433. [Google Scholar] [CrossRef]
- Amari, L.; Germain, M. Mitochondrial Extracellular Vesicles–Origins and Roles. Front. Mol. Neurosci. 2021, 14, 767219. [Google Scholar] [CrossRef]
- O’Brien, K.; Ughetto, S.; Mahjoum, S.; Nair, A.V.; Breakefield, X.O. Uptake, functionality, and re-release of extracellular vesicle-encapsulated cargo. Cell Rep. 2022, 39, 110651. [Google Scholar] [CrossRef]
- Ramirez-Garrastacho, M.; Bajo-Santos, C.; Line, A.; Martens-Uzunova, E.S.; de la Fuente, J.M.; Moros, M.; Soekmadji, C.; Tasken, K.A.; Llorente, A. Extracellular vesicles as a source of prostate cancer biomarkers in liquid biopsies: A decade of research. Br. J. Cancer 2022, 126, 331–350. [Google Scholar] [CrossRef]
- Gaglani, S.; Gonzalez-Kozlova, E.; Lundon, D.J.; Tewari, A.K.; Dogra, N.; Kyprianou, N. Exosomes as A Next-Generation Diagnostic and Therapeutic Tool in Prostate Cancer. Int. J. Mol. Sci. 2021, 22, 10131. [Google Scholar] [CrossRef]
- Chen, T.Y.; Gonzalez-Kozlova, E.; Soleymani, T.; La Salvia, S.; Kyprianou, N.; Sahoo, S.; Tewari, A.K.; Cordon-Cardo, C.; Stolovitzky, G.; Dogra, N. Extracellular vesicles carry distinct proteo-transcriptomic signatures that are different from their cancer cell of origin. iScience 2022, 25, 104414. [Google Scholar] [CrossRef]
- Chaiswing, L.; Yarana, C.; St. Clair, W.; Tovmasyan, A.; Batinic-Haberle, I.; Spasojevic, I.; St. Clair, D. A Redoxable Mn Porphyrin, MnTnBuOE-2-PyP5+, Synergizes with Carboplatin in Treatment of Chemoresistant Ovarian Cell Line. Oxidative Med. Cell. Longev. 2022, 2022, 9664636. [Google Scholar] [CrossRef]
- Chaiswing, L.; Zhong, W.; Oberley, T.D. Increasing discordant antioxidant protein levels and enzymatic activities contribute to increasing redox imbalance observed during human prostate cancer progression. Free Radic. Biol. Med. 2014, 67, 342–352. [Google Scholar] [CrossRef] [Green Version]
- Indo, H.P.; Ito, H.; Nakagawa, K.; Chaiswing, L.; Majima, H.J. Translocation of HSP47 and generation of mitochondrial reactive oxygen species in human neuroblastoma SK-N-SH cells following electron and X-ray irradiation. Arch. Biochem. Biophys. 2021, 703, 108853. [Google Scholar] [CrossRef]
- Wei, X.; Xu, Y.; Xu, F.F.; Chaiswing, L.; Schnell, D.; Noel, T.; Wang, C.; Chen, J.; St Clair, D.K.; St Clair, W.H. RelB Expression Determines the Differential Effects of Ascorbic Acid in Normal and Cancer Cells. Cancer Res. 2017, 77, 1345–1356. [Google Scholar] [CrossRef] [Green Version]
- Franken, N.A.P.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Tian, Y.; Gong, M.; Hu, Y.; Liu, H.; Zhang, W.; Zhang, M.; Hu, X.; Aubert, D.; Zhu, S.; Wu, L.; et al. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles 2020, 9, 1697028. [Google Scholar] [CrossRef]
- Bourdeau-Heller, J.; Oberley, T.D. Prostate carcinoma cells selected by long-term exposure to reduced oxygen tension show remarkable biochemical plasticity via modulation of superoxide, HIF-1alpha levels, and energy metabolism. J. Cell. Physiol. 2007, 212, 744–752. [Google Scholar] [CrossRef]
- Caicedo, A.; Fritz, V.; Brondello, J.-M.; Ayala, M.; Dennemont, I.; Abdellaoui, N.; de Fraipont, F.; Moisan, A.; Prouteau, C.A.; Boukhaddaoui, H.; et al. MitoCeption as a new tool to assess the effects of mesenchymal stem/stromal cell mitochondria on cancer cell metabolism and function. Sci. Rep. 2015, 5, 9073. [Google Scholar] [CrossRef] [Green Version]
- Farnie, G.; Sotgia, F.; Lisanti, M.P. High mitochondrial mass identifies a sub-population of stem-like cancer cells that are chemo-resistant. Oncotarget 2015, 6, 30472–30486. [Google Scholar] [CrossRef] [Green Version]
- McCann, E.; O’Sullivan, J.; Marcone, S. Targeting cancer-cell mitochondria and metabolism to improve radiotherapy response. Transl. Oncol. 2021, 14, 100905. [Google Scholar] [CrossRef]
