Abstract
The non-enzymatic interaction of polyunsaturated fatty acids with nitric oxide (NO) and derived species results in the formation of nitrated fatty acids (NO2-FAs). These signaling molecules can release NO, reversibly esterify with complex lipids, and modulate protein function through the post-translational modification called nitroalkylation. To date, NO2-FAs act as signaling molecules during plant development in plant systems and are involved in defense responses against abiotic stress conditions. In this work, the previously unknown storage biomolecules of NO2-FAs in Arabidopsis thaliana were identified. In addition, the distribution of NO2-FAs in storage biomolecules during plant development was determined, with phytosterol esters (SE) and TAGs being reservoir biomolecules in seeds, which were replaced by phospholipids and proteins in the vegetative, generative, and senescence stages. The detected esterified NO2-FAs were nitro-linolenic acid (NO2-Ln), nitro-oleic acid (NO2-OA), and nitro-linoleic acid (NO2-LA). The last two were detected for the first time in Arabidopsis. The levels of the three NO2-FAs that were esterified in both lipid and protein storage biomolecules showed a decreasing pattern throughout Arabidopsis development. Esterification of NO2-FAs in phospholipids and proteins highlights their involvement in both biomembrane dynamics and signaling processes, respectively, during Arabidopsis plant development.
1. Introduction
The non-enzymatic interaction of polyunsaturated fatty acids with nitric oxide (NO) and derived species, such as dioxide of nitrogen (NO2) and peroxynitrite (ONOO−), gives rise to the formation of nitrated fatty acids (NO2-FAs), also known as nitrolipids or nitroalkenes [1]. Although the formation process of NO2-FAs in vivo is still unknown, two possible mechanisms have been proposed to explain the nitration of fatty acids. One consists of a direct radical–radical reaction between the ·NO2 radical and an alkyl radical. Nevertheless, this process has no biological relevance [2]. This is not the case of the second mechanism, which involves the formation of carbon-centered radicals by the direct addition of a NO2 radical. This radical can react with another NO2 radical to form an unstable nitro-nitrite or dinitro compound, which will rapidly decompose to release nitrous acid (HNO2) and generate a NO2-FA [1,3].
To date, the presence of nitro-oleic acid (NO2-OA), nitro-linoleic acid (NO2-LA), conjugated nitro-linoleic acid (NO2-cLA), and nitro-arachidonic acid (NO2-AA) has been identified in animal systems [4,5,6]. The presence of NO2-OA has been very recently detected in Saccharomyces cerevisiae [7]. In plant systems, the endogenous presence of NO2-cLA and cysteine-adducted NO2-OA has been identified in both olive fruit and extra virgin olive oil (EVOO) [8]. Additionally, nitro-linolenic acid (NO2-Ln) and NO2-OA have been detected in different plant species, such as Arabidopsis thaliana [9], Pisum sativum, Oryza sativa [10], and Brassica napus [11].
It is important to highlight that NO2-FAs can release NO through two possible mechanisms: the modified Nef reaction [12] and nitroalkene rearrangement [13,14]. However, the exact mechanism by which NO2-FAs release NO is not yet known. Several studies in Arabidopsis thaliana roots and cell cultures have shown that NO2-Ln is capable of releasing NO in vivo [15,16]. It has been recently described how the NO released by NO2-Ln can modulate nitrosogluthation (GSNO) levels in vitro and in vivo in Arabidopsis plants [17].
Lack of electrons in the β-carbon adjacent to the carbon (α-carbon) attached to the nitro group (-NO2) converts NO2-FAs into potent electrophiles. These molecules can establish covalent adducts with glutathione (GSH) and nucleophilic residues, such as cysteine, histidine, and lysine, by generating a post-translational modification (PTM) in proteins. This PTM is known as nitroalkylation, and it modifies protein structure and function [1,2,18,19,20]. Nitroalkylation is a reversible PTM that acts as a selective signaling pathway in stress situations. Under these conditions, increased reactive oxygen species (ROS) and reactive nitrogen species (RNS) levels can lead to the oxidation of the NO2-FA-protein bond (Michael adduct), which results in nitroalquene being released and the protein initial state being recovered [7,18,20].
NO2-FAs are generally important signaling molecules in animal, plant, and yeast systems. In animals, they have shown therapeutic benefits given their potent anti-inflammatory and cytoprotective effects in several experimental models [21,22,23,24,25,26]. In yeast, NO2-OA regulates the antioxidant response under heat stress by the nitroalkylation of peroxyredoxin Tsa1 [7]. In plants, they play a signaling role during plant development and under different abiotic stress conditions because NO2-Ln induces heat-shock transcription factors that regulate the expression of antioxidant systems [9], and NO2-OA triggers ROS production [27].
Moreover, NO2-FAs are considered stable and ubiquitous deposits of NO. This is because NO and its derived species have a very short half-life and perform their function in the vicinity of the place where they are generated. In an aqueous environment, if the concentration of NO2-FAs exceeds the critical micellar concentration (CMC), monomers transform into self-assembled structures, frequently micelles or reversible lipid aggregations, which have lower reactivity and greater stability [28]. Knowledge about the interactions between NO2-FAs and lipid aggregations is generally lacking, specifically for biomembranes. However by means of different computer simulation techniques, it has been shown that NO2-FAs can modify the permeability of cell membranes to form clusters at the membrane–water interface, which can in turn affect the dynamic structure of integral proteins [29].
In plant systems, biomembranes are composed mainly of phospholipids whose amphipathic character allows them to establish lipid bilayers. Phospholipids contain a glycerol molecule esterified with two fatty acids and a phosphate group to which different types of alcoholic groups (choline, ethanolamine, serine, inositol, etc.) can be attached. These lipid molecules are unevenly distributed among different cell membranes, abound more in extraplastidic membranes, and are scarcer in photosynthetic membranes [30,31]. Phosphatidylcholine (PC) and phosphatidylethanolamine (PE) are the most predominant phospholipids and perform a structural function [32,33]. Phosphatidylserine (PS) and phosphatidylinositol (PI) are less abundant but display key functions in signaling, trafficking, cell division, and growth in cell membranes [34,35].
Sphingolipids and phytosterol esters (SE) are also components of biomembranes but are barely present [36]. Sphingolipids are molecules generated by the union of a sphingoid base with a fatty acid [37]. This lipid class contributes to structural membrane integrity [38] and the formation of membrane domains [39,40], and it also participates in different signaling processes [41,42,43]. SE are the result of the esterification of phytosterols with fatty acids. The main phytosterols in Arabidopsis are β-sitosterol, stigmasterol, and campesterol [44]. This lipid class participates in membrane homeostasis maintenance [45].
Other plant lipid classes are triacylglycerides (TAGs), which result from the union of three fatty acids with a glycerol molecule. These biomolecules act mainly as a source of energy in early seed germination stages [46]. Mono- and diacylglycerides (MAGs and DAGs) are the common precursors of TAGs and phospholipids [47].
In short, proteins and complex lipids can be considered NO2-FAs storage biomolecules [19,28]. In animal systems, a set of proteins able to bind NO2-FAs has been reported [22,48,49,50,51,52,53,54,55,56]. In addition, TAGs have been identified as the main NO2-FAs storage biomolecule in these organisms [55]. However in plant systems, the NO2-FAs reservoir biomolecules and their biodistribution during Arabidopsis development are completely unknown and are the subject of this work. To date, only the drop in NO2-OA and NO2-Ln levels during the development of Brassica napus and Arabidopsis thaliana, respectively, has been described [9,11]. Therefore, the presence in Arabidopsis thaliana of not only NO2-Ln but also of NO2-OA and NO2-LA was identified in this paper. The storage biomolecules of each NO2-FA was also characterized, as was the distribution and NO2-FAs levels in storage biomolecules during Arabidopsis thaliana development. Finally, the possible role of NO2-FAs esterified with lipid storage and adducted with protein reservoirs was studied.
2. Materials and Methods
2.1. Plant Material
In this work, the plant species A. thaliana of the Columbia ecotype was used. The vegetal material from the dried seeds of different aged plants was used to characterize the development of this plant.
2.1.1. Development Stages of Arabidopsis thaliana
The Bayer, BASF, Ciba-Geigy, and Hoechst (BBCH) scale, adapted to the Arabidopsis growth phenotypes, was used to select the development stages to be considered [57,58], and the plant vegetative, generative, and senescence stages were hence characterized. Specifically in the vegetative stage, the 5-, 14-, and 24-day-old plants were selected. From a phenotypical point of view, the 5-day-old seedlings showed completely open cotyledons, which represents the initial leaf development stage. In the 14-day-old seedlings, in addition to the two cotyledons, the circular arrangement of four leaves (rosette) was observed. This structure was completely configured in the 24-day-old plants, which presented a mature rosette constituted by a maximum of 13–14 leaves.
The 34- and 36-day-old plants represented the main events that took place in the generative stage. Particularly, the 34-day-old plants showed reproductive floral structures. Seed production was clearly observed inside the siliqua structures in the 36-day-old plants.
Finally, the senescence stage was represented by the 53-day-old plants, which phenotypically presented completely open, mature, yellow-brown siliques capable of releasing seeds.
The plant material used to characterize Arabidopsis development included the 5- and 14-day-old complete seedlings and the leaves from the 24-, 34-, 36-, and 53-day-old plants.
2.1.2. Growing Conditions of Arabidopsis thaliana
A. thaliana seeds were sterilized for 10 min in 70% (v/v) ethanol solution containing 0.1% (w/v) sodium dodecyl sulfate (SDS). Seeds were then immersed for 20 min in sterile water containing 20% bleach (v/v) and 0.1% SDS (w/v). The next several washes were performed with sterile water, followed by a 5-minute incubation in a 1/200 algaecide dilution. Finally, they were washed 4 times with sterile water (5 min. each) and left at 4 °C for 2 days to break dormancy.
The Arabidopsis seedlings from the 5- and 14-day-old plants were grown in Petri plates according to the method described by [59]. In contrast, the 24-, 34-, 36-, and 53-day-old plants were obtained from hydroponic cultures under aeration using a specific growth medium [60]. Plants were grown under conditions of 16 h of light at 22 °C, 8 h of darkness at 18 °C, 60% humidity, and 100 μE m–2 s–1 light intensity.
2.2. Reagents
MS medium for growing Arabidopsis thaliana plants, commercial oleic acid (OA), linoleic acid (LA), linolenic acid (Ln), and carbon 13-labelled OA (13C18-OA) as well as the different solvents herein used were purchased from Sigma-Aldrich (Saint Louis, MO, USA). The strata-NH2 solid-phase extraction columns were supplied by Phenomenex (Torrance, CA, USA).
2.3. Synthesis and Characterization of Standards NO2-OA, NO2-LA, and NO2-Ln and Internal Standard 13C18-NO2-OA by NMR Spectroscopy
The synthesis of standard NO2-OA was carried out by nitroselenation, oxidation, and hydroselenoxide elimination similarly to that previously described by [7]. Briefly, in a tetrahydrofuran–acetonitrile mixture (1:1, v/v, 24 mL), commercial OA (1 g, 3.05 mmol), solid mercury chloride (1.15 g, 4.23 mmol), phenylselenyl bromide (0.93 g, 3.93 mmol), and sodium nitrite (0.49 g, 7.19 mmol) were dissolved. This mixture was incubated in an argon atmosphere for 12 h.
After incubation, the mixture was filtered and evaporated. The residue was dissolved in 12 mL of tetrahydrofuran, and after adding 33% (v/v) hydrogen peroxide (3.63 mL), the mixture was stirred for 1 h in an ice bath. This step was followed by extraction with hexane. The solvent was then washed with saturated sodium chloride solution, and any water was removed with anhydrous magnesium sulfate. Next, the sample was evaporated. NO2-OA purification was performed by flash column chromatography (silica gel 60) with a mixture of hexane/diethyl ether/acetic acid (80:20:0.5, v/v/v) as the mobile phase. The selection of the fractions of interest was carried out on TLC plates (Sigma-Aldrich, Saint Louis, MO, USA) and developed with the same solvent mixture used in the previous chromatography. Finally, the structure of the synthesized compound was analyzed by NMR using a Bruker Avance 400 spectrometer (Billerica, MA, USA) that operated at 400.13 MHz for 1H and 100.61 MHz for 13C.
Carbon 13-labelled NO2-OA (13C18-NO2-OA), which was used as an internal standard in the NO2-FAs quantification protocol, was synthesized as described above.
For the synthesis of standards NO2-LA and NO2-Ln, the above protocol was also followed, and only the incubation period of the nitroselenation process was changed, which was shortened to 4 h in an argon atmosphere instead of 12 h.
2.4. Detection of NO2-FAs from Lipid Storages
2.4.1. Lipid Extraction
The plant samples that came from the different developmental stages were homogenized with liquid nitrogen to achieve greater cell wall and tissue disintegration. The lipid material was then extracted by the Bligh and Dyer technique using a mixture of hexane/isopropanol/1M formic acid (30:20:2, v/v/v) [61] before performing a solid-phase extraction.
2.4.2. Solid-Phase Extraction
The lipid material obtained in the Section 2.4.1. was chromatographically separated following the protocol described by [55] with a few modifications. This protocol allows the individual separation and collection of each of the fractions according to their polarity using solvents of increasing polarity. For this purpose, Strata-NH2 solid-phase extraction (SPE) columns were used, and SE, TAGs, MAGs-DAGs, free fatty acids (FFA), PC, PE, PS, PS, and PI were sequentially eluted with 12 mL of hexane, hexane/chloroform/ethyl acetate (100:5:5, v/v/v), chloroform/isopropanol (2:1, v/v), diethyl ether/2% acetic acid, acetonitrile/1-propanol (2:1, v/v), methanol, isopropanol/methanolic HCl (4:1, v/v), and methanol/methanolic HCl (9:1, v/v), respectively. The chromatographic fractions were then evaporated and later resuspended in 50 µL methanol, and 10 nM 13C18-NO2-OA was added as internal standard.
2.4.3. Identification and Confirmation of Complex Lipid Class
The detection and confirmation of the different complex lipids obtained after the chromatography with Strata-NH2 columns was carried out following the protocol described by Baker et al. [62] by direct infusion MS/MS spectrometry. For this purpose, the all fractions were solubilized in 100 μL of a 300 mM methanol/chloroform/ammonium acetate mixture (665:300:35, v/v/v) with the exception of PS, to which 8 μL of acetic acid was also added. MS/MS analysis was carried out on an Agilent QTOF mass spectrometer sending the samples directly to the ion source (operating in positive mode). All lipids were measured using the following equipment settings: drying gas flow: 4 l/min nitrogen; fragmentation voltage: 100 V; gas temperature: 325 °C; scan rate: 1 spectra/s.
To identify each lipid class, a representative molecular specie of each type of complex lipid, for which we knew the molecular adduct ion [M + H]+ or [M + NH4]+ and the molecular mass corresponding to the adduct ion (m/z), were used as a standard. Fragmentation of TAGs and DAGs was characterized by the neutral loss of the adduct corresponding to the fatty acid + NH3, which could be detected by a neutral loss scan for the indicated mass. Fragmentation of phospholipids, with the exception of PC, was also detected by scanning for the difference of the neutral loss corresponding to the adduct formed by the phospholipid head group. In the case of PC, detection was carried out by scanning the precursor ion corresponding to its head group.
2.4.4. Acid Hydrolysis
In order to proceed to the detection and quantification of NO2-FAs from the lipid storages, the chromatographic fractions obtained following the protocol described in Section 2.4.2, SE, TAG, MAG-DAG, PC, PE, PS, and PI, with the exception of the FFA fraction, were subjected to acid hydrolysis to separate NO2-FAs from their corresponding complex lipid. To limit artificial nitration reactions catalyzed by the subsequent acid hydrolysis process, 250 µL of methanolic sulfanilamide (1 g/10 mL) were added to all fractions and incubated for 20 min. Next, the sample was evaporated and incubated in 2.5 mL of acetonitrile/hydrochloric acid (9:1, v/v) for 1 h at 90 °C. After acid hydrolysis, NO2-FAs were extracted with hexane/H2O (2:1, v/v). Finally, the hexane fraction was isolated, evaporated, and resuspended in methanol to be analyzed by LC-MS/MS.
2.5. Detection of NO2-FAs from Protein Storage
The plant sample was homogenized in liquid nitrogen with a specific buffer at the 1:2 (w/v) ratio (0.1 M Tris-HCl, pH 7.6, 7% sucrose (w/v); 7% PVPP (w/v); 0.05% Triton X-100 (v/v) and 0.1 mM EDTA). The sample was then centrifuged twice at 10,000 xg for 15 min at 4 °C. The obtained supernatant was precipitated with 70% (v/v) cold acetone (−20 °C) and stored overnight at −20 °C. After this time, the sample was recentrifuged at 10,000× g for 15 min at 4 °C.
The protein extracts obtained from acetone precipitation were dissolved in 2 mL of methanol. Next, the oxidation of the NO2-FAs-proteins adducts was carried out following the method described by [20] with some modifications. Firstly, the sample was divided into two equal parts. One part was non-oxidized and used to quantify the free NO2-FAs levels that could result from the acetone precipitation step. The other part of the sample was incubated with 50 mM of H2O2, which were added under shaking the infusion every 20 min for 3 h and 20 min. The treatment with H2O2 oxidizes Michael adducts with the subsequent NO2-FAs release.
A known concentration of the internal standard, 13C18-NO2-OA (10 nM), was added to both the non-oxidized and oxidized samples. Next, the FFA were extracted by adding 7.5 mL of hexane and vigorously shaking for 2 min. To properly separate phases, the mixture was centrifuged at 1000× g for 1 min at 4 °C. The upper apolar phase corresponding to hexane, rich in fatty acids, was collected, evaporated, resuspended in methanol, and analyzed by LC-MS/MS.
2.6. Detection and Identification of NO2-FAs by LC-MS/MS
Characterization and quantification of NO2-FAs obtained from both the FFA fraction and the complex lipid fractions after acid hydrolysis were performed following the protocol described in [7] with some modifications. A Sciex Triple Quadrupole Linear Trap Mass Spectrometer (QTRAP 6500+), coupled to a Sciex ExionLC AD ultrahigh-performance liquid chromatograph (UHPLC), was used. Lipid extracts were separated using a Kinetex 1.7 μm C18 100 Å (150 mm × 2.1 mm) column (Phenomenex, Torrance, CA, USA) in a solvent system consisting of water with 0.1% formic acid (A) and methanol with 0.1% formic acid (B).
The MS/MS analysis was performed in the negative ion mode. The presence of the different NO2-FAs was detected by applying the multiple reaction monitoring (MRM) scanning mode with the specific MRM transitions corresponding to NO2-OA (326/46 and 326/279 m/z), 13C18-NO2-OA (344.1/46 m/z), NO2-LA (324/46 and 324/277 m/z), and NO2-Ln (322/46 and 322/275 m/z) [63,64].
The quantification of the NO2-FAs levels was performed using a calibration curve prepared with the reference standard for each NO2-FA and with the presence of 13C18-NO2-OA as the internal standard. In all these cases, the obtained data were analyzed and processed with the Sciex OS software version 1.7.0.36606 (Ab Sciex, Redwood City, CA, USA).
2.7. Statistical Analysis
Statistical procedures were carried out by the Student’s t-test. Values represent the mean ± SEM of at least three biological replicates with significance when at least p < 0.05.
3. Results
3.1. Identification of the Main Endogenous NO2-FAs in Arabidopsis thaliana
NO2-FAs such as NO2-OA and NO2-LA play an important signaling role in animal systems [48,51,65]. However, in plant systems, only the presence of NO2-OA and NO2-Ln has been identified during the development of Brassica napus [11] and Arabidopsis thaliana [9], respectively. Additionally, the participation of NO2-Ln in the response to different stress situations has been described [9]. Given the relevance of NO2-OA, NO2-LA, and NO2-Ln in both animal and plant systems, this work applied mass spectrometry techniques and detected the endogenous presence of NO2-OA and NO2-LA in 14-day-old Arabidopsis thaliana seedlings for the first time. The existence of NO2-Ln was also corroborated. The 14-day-old seedlings were employed as a representative example of Arabidopsis plants because they are juvenile organisms in which flowering and gametogenesis processes have not yet occurred.
For this purpose, MRM transitions corresponding to NO2-OA (m/z 326/46), NO2-LA (m/z 324/46), and NO2-Ln (m/z 322/46) were monitored. Otherwise, m/z 326, 324, and 322 were the main ion products that corresponded to the complete ion mass spectra (MS) of standards NO2-OA, NO2-LA, and NO2-Ln, respectively, when analyzed in the negative ion mode. In contrast, the MS/MS (MS2) spectra showed a major fragment with m/z 46 corresponding to the NO2 molecule.
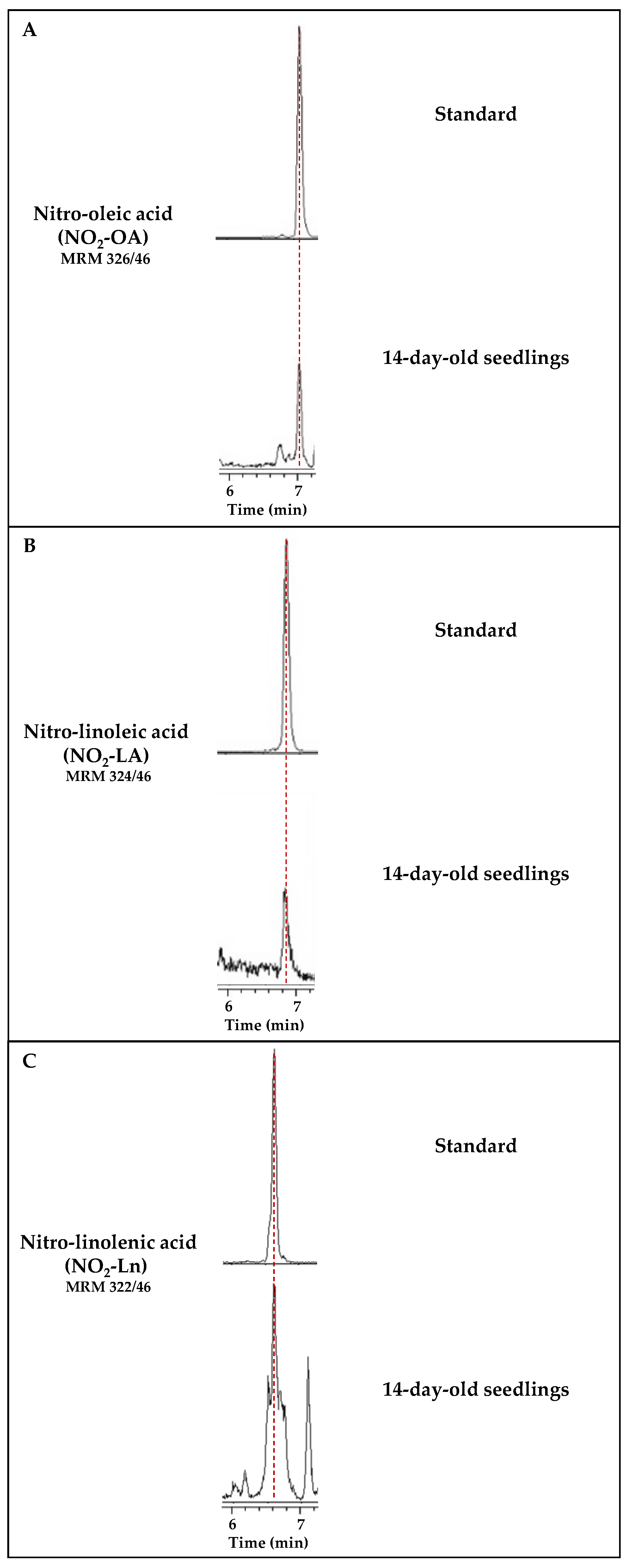
Figure 1 shows the chromatographic peaks for the MRM transitions of standards NO2-FAs and the 14-day-old seedlings. In this case, the sample showed a chromatographic peak coincident with the MRM 326/46 transition, which shared the same retention time as standard NO2-OA (Figure 1A). The same behavior was observed for the MRM 324/46 transition corresponding to NO2-LA (Figure 1B). In this work, the presence of NO2-Ln, already published in Arabidopsis [9], was also corroborated (Figure 1C).