- Liu, Y.e.; Shi, Y. Mitochondria as a target in cancer treatment. MedComm 2020, 1, 129–139. [Google Scholar] [CrossRef]
- Kumar, P.R.; Moore, J.A.; Bowles, K.M.; Rushworth, S.A.; Moncrieff, M.D. Mitochondrial oxidative phosphorylation in cutaneous melanoma. Br. J. Cancer 2021, 124, 115–123. [Google Scholar] [CrossRef]
- Holl, M.; Koziel, R.; Schafer, G.; Pircher, H.; Pauck, A.; Hermann, M.; Klocker, H.; Jansen-Durr, P.; Sampson, N. ROS signaling by NADPH oxidase 5 modulates the proliferation and survival of prostate carcinoma cells. Mol. Carcinog. 2016, 55, 27–39. [Google Scholar] [CrossRef]
- Vayalil, P.K. Mitochondrial oncobioenergetics of prostate tumorigenesis (Review). Oncol. Lett. 2019, 18, 4367–4376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petros, J.A.; Baumann, A.K.; Ruiz-Pesini, E.; Amin, M.B.; Sun, C.Q.; Hall, J.; Lim, S.; Issa, M.M.; Flanders, W.D.; Hosseini, S.H.; et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 719–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; St Clair, D.K.; Xu, Y.; Crooks, P.A.; St Clair, W.H. A NADPH oxidase-dependent redox signaling pathway mediates the selective radiosensitization effect of parthenolide in prostate cancer cells. Cancer Res. 2010, 70, 2880–2890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eidelman, E.; Twum-Ampofo, J.; Ansari, J.; Siddiqui, M.M. The Metabolic Phenotype of Prostate Cancer. Front. Oncol. 2017, 7, 131. [Google Scholar] [CrossRef] [Green Version]
- Lennicke, C.; Rahn, J.; Lichtenfels, R.; Wessjohann, L.A.; Seliger, B. Hydrogen peroxide–production, fate and role in redox signaling of tumor cells. Cell Commun. Signal. 2015, 13, 39. [Google Scholar] [CrossRef] [Green Version]
- Candas, D.; Li, J.J. MnSOD in oxidative stress response-potential regulation via mitochondrial protein influx. Antioxid. Redox Signal. 2014, 20, 1599–1617. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, K.I.; Ueno, M.; Nyui, M.; Shoji, Y.; Nakanishi, I. Effects of LET on oxygen-dependent and-independent generation of hydrogen peroxide in water irradiated by carbon-ion beams. Free Radic. Res. 2021, 55, 714–719. [Google Scholar] [CrossRef]
- Wagner, B.A.; Evig, C.B.; Reszka, K.J.; Buettner, G.R.; Burns, C.P. Doxorubicin increases intracellular hydrogen peroxide in PC3 prostate cancer cells. Arch. Biochem. Biophys. 2005, 440, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef] [Green Version]
- Guescini, M.; Genedani, S.; Stocchi, V.; Agnati, L.F. Astrocytes and Glioblastoma cells release exosomes carrying mtDNA. J. Neural Transm. 2010, 117, 1–4. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, J.; Miao, Y.; Zhang, Q. The effect of extracellular vesicles on the regulation of mitochondria under hypoxia. Cell Death Dis. 2021, 12, 358. [Google Scholar] [CrossRef] [PubMed]
- Stewart, L.M.; Gerner, L.; Rettel, M.; Stein, F.; Burrows, J.F.; Mills, I.G.; Evergren, E. CaMKK2 facilitates Golgi-associated vesicle trafficking to sustain cancer cell proliferation. Cell Death Dis. 2021, 12, 1040. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.C.; Crescitelli, R.; Cvjetkovic, A.; Belgrano, V.; Olofsson Bagge, R.; Sundfeldt, K.; Ochiya, T.; Kalluri, R.; Lotvall, J. Mitochondrial protein enriched extracellular vesicles discovered in human melanoma tissues can be detected in patient plasma. J. Extracell. Vesicles 2019, 8, 1635420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorrentino, I.; Galli, M.; Medrano-Fernandez, I.; Sitia, R. Transfer of H2O2 from Mitochondria to the endoplasmic reticulum via Aquaporin-11. Redox Biol. 2022, 55, 102410. [Google Scholar] [CrossRef] [PubMed]