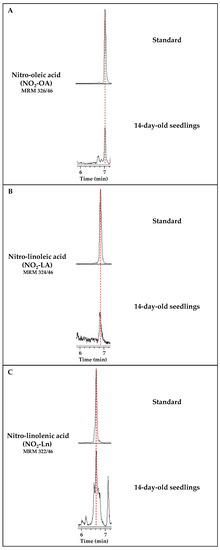
Figure 1.
Detection of the main NO2-FAs present in Arabidopsis thaliana. Each panel shows the chromatogram corresponding to the standard of each nitrated fatty acid, NO2-OA (A), NO2-LA (B), and NO2-Ln (C). Their retention times coincide with the chromatographic peaks detected in the Arabidopsis lipid extracts from the 14-day-old seedlings. Peaks refer to a total ion intensity of 1.2 × 104 for the m/z transition of NO2-OA (326/46), 4 × 103 for NO2-LA (m/z 324/46), and 1.4 × 104 for NO2-Ln (m/z 322/46). The dotted vertical line indicates the peaks with the same retention time.
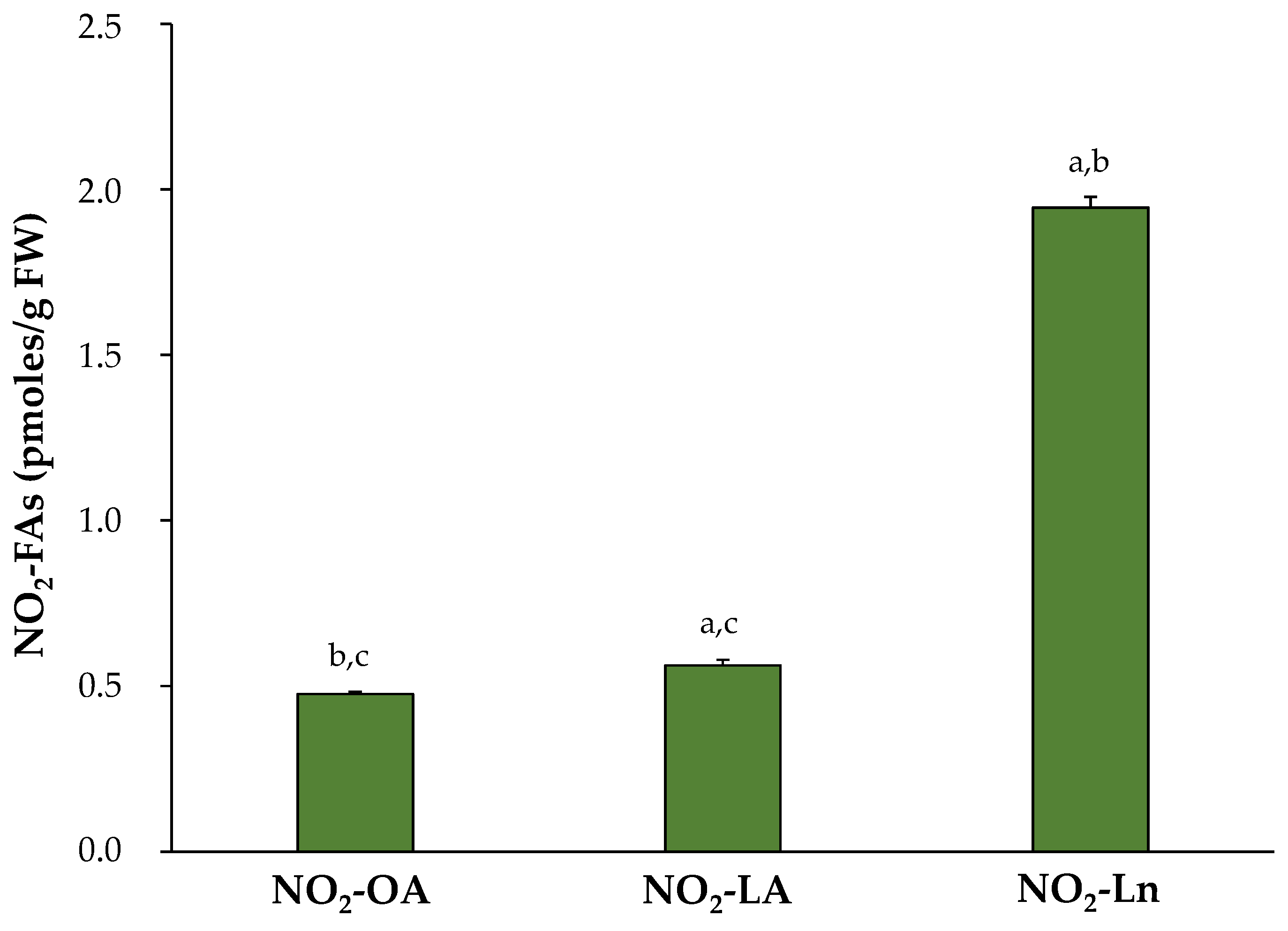
After identifying and confirming the presence of the three NO2-FAs in Arabidopsis, their levels were quantified in the 14-day-old seedlings. The most abundant NO2-FA was NO2-Ln (1.945 ± 0.032 pmol/g FW), followed by NO2-LA (0.563 ± 0.016 pmol/g FW) and NO2-OA (0.476 ± 0.007 pmol/g FW) (Figure 2).
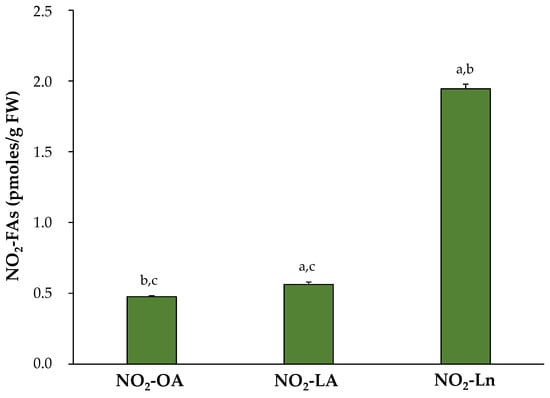
Figure 2.
Endogenous NO2-FAs levels in the 14-day-old Arabidopsis thaliana seedlings. The NO2-FAs values (pmoles/g FW) are those obtained from the mean ± SEM of at least ten independent experiments. The statistical significance between means was analyzed by the Student’s t-test. Letters a, b, and c denote significant differences (p < 0.05) with NO2-OA, NO2-LA, and NO2-Ln, respectively. FW, fresh weight.
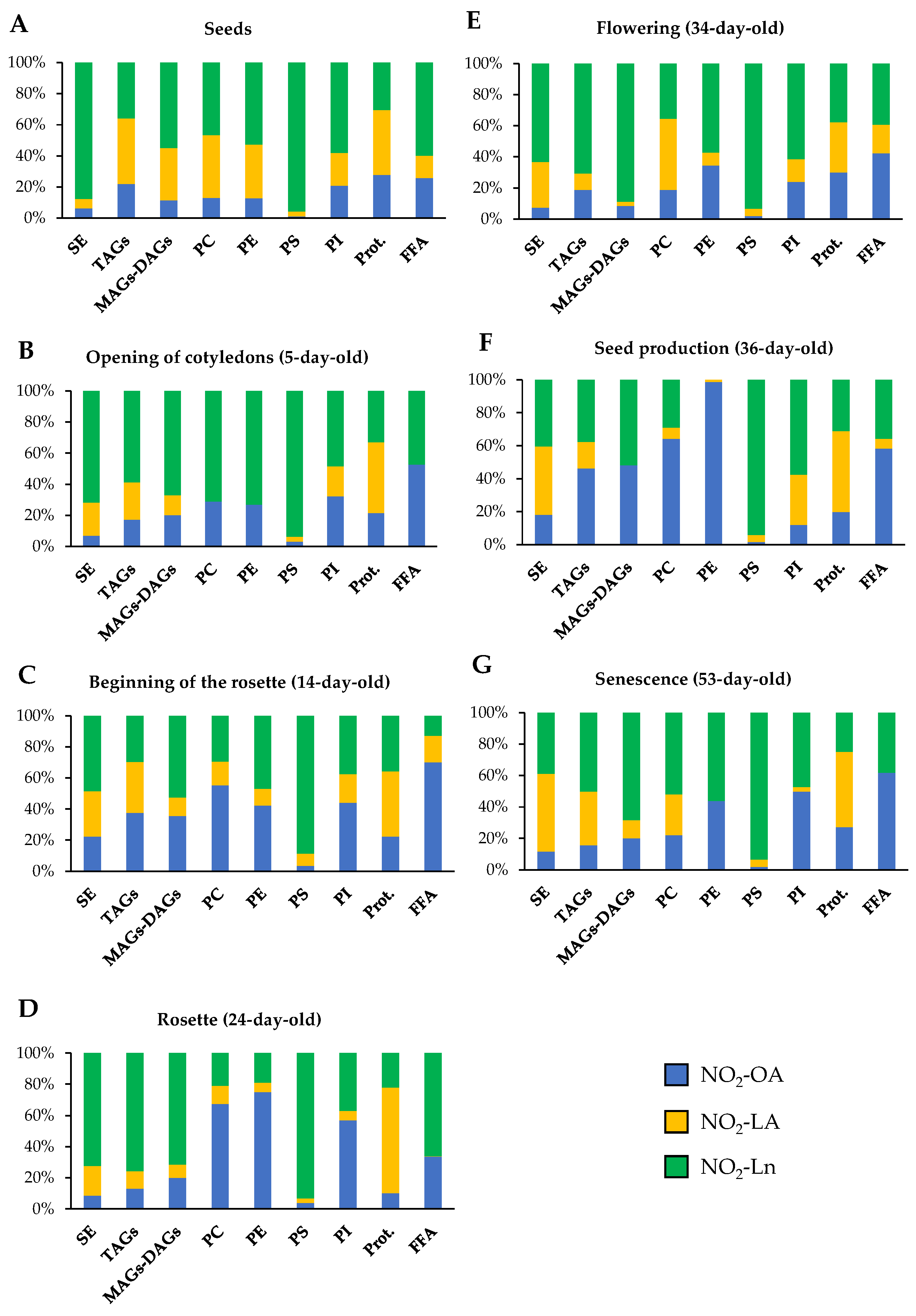
3.2. Characterization of the Main NO2-FAs Storage Biomolecules in Arabidopsis thaliana
The physico-chemical properties of NO2-FAs allow their esterification with proteins and complex lipids, molecules that thus become NO2-FAs storage biomolecules. In animal systems, the esterification of NO2-FAs to complex lipids has been reported, where TAGs are the main reservoir [54,56]. In parallel, different proteins susceptible to bind NO2-FAs have also been characterized in animals [22,48,49,50,51,52,53,54,55,56]. However, in plant systems, all these aspects are completely unknown and constitute the purpose of this work.
The identification of NO2-FAs storage biomolecules was carried out in the 14-day-old Arabidopsis seedlings, which are a representative example of vegetative Arabidopsis growth. The characterization of the storage of NO2-FAs in lipid biomolecules was performed by modifying a chromatographic method previously used in animal systems [54]. This method allows complex lipids to be separated into different fractions according to their polarity, such as SE, TAGs, MAGs-DAGs, PC, PE, PS, and PI. We developed a new methodology for the detection of NO2-FAs from the adduction of proteins to look into the protein reservoir.
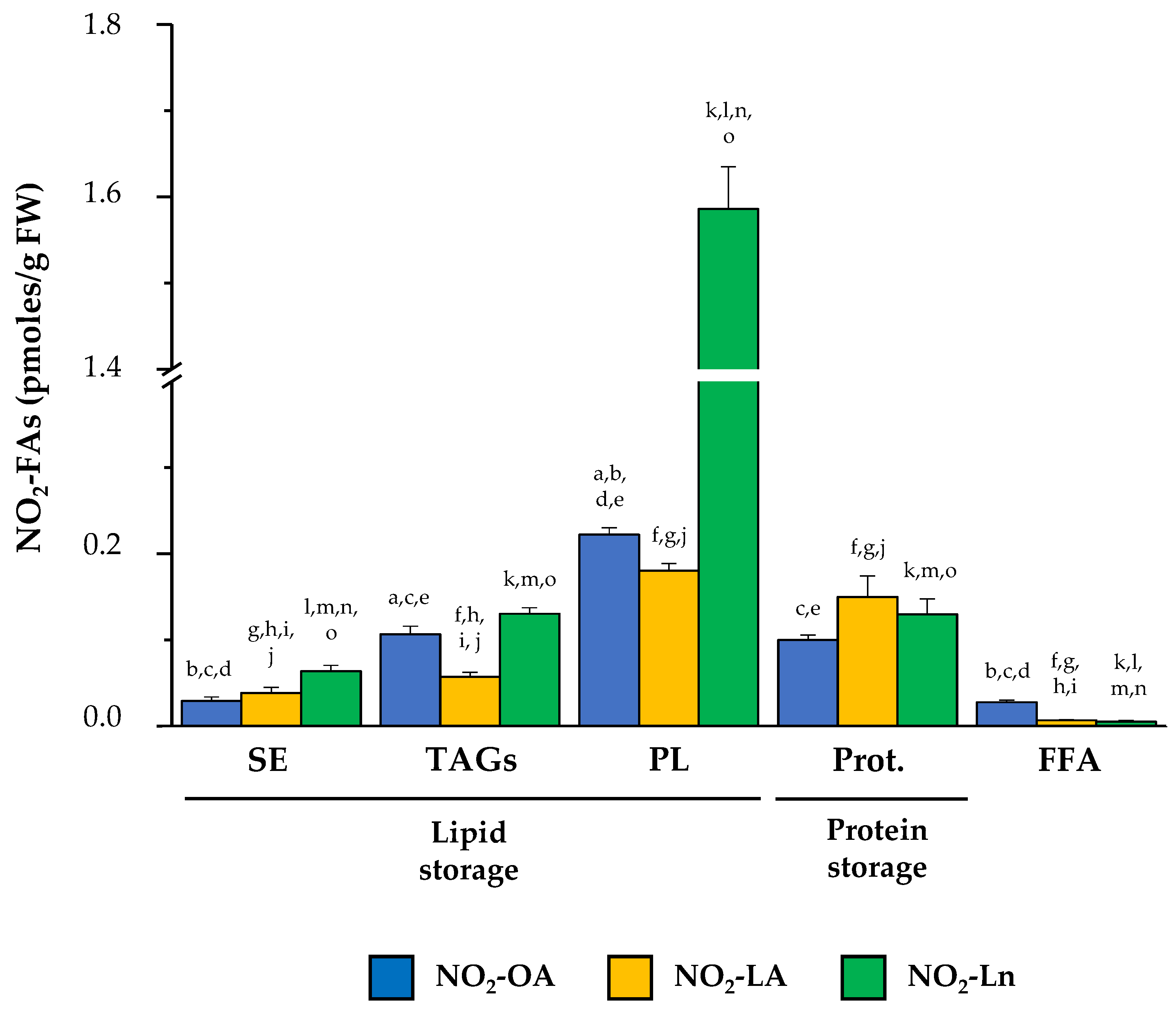
As shown in Supplementary Figure S1, all three NO2-FAs are present in both the protein and lipid storages. A detailed analysis of the NO2-FAs distribution in the storage biomolecules showed that in SE, NO2-Ln was the most abundant (0.064 ± 0.007 pmol/g FW), followed by NO2-LA (0.039 ± 0.007 pmol/g FW) and NO2-OA (0.029 ± 0.005 pmol/g FW) (Figure S1A). In TAGs, the three NO2-FAs were present at similar levels: 0.04 ± 0.007 pmol/g FW for NO2-OA, 0.035 ± 0.008 pmol/g FW for NO2-LA, and 0.032 ± 0.003 pmol/g FW for NO2-Ln (Figure S1B). In MAGs-DAGs storage, the amounts of both NO2-Ln (0.099 ± 0.011 pmol/g FW) and NO2-OA (0.067 ± 0.012 pmol/g FW) were considerable, but NO2-LA was less represented (0.022 ± 0.003 pmol/g FW) (Figure S1C). Abundance patterns differed for phospholipids. In PC, the presence of NO2-OA (0.057 ± 0.009 pmol/g FW) predominated, followed by NO2-Ln (0.03 ± 0.006 pmol/g FW) and NO2-LA (0.016 ± 0.002 pmol/g FW) (Figure S1D). In PE, NO2-OA (0.045 ± 0.007 pmol/g FW) and NO2-Ln (0.05 ± 0.005 pmol/g FW) showed similar quantities, while the participation of NO2-LA (0.011 ± 0.002 pmol/g FW) was lesser (Figure S1E). NO2-Ln (1.453 ± 0.18 pmol/g FW) was located mainly in the PS reservoir biomolecule where the other two NO2-FAs were the minority (NO2-LA: 0.127 ± 0.024 pmol/g FW; NO2-OA: 0.059 ± 0.009 pmoles/g FW) (Figure S1F). Finally, the levels of the three NO2-FAs in PI showed the same abundance pattern as that described for PE; that is, 0.062 ± 0.007 pmoles/g FW for NO2-OA, 0.053 ± 0.007 pmoles/g FW for NO2-Ln, and 0.026 ± 0.006 pmoles/g FW for NO2-LA (Figure S1G).
Proteins are other NO2-FAs storage biomolecules to consider. We developed a new methodology that allowed us to break the nitroalkylation adduct between NO2-FAs and proteins. In the 14-day-old seedlings, we detected similar levels for the three NO2-FAs: 0.1 ± 0.01 pmol/g FW for NO2-OA, 0.15 ± 0.02 pmol/g FW for NO2-LA, and 0.13 ± 0.02 pmol/g FW for NO2-Ln (Figure S1H). Finally, it should be noted that a small amount of each NO2-FA was neither adducted with proteins nor esterified with complex lipids, which constitutes the free nitrated fatty acids fraction (FFA) (Figure S1I).
After combining the information obtained in our experiments about the distribution of NO2-FAs in both biological reservoir types (Figure 3), we conclude that the lipid reservoir, mainly represented by phospholipids, can be considered the main reservoir of NO2-OA and NO2-Ln. With NO2-LA, proteins and phospholipids appeared as the main storage biomolecules because NO2-LA presented similar levels in both reservoirs. The three NO2-FAs showed a different distribution in the storage biomolecules. The NO2-Ln reservoirs that were ordered from greater to lesser abundance were phospholipids, proteins together with TAGs, and finally SE. For NO2-LA, both phospholipids and proteins were preferential reservoirs, followed by TAGs and SE. Finally, the participation of NO2-OA in phospholipids was greater than in TAGs and proteins, while SE appeared as a minority deposit. The NO2-FAs levels in their free form were practically undetectable (Figure 3).
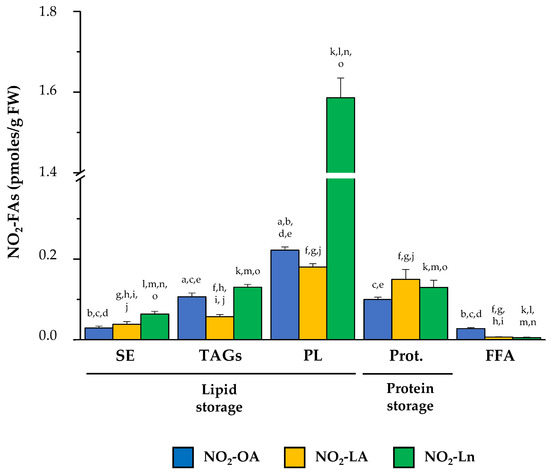
Figure 3.
The main NO2-FAs storage biomolecules in Arabidopsis thaliana. This figure shows the different lipid and protein reservoirs of NO2-FAs in the 14-day-old Arabidopsis seedlings. The lipid deposit consisted of SE, TAGs (including MAGs-DAGs), and the four phospholipid types (PC, PE, PS, and PI). The protein deposit is represented by the NO2-FAs that were experimentally released from adducts with proteins. The FFA levels, which were neither esterified nor adducted, are also shown. The NO2-FAs values are the mean ± SEM of at least ten independent experiments. The statistical significance between means was analyzed by the Student’s t-test. Letters a, b, c, d, and e indicate the significant differences (p < 0.05) between the NO2-OA levels found in SE (a), TAGs (b), PL (c), Prot. (d), and FFA (e). Letters f, g, h, i, and j denote the significant differences (p < 0.05) between the NO2-LA levels found in SE (f), TAGs (g), PL (h), Prot. (i), and FFA (j). Letters k, l, m, n, and o represent the significant differences (p < 0.05) between the NO2-Ln levels found in SE (k), TAGs (l), PL (m), Prot. (n), and FFA (o). PL, phospholipids; Prot., proteins.
3.3. Endogenous Levels of NO2-FAs during Arabidopsis thaliana Development
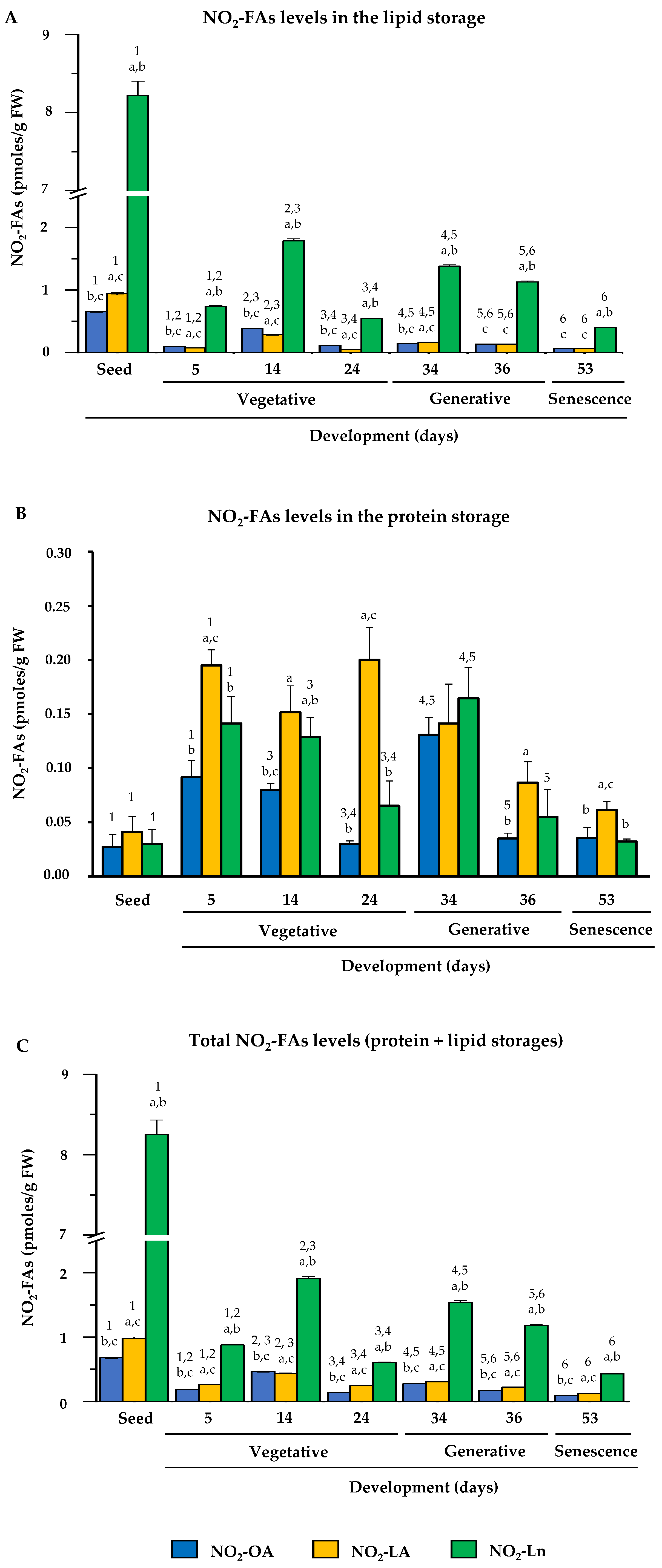
After identifying the endogenous presence of NO2-OA, NO2-LA, and NO2-Ln in Arabidopsis, the NO2-FAs levels were quantified in different plant developmental stages. Figure 4 shows the endogenous levels of the NO2-FAs esterified with complex lipids (panel A), those which came from the adduction with proteins (panel B), and the total levels when considering both protein and lipid storages (panel C) in each plant development stage.