- Takata, T.; Kimura, J.; Ihara, H.; Hatano, N.; Tsuchiya, Y.; Watanabe, Y. Redox regulation of Ca(2+)/calmodulin-dependent protein kinase IV via oxidation of its active-site cysteine residue. Free Radic. Biol. Med. 2019, 130, 99–106. [Google Scholar] [CrossRef]
- Ariotti, N.; Wu, Y.; Okano, S.; Gambin, Y.; Follett, J.; Rae, J.; Ferguson, C.; Teasdale, R.D.; Alexandrov, K.; Meunier, F.A.; et al. An inverted CAV1 (caveolin 1) topology defines novel autophagy-dependent exosome secretion from prostate cancer cells. Autophagy 2021, 17, 2200–2216. [Google Scholar] [CrossRef]
- van der Pol, E.; Coumans, F.A.W.; Grootemaat, A.E.; Gardiner, C.; Sargent, I.L.; Harrison, P.; Sturk, A.; van Leeuwen, T.G.; Nieuwland, R. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J. Thromb. Haemost. 2014, 12, 1182–1192. [Google Scholar] [CrossRef]
- Erdbrügger, U.; Lannigan, J. Analytical challenges of extracellular vesicle detection: A comparison of different techniques. Cytom. Part A 2016, 89, 123–134. [Google Scholar] [CrossRef]
- He, D.; Zhao, Z.; Fu, B.; Li, X.; Zhao, L.; Chen, Y.; Liu, L.; Liu, R.; Li, J. Exosomes Participate in the Radiotherapy Resistance of Cancers. Radiat Res. 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Kahroba, H.; Davatgaran-Taghipour, Y. Exosomal Nrf2: From anti-oxidant and anti-inflammation response to wound healing and tissue regeneration in aged-related diseases. Biochimie 2020, 171–172, 103–109. [Google Scholar] [CrossRef]
- Chowdhury, A.R.; Zielonka, J.; Kalyanaraman, B.; Hartley, R.C.; Murphy, M.P.; Avadhani, N.G. Mitochondria-targeted paraquat and metformin mediate ROS production to induce multiple pathways of retrograde signaling: A dose-dependent phenomenon. Redox Biol. 2020, 36, 101606. [Google Scholar] [CrossRef]
- Corbet, C.; Pinto, A.; Martherus, R.; Santiago de Jesus, J.P.; Polet, F.; Feron, O. Acidosis Drives the Reprogramming of Fatty Acid Metabolism in Cancer Cells through Changes in Mitochondrial and Histone Acetylation. Cell Metab. 2016, 24, 311–323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herzig, S.; Shaw, R.J. AMPK: Guardian of metabolism and mitochondrial homeostasis. Nat. Rev. Mol. Cell Biol. 2018, 19, 121–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Roves, P.M.; Osler, M.E.; Holmstrom, M.H.; Zierath, J.R. Gain-of-function R225Q mutation in AMP-activated protein kinase gamma3 subunit increases mitochondrial biogenesis in glycolytic skeletal muscle. J. Biol. Chem. 2008, 283, 35724–35734. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, C.E.; Xu, F.; Zhao, Y.; Luo, W.; Zhong, W.; Meyer, K.; Jayswal, R.; Weiss, H.L.; St. Clair, W.H.; St. Clair, D.K.; et al. Hydrogen Peroxide Promotes the Production of Radiation-Derived EVs Containing Mitochondrial Proteins. Antioxidants 2022, 11, 2119. https://doi.org/10.3390/antiox11112119
Miller CE, Xu F, Zhao Y, Luo W, Zhong W, Meyer K, Jayswal R, Weiss HL, St. Clair WH, St. Clair DK, et al. Hydrogen Peroxide Promotes the Production of Radiation-Derived EVs Containing Mitochondrial Proteins. Antioxidants. 2022; 11(11):2119. https://doi.org/10.3390/antiox11112119
Chicago/Turabian StyleMiller, Caitlin E., Fangfang Xu, Yanming Zhao, Wei Luo, Weixiong Zhong, Kristy Meyer, Rani Jayswal, Heidi L. Weiss, William H. St. Clair, Daret K. St. Clair, and et al. 2022. "Hydrogen Peroxide Promotes the Production of Radiation-Derived EVs Containing Mitochondrial Proteins" Antioxidants 11, no. 11: 2119. https://doi.org/10.3390/antiox11112119
APA StyleMiller, C. E., Xu, F., Zhao, Y., Luo, W., Zhong, W., Meyer, K., Jayswal, R., Weiss, H. L., St. Clair, W. H., St. Clair, D. K., & Chaiswing, L. (2022). Hydrogen Peroxide Promotes the Production of Radiation-Derived EVs Containing Mitochondrial Proteins. Antioxidants, 11(11), 2119. https://doi.org/10.3390/antiox11112119