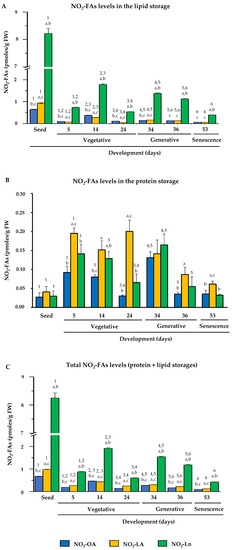
Figure 4.
NO2-FAs endogenous levels during Arabidopsis development. This figure shows the quantification of the NO2-OA, NO2-LA, and NO2-Ln contents in the different plant development stages, which ranged from seed and vegetative stages (seed germination (at 5 days) and initial and complete rosette development (at 14 and 24 days, respectively)), generative stage (flowering (at 34 days), and seed production (at 36 days)) and senescence stage (at 53 days). Panel (A) displays the NO2-FAs endogenous levels esterified with the lipid storage biomolecules. Panel (B) shows the levels of the NO2-FAs from the adduction with the protein reservoir biomolecules. Panel (C) represents the total levels from the lipid and protein storages. The NO2-FAs values are the mean ± SEM of at least ten independent experiments. The statistical significance between means was analyzed by the Student’s t-test. Letter a indicates significant differences (p < 0.05) between the NO2-OA levels to the other two NO2-FAs in each developmental stage. This same statistical comparison was also made for NO2-LA and NO2-Ln. Significant differences (p < 0.05) are denoted by letters b and c, respectively. A comparison of each amount of NO2-FA in the different developmental stages was also made. Numbers 1, 2, 3, 4, 5, and 6 respectively indicate the significant differences (p < 0.05) between: the NO2-FAs levels in the seedlings and 5-day-old plants; the 5- and 14-day-old plants; the 14- and 24-day-old plants; the 24- and 34-day-old plants; the 34- and 36-day-old plants; and the 36- and 53-day-old plants.
In the storage lipids, NO2-Ln was the most abundant, followed by NO2-OA and NO2-LA, both of which had a similar abundance. The NO2-FAs levels generally lowered during development, albeit with some fluctuations in certain developmental stages. Specifically in the stages when plants presented fully open cotyledons (5-day-old seedlings) and a mature rosette (24-day-old plants), the levels of the three NO2-FAs lowered (Figure 4A).
In the protein reservoir, the three NO2-FAs appeared at similar levels in the seed stage. However in the vegetative, generative, and senescence stages, NO2-LA was the major NO2-FA, followed by NO2-Ln and NO2-OA, and the levels of both were generally similar. When analyzing behavior on the whole, the NO2-FAs levels in seeds were a minority, but their content increased for the three NO2-FAs in the seedlings with open cotyledons (5-day-old seedlings). These high levels were similar during vegetative rosette growth (14- and 24-day-old plants) and during flowering (34-day-old plants). However, when plants initiated seed production (36 day-old plants), the NO2-FAs from the protein reservoir lowered, and these levels were similar in the senescent plants (53-day-old plants) (Figure 4B).
The compilation of previous data (Panels 4A and 4B) allowed us to establish that the total levels of the three NO2-FAs from the protein and lipid storages during development displayed similar behavior to that described above for the NO2-FAs esterified with complex lipids (Figure 4C).
3.4. Nitro-Fatty Acids Distribution in Storage Biomolecules during Arabidopsis thaliana Development
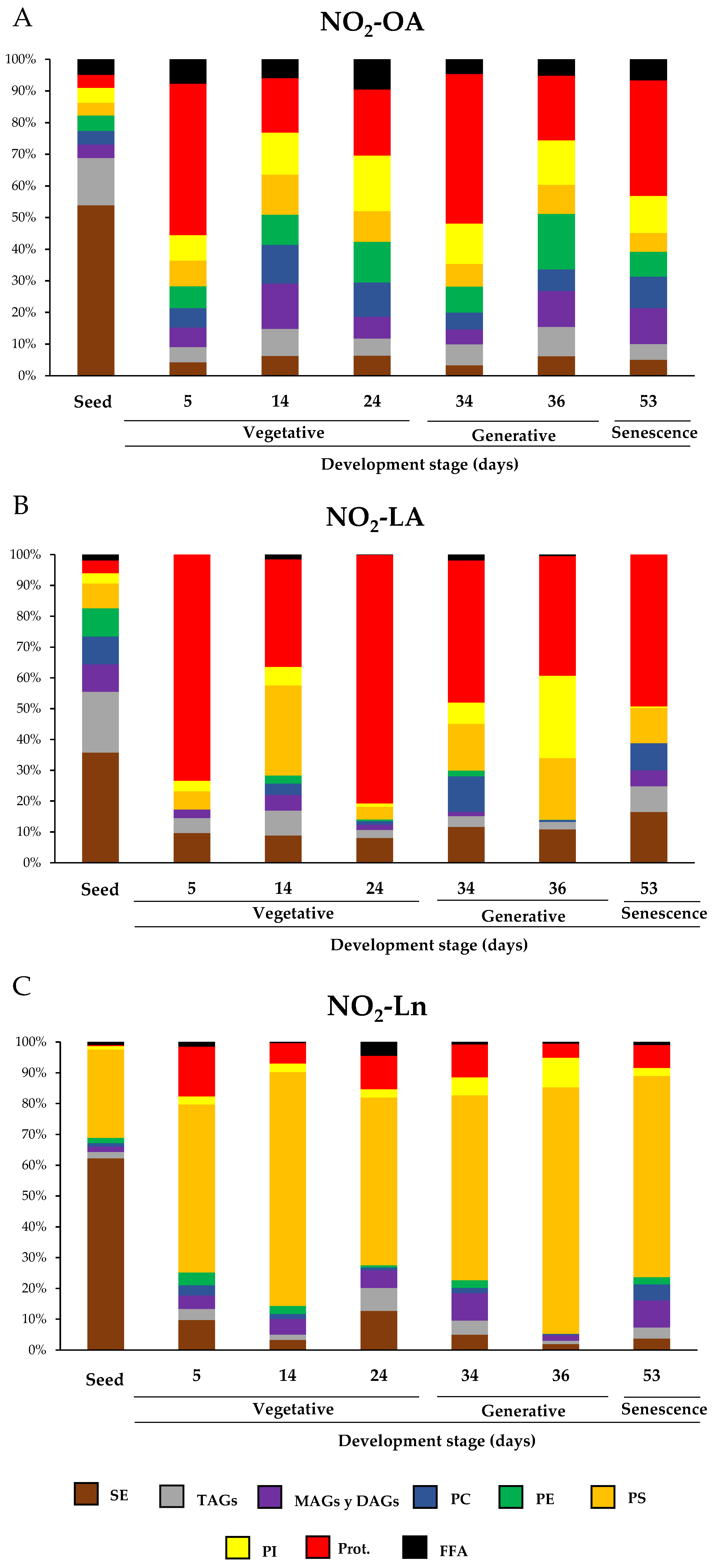
In this work, we identified the main NO2-FAs storage biomolecules implicated in the different analyzed growth stages. We selected different phases to cover the main events that take place during Arabidopsis development where the NO2-FAs levels in each reservoir were quantified. Figure 5 specifically shows the storage biomolecules of each NO2-FA in all the development stage. Hence, it indicates which storage biomolecule is the richest in NO2-OA (Panel A), NO2-LA (Panel B), or NO2-Ln (Panel C) in each development stage and how this abundance changes as plants grow and develop. Supplementary Table S1 displays the absolute values for the NO2-FAs levels for each reservoir biomolecule and development stage.
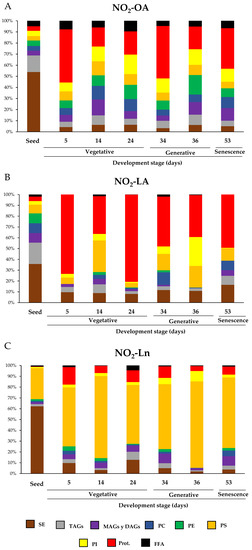
Figure 5.
The relative distribution of NO2-FAs in the different storage biomolecules during Arabidopsis development. Graphs show NO2-FAs as the storage biomolecules (NO2-OA in panel (A); NO2-LA in panel (B); NO2-Ln in panel (C)) for each developmental stage and how this distribution is modulated during development. The seeds and plants corresponding to the vegetative (5-, 14-, and 24-day-old plants), generative (34- and 36-day-old plants) and senescence stages (53-day-old plants) were used to characterize Arabidopsis development. The 100% corresponds to the total content of each NO2-FA and its distribution in biomolecule storages in each of the development situations studied.
Our results showed that, during Arabidopsis development, the distribution of NO2-FAs in the lipid and protein storages was wide and ubiquitous. However, a detailed analysis of the NO2-FAs distribution in the different storage biomolecules evidenced for seeds that the three NO2-FAs were esterified mainly with SE, with levels of 0.366 ± 0.038 pmol/g FW for NO2-OA, 0.352 ± 0.072 pmol/g FW for NO2-LA, and 5.137 ± 1.002 pmol/g FW for NO2-Ln (Figure 5 and Supplementary Table S1). NO2-OA and NO2-LA were significantly stored in TAGs with absolute values of 0.101 ± 0.019 pmol/g FW and 0.194 ± 0.032 pmol/g FW, respectively, while PS (2.367 ± 0.329 pmol/g FW) was the preferential deposit for NO2-Ln and thus was the second NO2-FAs storage biomolecule in seeds (Figure 5 and Supplementary Table S1). In the vegetative stage, specifically when seed germination progressed (5-day-old seedlings), proteins were the main reservoir biomolecule of NO2-OA (0.1 ± 0.016 pmol/g FW), NO2-LA (0.2 ± 0.014 pmol/g FW), and, to a lesser extent, of NO2-Ln because the main storage for this NO2-FA was located in PS (0.479 ± 0.092 pmol/g FW) (Figure 5 and Supplementary Table S1). During rosette development, which occurs in the vegetative stage (14- and 24-day-old plants), phospholipids and proteins become the main storage biomolecules for NO2-OA (Figure 5). In contrast, NO2-LA and NO2-Ln showed the same abundance pattern as that described above; that is, they were essentially located in proteins and PS, respectively (Figure 5). Finally, in the generative (34- and 36-day-old plants) and senescence (53-day-old plants) stages, phospholipids and proteins were the most representative storage biomolecules for NO2-OA, proteins for NO2-LA, and PS for NO2-Ln (Figure 5).
In order to obtain information about NO2-FAs abundance in each storage biomolecule in all they development stages, we looked for the preponderant NO2-FA in each storage biomolecule in the different development stages.
In the seed stage, NO2-OA presented relatively low abundance in each storage biomolecule compared to the other NO2-FAs, especially in PS. NO2-LA distribution was homogeneous except in the SE and PS reservoirs, where its presence was scarce. For NO2-Ln, its presence in SE and PS was predominantly noted although this NO2-FA was also found in the other storage biomolecules (Figure 6A and Supplementary Table S1). In the next growth stage, plants exhibited completely open cotyledons (5-day-old seedlings). In this case, the quantification of the MS/MS spectra showed that NO2-Ln was the predominant NO2-FA in most of the analyzed lipid storage biomolecules, which highlights its high esterification in PS. On the contrary, NO2-LA disappeared from the PC and PE deposits, and its contribution in the protein reservoir was marked. The participation of NO2-OA was homogeneous in the different storage biomolecules except for PS, where it was minimal (Figure 6B and Supplementary Table S1). In the 14-day-old seedlings, which phenotypically showed the onset of rosette formation, NO2-FAs were present in all the reservoirs, but their abundances differed. NO2-OA predominated in PC, NO2-LA in proteins, and PS remained as the main NO2-Ln storage biomolecule (Figure 6C and Supplementary Table S1). When plants presented a completely developed rosette (24-day-old plants), phospholipids accumulated mainly NO2-OA, which was not applicable to PS for being mostly esterified with NO2-Ln. NO2-LA was predominant in the protein deposit. NO2-Ln participation was greater in the other analyzed storage biomolecules (Figure 6D and Supplementary Table S1). The flowering 34-day-old seedlings played a predominant role for NO2-Ln, especially in PS and MAGs + DAGs. However, NO2-LA also played a major role in PC. NO2-OA was present in all the storage biomolecules but did not play a predominant role in any of them (Figure 6E and Supplementary Table S1). After flowering, the plant started seed production (36-day-old plants). In this phase, the dominance of NO2-OA in most storage biomolecules, especially in PC and PE, was noteworthy. For the other NO2-FAs, the abundance of NO2-Ln in PS and lack of NO2-LA in MAGs + DAGs were remarkable (Figure 6F and Supplementary Table S1). In the last plant development stage (senescence at 53 days), NO2-FAs presented a homogeneous abundance pattern, albeit with some exceptions: i.e., PE, where only NO2-OA and NO2-Ln were present; the observed MAGs + DAGs and PS reservoirs, where NO2-Ln almost completely dominated; and the quantities detected in PI where NO2-OA is highlighted (Figure 6G and Supplementary Table S1).
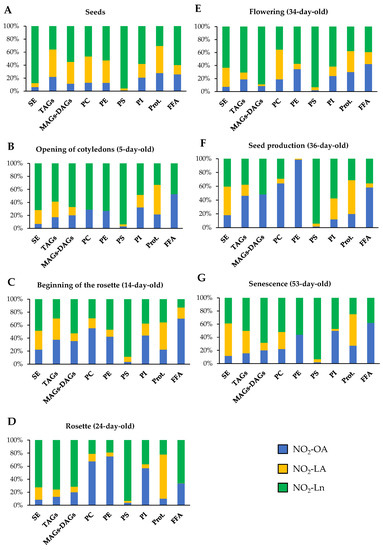
Figure 6.
NO2-FAs relative abundance in the different storage biomolecules in each Arabidopsis development stage. This figure shows the relative concentration (expressed as a percentage) of each NO2-FA in the different lipid and protein storages present in seeds (A) and in the 5- (B), 14- (C), 24- (D), 34- (E), 36- (F), and 53-day-old plants (G). The 100% represents the sum of the total NO2-OA, NO2-LA, and NO2-Ln content in each of the biomolecules storages studied for each stage of development.
In short, PC and PE were the main reservoir biomolecules for NO2-OA, proteins for NO2-LA, and TAGs and SE and PS for NO2-Ln.
The joint analysis of Figure 5 and Figure 6 showed that the SE storage in seeds was the main biomolecule for the three NO2-FAs (Figure 5), with NO2-Ln being the main one in this reservoir biomolecule (Figure 6A). In the vegetative stage, NO2-OA was distributed mostly in phospholipids and proteins (Figure 5). NO2-OA dominated only in the 14- and 24-day-old plants in PC and PE (Figure 6C,D). The distribution and abundance of NO2-LA were both located mostly in the protein storage (Figure 5 and Figure 6B–D). This same behavior was observed for NO2-Ln, but in this case, the main storage biomolecule was PS (Figure 5 and Figure 6B–D). Finally, in the generative and senescence stages, phospholipids and proteins continued to be the main NO2-OA reservoir biomolecules. However, there were some differences depending on the storage biomolecule and development stage. That was the case of the 36-day-old plants, where NO2-OA was exclusively detected in PC and PE, while NO2-LA and NO2-Ln presented the same behavior pattern as previously described in the vegetative stage (Figure 5 and Figure 6E–G).
4. Discussion
NO2-FAs are important signaling molecules that have been widely studied in animal systems. The actions mediated by NO2-OA, NO2-LA, NO2-cLA, and NO2-AA have been the focus of different works [4,5,6]. In plant systems, the first indications of the existence of NO2-FAs were obtained in olive fruit and EVOO, where the presence of NO2-cLA and cysteine-adducted-NO2-OA was specifically identified [8]. Currently, NO2-FAs have an ubiquitous distribution in plant organisms since the NO2-Ln endogenous presence has been identified in different plant species such as Arabidopsis thaliana [9], Pisum sativum, and Oryza sativa [10]. Moreover, NO2-OA has been recently described in Brassica napus plants [11]. The presence of NO2-FAs in plants is related to developmental signaling processes and stress responses [9,11]. Given the important signaling role of NO2-OA, NO2-LA, and NO2-Ln in both animal and plant systems, this work applied high-sensitivity technologies and detected the endogenous presence of NO2-OA and NO2-LA for the first time in Arabidopsis thaliana. In addition, the presence of the previously described NO2-Ln was confirmed (Figure 1). The quantification of the three NO2-FAs levels identified in Arabidopsis highlighted NO2-Ln as the most abundant, whereas NO2-LA and NO2-OA had lower but similar concentrations (Figure 2). These results are consistent with the availability of the corresponding unsaturated precursors in Arabidopsis, which may be susceptible to nitrate and might give rise to NO2-FAs. Thus, in the lipid profile of this plant species, Ln appeared as the majority in leaves and represents almost 50% of the total fatty acid content. The second most abundant was LA, followed by OA [66,67].
The addition of the nitro-group to unsaturated fatty acids converts NO2-FAs into potent electrophilic molecules that can establish adducts with proteins. In addition, NO2-FAs, like its non-nitrated forms, can also be esterified with complex lipids. Therefore, proteins and complex lipids can be considered NO2-FAs storage biomolecules [19,28]. In animal systems, a large group of proteins capable of binding to NO2-FAs has been identified [22,48,49,50,51,52,53,54,55,56]. In addition, TAGs have been identified as the main NO2-FAs storage lipid in these organisms [55]. However, the NO2-FAs storage biomolecules are completely unknown in plant systems. In this work, the MS/MS analyses performed on the 14-day-old seedlings, which represent a clear example of Arabidopsis vegetative growth, allowed NO2-FAs storage biomolecules to be identified in this plant species. Hence, the presence of NO2-OA, NO2-LA, and NO2-Ln bound to proteins and complex lipids, such as SE, TAGs, and different kinds of phospholipids, was identified (Figure 3 and Supplementary Figure S1).
The results obtained in this work revealed that phospholipids were the main reservoir of the three NO2-FAs. This was especially the case of NO2-Ln (Figure 3). Phospholipids are essential components of lipid bilayers for their amphipathic character [30,31]. Different types of phospholipids exist depending on the molecule to which the phosphate group is attached, such as PC, PE, PS, and PI. Based on our results, NO2-FAs were present in all the afore-mentioned phospholipid classes although they showed different abundance patterns. In general, PS were highlighted as the main NO2-FAs storage biomolecule, and this is especially applicable to NO2-Ln, whose esterification levels in PS were very high. For the other phospholipids, lower levels than those detected in PS were found but were similar for the three NO2-FAs (Supplementary Figure S1). Interestingly, the abundance of NO2-FAs in each phospholipid class did not correspond to their content in cell membranes. Indeed PC and PE were the main phospholipids in plant membranes, while PS and PI represented between 10% and 20% of the total phospholipid content [32,33].
Other albeit less representativeNO2-FAs storage biomolecules were MAGs, DAGs, and TAGs (Supplementary Figure S1). These molecules act mainly as a source of energy for young seedling growth, especially in early germination stages [46]. Furthermore, MAGs and DAGs are the common precursors of TAGs and membrane lipids (phospholipids). In leaves, most DAGs participate in the synthesis of membrane lipids that will be used for cell growth and proliferation and also for the maintenance of membranes [47]. SE were the most residual NO2-FAs storage lipids in juvenile plants (14-day-old) (Supplementary Figure S1). SE involve the esterification of a fatty acid with phytosterols, such as β-sitosterol, stigmasterol, campesterol, and cholesterol [44]. These molecules are found in plasma membranes and are implicated in the maintenance of the homeostasis of the phytosterols in them because, together with phospholipids and sphingolipids, they are essential plasma membrane components [36].
In addition to NO2-FAs lipid storages, NO2-FAs can also appear adducted with proteins. This makes them an important source of NO2-FAs because the levels detected in proteins were similar to those quantified in TAGs in the 14-day-old Arabidopsis plants (Supplementary Figure S1).
In summary, NO2-FAs are mainly localized in biomembranes, where they are esterified with phospholipids and SE. The presence of NO2-FAs in biomembranes can modulate physical bilayer properties and can even indirectly influence transmembrane proteins. In a recent study, the effect of the presence of NO2-FAs in biomembranes was evaluated by treating a model membrane with NO2-OA. The researchers found that NO2-FA altered the organization of membrane lipids, which led to not only lipid bilayer reorganization but also to the modulation of the structure and function of membrane-associated proteins [29]. The marked presence of NO2-FAs is also highlighted, especially NO2-Ln in PS. This phospholipid kind is considered a signaling lipid that regulates membrane-associated processes [35]. Therefore, the presence of NO2-FAs in PS could implicate them in these signaling processes.
After identifying the main NO2-FAs storage biomolecules in Arabidopsis thaliana, a study of the distribution of the NO2-FAs inside them during plant development was carried out. The starting point for plant development is seeds. The highest total NO2-OA, NO2-LA, and NO2-Ln levels were found in this plant material type (Figure 4C and Supplementary Table S1). However, when the NO2-FAs levels stored in lipid and protein reservoirs were analyzed separately, opposite behaviors were observed. High levels of complex lipid-esterified NO2-FAs and lesser abundance of protein-adducted NO2-FAs were quantified (Figure 4A,B). This NO2-FAs storage pattern was expected because seeds contain more than 60% lipids but also contain carbohydrates and proteins in smaller proportions [68]. The most abundant lipids in seeds are TAGs and SE, and both storage biomolecules are mobilized during germination to facilitate seedling growth in early stages [69]. In our study, these lipids (TAGs and SE), together with PS, contained the majority of NO2-FAs. Specifically, 50% NO2-OA, 35% NO2-LA, and 60% NO2-Ln were found to be esterified with SE, which proved to be the main NO2-FAs storage biomolecule in seeds. The contents of NO2-OA (15%) and NO2-LA (20%) in TAGs and the abundance of NO2-Ln (30%) in PS were also noteworthy (Figure 5 and Supplementary Table S1). Therefore, generally in seeds, NO2-FAs were mainly esterified with lipids with an energy function (TAGs) by acting as a source of sterols for membrane generation (SE) or by forming part of membranes (PS). In addition, the NO2-FAs esterified with complex lipids in seeds could act as stable NO reservoirs with the ability to donate NO to favor the germination process. In line with this, it has been reported that ABI5 transcription factor degradation by S-nitrosylation initiates seed germination [70].
Next, the Arabidopsis development stages consisting of seed germination and vegetative plant growth were studied. For this purpose, the newly germinated seedlings showing a radicle, a hypocotyl, and two completely open cotyledons (at 5 days), along with the seedlings in which rosette arrangement began to be noted (at 14 days) and the plants displaying a mature rosette (at 24 days), were analyzed. The total NO2-FAs levels progressively lowered, which was mainly motivated by the reduction of NO2-FAs esterification with complex lipids. However, adduction with proteins increased in these stages (Figure 4). The decrease observed in NO2-FAs content in the lipid storage, especially in relation to TAGs and SE (Figure 5), can be associated with the hydrolysis of TAGs to FFA and glycerol by the action of lipase enzymes [71]. Glycerol can be phosphorylated to undergo gluconeogenesis after its conversion into dihydroxyacetone phosphate (DHAP) [72]. FFA can be transported to the peroxisome, where they are activated to acyl-CoA to initiate β-oxidation. The β-oxidation product, acetyl-CoA, enters the glyoxylate cycle and, via the gluconeogenic pathway, provides the glucose required by the embryo as a source of energy during germination [73,74]. The scientific literature also includes SE mobilization processes during membrane development [69]. Something similar occurs with DAGs, which are precursor molecules of both TAGs and phospholipids, as indicated above. During germination and vegetative growth, most DAGs are used for membrane lipid assembly in areas where cell biogenesis, membrane expansion, and maintenance take place [47]. Consequently, significant amounts of TAGs do not accumulate in these plant growth phases [46].
Phospholipases, which are capable of hydrolyzing several types of phospholipid bonds, also play an important role in the vegetative stage [75]. Specifically, phospholipases act as key regulators during membrane organization by remodeling their lipids component, including NO2-FA. Consequently, they influence the physical properties of membranes [76]. By way of example, it has been described that phospholipase A1 (PLA1), which is expressed at high levels in juvenile plants, hydrolyses the acyl group at the sn-1 position of phospholipids and produces FFA and lysophospholipids. PLA1 shows increased expression during young rosette development [77]. Moreover, the scientific literature points out that these enzymes catalyze the release of Ln, which is the precursor of jasmonic acid, an organic compound involved in plant growth and development [78]. Another protein type that hydrolyses phospholipids, specifically at the sn-2 position, is phospholipase A2 (PLA2), and it also interacts with auxins. In particular, PLA2 and the product of its activity, lysophosphatidic ethanolamine, are required for the trafficking of PINs (auxin exit transporters) to the plasma membrane. This interaction is necessary for proper root growth [79] and correct endomembrane system organization during development and germination processes [80,81].
Generally, during seed germination and in the vegetative stage, phospholipids, as the main constituents of membranes, and proteins play a major role in plant growth [46]. They are also the main NO2-FAs storage biomolecules. Specifically, in this work, the distribution of NO2-OA was homogeneous between phospholipids and proteins, and NO2-LA was stored mostly in proteins, which associated both NO2-FAs with biological signaling processes. In contrast, NO2-Ln abundance in phospholipids was remarkable, especially in PS (Figure 5). At this point, it is important to mention that plant cell membranes are composed mainly of three lipid classes, namely glycerolipids, sphingolipids, and sterols, of which glycerolipids are the most abundant [33]. Glycerolipids are divided, in turn, into galactolipids, sulfolipids, TAGs, and phospholipids. The last are the most abundant in membranes [82]. PC and PE are the major phospholipids in membranes and are often considered “structural” lipids because they are essential for establishing a hydrophobic barrier in the membrane. In contrast, PS and PI are less abundant in membranes and act as “signaling” molecules because they mostly have regulatory effects on membrane functions. As these signaling lipids are negatively charged, they can interact with specific proteins and regulate almost all membrane-associated events [34,35,83]. Hence, the presence of NO2-FAs in different membrane lipid components during germination and in the vegetative stage was analyzed. Our results showed that approximately more than 30% NO2-OA, 10% NO2-LA and, above all, more than 70% NO2-Ln were esterified with some phospholipid type (Figure 5). Although very little is known about the role of NO2-FAs in biomembranes, different computer simulation techniques have shown that NO2-OA can form domains that cause membrane reorganization with the concomitant modification of the dynamic structure of their integral proteins [29]. In particular, the presence of NO2-Ln in a signaling complex lipid such as PS (Figure 5) could be related to its involvement in some processes, such as cell signaling, molecular trafficking, and cell division and growth. All these possibilities are supported by the presence of the nitro-group, which makes NO2-Ln a very reactive molecule that can carry out the PTM of proteins by nitroalkylation [7,84] or can act as a NO donor by giving rise to the S-nitrosylation PTM [15,17].
In short, NO2-FAs levels are lowered in the vegetative stage, especially in the TAGs and SE storages. This behavior could be justified by TAGs degradation to obtain energy and SE hydrolysis to generate the membranes needed to develop photosynthetic structures, root systems, stems, and leaves. In these growth stages, the remodeling of NO2-FAs storage biomolecules also occurs because, although TAGs and SE are major reservoirs in seeds, they are minor ones during seed germination and in the vegetative stage when phospholipids and proteins acquire a more relevant role and confer NO2-FAs functional and signaling implications.
Once the plant presents a complete mature rosette (24-day-old plants), it starts the generative stage with flowering. It is noteworthy that the NO2-FAs levels stored in PC were lower in plants with a complete rosette (at 24 days) in relation to the plants displaying a juvenile rosette (14-day-old seedlings). Specifically a 72% decrease was detected for NO2-OA, one of 81% for NO2-LA, and one of 83% for NO2-Ln (Supplementary Table S1). The diminished presence of NO2-FAs in PC, which was more pronounced in NO2-Ln in the plants whose rosette development had been completed, could be related to the next flowering step because such a process is initiated by the transcription of the FLOWERING LOCUS T (FT) gene, which gives rise to a mobile protein that induces flowering in plants [85]. Despite PC being a “structural” lipid, it participates in the flowering process because different works conducted with mutant plants have established that seedlings with high PC levels show early flowering, while the opposite occurs in plants with lower PC contents [86,87]. Interestingly, the flowering phenotype observed in the plants with higher PC contents depended on the expression of FT, which was directly influenced by the high saturated fatty acids composition of PC. The saturated or unsaturated fatty acids content in PC is influenced by photoperiod, with saturated fatty acids predominating in the daytime and unsaturated content at night [86,88]. Therefore, the punctual decrease in the NO2-FAs content in PC detected in the 24-day-old plants could modulate the expression of the gene to favor flowering (FT). This decrease was restricted to that phase because in those plants, in a more advanced flowering stage with open flowers (34-day-old plants), the NO2-FAs levels in PC recovered, whose values were similar to those detected in the seedlings with a juvenile rosette (14-day-old seedlings) (Supplementary Table S1).
Next, the plant enters a generative period when it spends its energy on reproduction by developing flowers (34-day-old plants) from which fruit and seeds are generated (36-day-old plants). In this stage, the main NO2-FAs storage biomolecules continued to be proteins and phospholipids (Figure 5). The marked presence of NO2-Ln in PS should be particularly underlined (Figure 6), which is a phospholipid that is relevant in reproductive organ formation. Specifically, different studies performed in Arabidopsis plants deficient in phosphatidylserine synthase 1 (PSS1), a key protein in the synthesis of PS from PE via a base exchange mechanism, have demonstrated a reduction in the inflorescence meristem and evidenced plants’ inability to develop new floral meristems. Therefore, the presence of PSS1 and thus PS plays an essential role in floral meristem maintenance and development through the activation of the CLAVATA (CLV)-WUSCHEL (WUS) signaling pathway [89,90]. These results suggest the possible involvement of NO2-FAs to signal flowering initiation and also in the maintenance and development of secondary floral structures.
Furthermore, in the generative stage, specifically during flowering, phospholipids and proteins continued to be the main NO2-FAs storage biomolecules. However, when the plant obtained its first fruit, which are called siliques in Arabidopsis, the total NO2-FAs levels slightly lowered in both the lipid and protein reservoirs although the reduction in the latter was more pronounced (Figure 4). This behavior could be associated with the beginning of the senescence stage because the 36-day-old plants showed mature siliques and represented an intermediate stage between the generative and senescence stages. When senescence starts, plants enter a state of programmed cell death because photosynthetic capacity reduces when chloroplast thylakoid membranes are degraded. These membranes are formed by glycolipids, and an increase in fatty acids release occurs as a consequence of glycolipid degradation. These processes cause TAGs to accumulate in senescent leaves [91,92]. The phospholipids and sterols located in extraplasmatic membranes, such as the plasma membrane, tonoplast, and endoplasmic reticulum, also decrease during senescence. Phospholipase activity increases in phospholipids, which leads to membrane degradation, and in the case of sterols, senescence generates SE accumulation [93,94]. Briefly, senescence brings about loss of plants’ photosynthetic capacity because chloroplasts’ thylakoid membranes degrade, and chlorophyll levels decrease. Extraplasmatic membranes also degrade, which reduces membrane fluidity. All this leads to the mobilization of nutrients from leaves, which causes lipids accumulation in seeds, such as TAGs and SE [94,95].
The last stage development to be characterized was senescence. To study this phase, we used plants with advanced senescence (at 53 days), in which very low NO2-FAs levels were quantified in both the lipid and protein deposits (Figure 4). The drop in NO2-FAs levels could be a consequence of membrane phospholipids hydrolysis because it was in these complex lipids where NO2-FAs were localized mainly in earlier growth stages. It is well-established in the senescence stage that ROS levels increase [80] and can cause the oxidation of the Michael adducts generated between proteins and NO2-FAs, which would lead to NO2-FA release [20,48]. All these mechanisms would cause NO2-FAs release from the membranous structures present in leaves and their mobilization towards seeds, where they would be integrated into lipids that can act as a source of either energy (TAGs) or sterols for membrane generation (SE). All this would explain the high NO2-FAs levels quantified in seeds, to which the possible nitration of unsaturated fatty acids as a consequence of the high oxidative state in senescent cells would contribute.
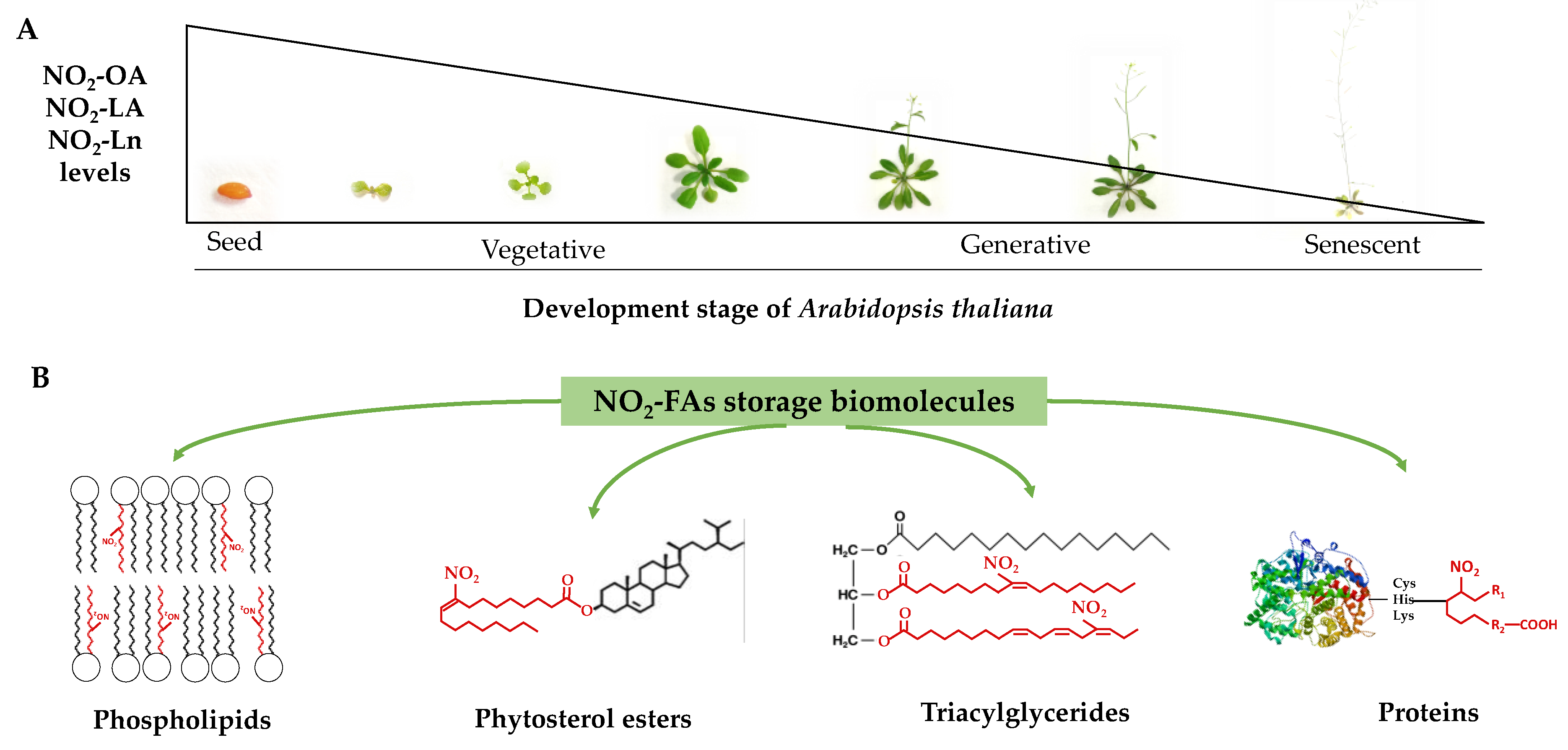
5. Conclusions
In this work, the presence of nitro-oleic acid and nitro-linoleic acid in Arabidopsis thaliana was identified and quantified for the first time. Additionally, the presence of nitro-linolenic acid in this plant species was corroborated, which played a predominant role in the analyzed processes (Figure 7). The characterization of the distribution of nitrated fatty acids at the cellular level, unknown to date, allowed the establishment of phytosterol esters; mono-, di-, and triacylglycerides; phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, and phosphatidylinositol as the main lipid reservoirs of these molecules. Our results reflect a heterogeneous distribution of nitro-fatty acids in cellular compartments and their preferential localization in lipid storage upon the protein reservoir (Figure 7).
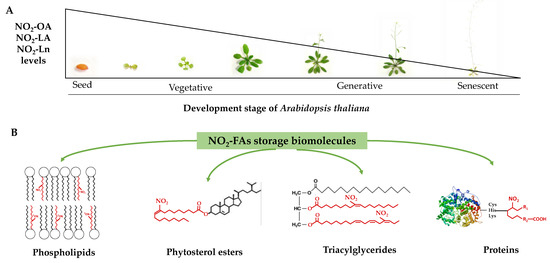
Figure 7.
The NO2-OA, NO2-LA, and NO2-Ln levels during Arabidopsis thaliana development (panel (A)) and the identification of phospholipids, phytosterol esters, triacylglycerides, and proteins as NO2-FAs storage biomolecules in Arabidopsis (panel (B)).
The study carried out throughout Arabidopsis development showed a decreasing pattern in the nitro-fatty acids content stored in both the protein and lipid reservoirs (Figure 7). Phytosterol esters and triacylglycerides were the preferential lipid storages in seeds, which were replaced with phospholipids and proteins in the vegetative, reproductive, and senescence stages. The majority esterification of the nitro-fatty acids in phospholipids, specifically in phosphatidylserine, suggested that nitro-fatty acids were involved in biomembrane dynamics by directly influencing their biophysical properties and also in phosphatidylserine-mediated signaling phenomena. The decreased content of nitro-fatty acids in phosphatidylcholine would limit the inhibitory effect exerted by unsaturated fatty acids on the flowering process initiation.
Therefore, this work provides a detailed study of the role of nitro-fatty acids during plant development by highlighting the possible implications of these molecules in decisive events for plant development. It also opens up a new research field to analyze and study the possible role and interactions between nitro-fatty acids and phospholipids and with the proteins in the signaling processes that occur during plant development.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antiox11101869/s1, Figure S1: Distribution and quantification of the endogenous levels of NO2-FAs in the different storage biomolecules; Table S1: The endogenous levels of NO2-OA, NO2-LA, and NO2-Ln in the different storage biomolecules in the selected Arabidopsis thaliana development stages.
Author Contributions
Conceptualization, J.B.B.; methodology, L.A.-C., J.B.B. and R.V.; investigation, L.A.-C., R.V., J.C.B.-M., M.C., J.B.B. and M.M.; writing—original draft, L.A.-C.; writing—review and editing, L.A.-C., R.V. and J.B.B.; funding acquisition, J.B.B.; supervision, J.B.B. and R.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by ERDF grants co-financed by the Spanish Ministry of Economy and Competitiveness (Project PGC2018-096405-B-I00); the Junta de Andalucía (group BIO286); the I+D+I project within the framework Programme of FEDER Andalucía 2014–2020 (Reference 1380901); the grants for I+D+I projects, on a competitive basis, within the scope of the Andalusian plan for research, development and innovation (Junta de Andalucía, PAIDI 2020, Reference: PY20_01002); and the funding for the recruitment of researchers under Action 9 and 10 of the Research Support Plan of the University of Jaén (2019–2020, R.02/10/2020; 2020–2021, R.01/01/2022).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Material.
Acknowledgments
L.A.-C. wishes to thank the University of Jaén for funding her Ph.D. fellowship. The LC-MS/MS technical and human support provided by the Central Research Support Services (UJA-CICT) of the University of Jaén is gratefully acknowledged.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schopfer, F.J.; Khoo, N.K.H. Nitro-Fatty Acid Logistics: Formation, Biodistribution, Signaling, and Pharmacology. Trends Endocrinol. Metab. 2019, 30, 505–519. [Google Scholar] [CrossRef] [PubMed]
- Buchan, G.J.; Bonacci, G.; Fazzari, M.; Salvatore, S.R.; Gelhaus Wendell, S. Nitro-fatty acid formation and metabolism. Nitric Oxide 2018, 79, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Begara-Morales, J.C.; Mata-Pérez, C.; Padilla, M.N.; Chaki, M.; Valderrama, R.; Aranda-Caño, L.; Barroso, J.B. Role of electrophilic nitrated fatty acids during development and response to abiotic stress processes in plants. J. Exp. Bot. 2021, 72, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.R.; Schopfer, F.J.; Sweeney, S.; Freeman, B.A. Red cell membrane and plasma linoleic acid nitration products: Synthesis, clinical identification, and quantitation. Proc. Natl. Acad. Sci. USA 2004, 101, 11577–11582. [Google Scholar] [CrossRef] [PubMed]
- Balazy, M.; Poff, C.D. Biological nitration of arachidonic acid. Curr. Vasc. Pharmacol. 2004, 2, 81–93. [Google Scholar] [CrossRef]
- Tsikas, D.; Zoerner, A.A.; Jordan, J. Oxidized and nitrated oleic acid in biological systems: Analysis by GC-MS/MS and LC-MS/MS, and biological significance. Biochim. Biophys. Acta 2011, 1811, 694–705. [Google Scholar] [CrossRef]
- Aranda-Caño, L.; Valderrama, R.; Pedrajas, J.R.; Begara-Morales, J.C.; Chaki, M.; Padilla, M.N.; Melguizo, M.; López-Jaramillo, F.J.; Barroso, J.B. Nitro-Oleic Acid-Mediated Nitroalkylation Modulates the Antioxidant Function of Cytosolic Peroxiredoxin Tsa1 during Heat Stress in Saccharomyces cerevisiae. Antioxidants 2022, 11, 972. [Google Scholar] [CrossRef]
- Fazzari, M.; Trostchansky, A.; Schopfer, F.J.; Salvatore, S.R.; Sánchez-Calvo, B.; Vitturi, D.; Valderrama, R.; Barroso, J.B.; Radi, R.; Freeman, B.A.; et al. Olives and olive oil are sources of electrophilic fatty acid nitroalkenes. PLoS ONE 2014, 9, e84884. [Google Scholar] [CrossRef]
- Mata-Pérez, C.; Sánchez-Calvo, B.; Padilla, M.N.; Begara-Morales, J.C.; Luque, F.; Melguizo, M.; Jiménez-Ruiz, J.; Fierro-Risco, J.; Peñas-Sanjuán, A.; Valderrama, R.; et al. Nitro-Fatty Acids in Plant Signaling: Nitro-Linolenic Acid Induces the Molecular Chaperone Network in Arabidopsis. Plant Physiol. 2016, 170, 686–701. [Google Scholar] [CrossRef]
- Mata-Pérez, C.; Sánchez-Calvo, B.; Padilla, M.N.; Begara-Morales, J.C.; Valderrama, R.; Corpas, F.J.; Barroso, J.B. Nitro-fatty acids in plant signaling: New key mediators of nitric oxide metabolism. Redox Biol. 2017, 11, 554–561. [Google Scholar] [CrossRef]
- Vollár, M.; Feigl, G.; Oláh, D.; Horváth, A.; Molnár, Á.; Kúsz, N.; Ördög, A.; Csupor, D.; Kolbert, Z. Nitro-Oleic Acid in Seeds and Differently Developed Seedlings of Brassica napus L. Plants 2020, 9, 406. [Google Scholar] [CrossRef] [PubMed]
- Schopfer, F.J.; Baker, P.R.; Giles, G.; Chumley, P.; Batthyany, C.; Crawford, J.; Patel, R.P.; Hogg, N.; Branchaud, B.P.; Lancaster, J.R., Jr.; et al. Fatty acid transduction of nitric oxide signaling. Nitrolinoleic acid is a hydrophobically stabilized nitric oxide donor. J. Biol. Chem. 2005, 280, 19289–19297. [Google Scholar] [CrossRef] [PubMed]
- Lima, E.S.; Bonini, M.G.; Augusto, O.; Barbeiro, H.V.; Souza, H.P.; Abdalla, D.S. Nitrated lipids decompose to nitric oxide and lipid radicals and cause vasorelaxation. Free Radic. Biol. Med. 2005, 39, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.R.; Schopfer, F.J.; O’Donnell, V.B.; Freeman, B.A. Convergence of nitric oxide and lipid signaling: Anti-inflammatory nitro-fatty acids. Free Radic. Biol. Med. 2009, 46, 989–1003. [Google Scholar] [CrossRef] [PubMed]
- Mata-Pérez, C.; Sánchez-Calvo, B.; Begara-Morales, J.C.; Carreras, A.; Padilla, M.N.; Melguizo, M.; Valderrama, R.; Corpas, F.J.; Barroso, J.B. Nitro-linolenic acid is a nitric oxide donor. Nitric Oxide 2016, 57, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Mata-Pérez, C.; Sánchez-Calvo, B.; Begara-Morales, J.C.; Padilla, M.N.; Valderrama, R.; Corpas, F.J.; Barroso, J.B. Nitric oxide release from nitro-fatty acids in Arabidopsis roots. Plant Signal. Behav. 2016, 11, e1154255. [Google Scholar] [CrossRef]
- Mata-Pérez, C.; Padilla, M.N.; Sánchez-Calvo, B.; Begara-Morales, J.C.; Valderrama, R.; Chaki, M.; Aranda-Caño, L.; Moreno-González, D.; Molina-Díaz, A.; Barroso, J.B. Endogenous Biosynthesis of S-Nitrosoglutathione From Nitro-Fatty Acids in Plants. Front. Plant Sci. 2020, 11, 962. [Google Scholar] [CrossRef]
- Rudolph, T.K.; Freeman, B.A. Transduction of redox signaling by electrophile-protein reactions. Sci. Signal. 2009, 2, re7. [Google Scholar] [CrossRef] [PubMed]
- Schopfer, F.J.; Cipollina, C.; Freeman, B.A. Formation and signaling actions of electrophilic lipids. Chem. Rev. 2011, 111, 5997–6021. [Google Scholar] [CrossRef]
- Padilla, M.N.; Mata-Pérez, C.; Melguizo, M.; Barroso, J.B. In vitro nitro-fatty acid release from Cys-NO(2)-fatty acid adducts under nitro-oxidative conditions. Nitric Oxide 2017, 68, 14–22. [Google Scholar] [CrossRef]
- Schopfer, F.J.; Vitturi, D.A.; Jorkasky, D.K.; Freeman, B.A. Nitro-fatty acids: New drug candidates for chronic inflammatory and fibrotic diseases. Nitric Oxide 2018, 79, 31–37. [Google Scholar] [CrossRef]
- Turell, L.; Steglich, M.; Alvarez, B. The chemical foundations of nitroalkene fatty acid signaling through addition reactions with thiols. Nitric Oxide 2018, 78, 161–169. [Google Scholar] [CrossRef]
- Khoo, N.K.H.; Schopfer, F.J. Nitrated fatty acids: From diet to disease. Curr. Opin. Physiol. 2019, 9, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Rom, O.; Xu, G.; Guo, Y.; Zhu, Y.; Wang, H.; Zhang, J.; Fan, Y.; Liang, W.; Lu, H.; Liu, Y.; et al. Nitro-fatty acids protect against steatosis and fibrosis during development of nonalcoholic fatty liver disease in mice. EBioMedicine 2019, 41, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Panati, K.; Thimmana, L.V.; Narala, V.R. Electrophilic nitrated fatty acids are potential therapeutic candidates for inflammatory and fibrotic lung diseases. Nitric Oxide 2020, 102, 28–38. [Google Scholar] [CrossRef]
- Piesche, M.; Roos, J.; Kühn, B.; Fettel, J.; Hellmuth, N.; Brat, C.; Maucher, I.V.; Awad, O.; Matrone, C.; Comerma Steffensen, S.G.; et al. The Emerging Therapeutic Potential of Nitro Fatty Acids and Other Michael Acceptor-Containing Drugs for the Treatment of Inflammation and Cancer. Front. Pharmacol. 2020, 11, 1297. [Google Scholar] [CrossRef] [PubMed]
- Di Fino, L.M.; Cerrudo, I.; Salvatore, S.R.; Schopfer, F.J.; García-Mata, C.; Laxalt, A.M. Exogenous Nitro-Oleic Acid Treatment Inhibits Primary Root Growth by Reducing the Mitosis in the Meristem in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 1059. [Google Scholar] [CrossRef]
- Grippo, V.; Mojovic, M.; Pavicevic, A.; Kabelac, M.; Hubatka, F.; Turanek, J.; Zatloukalova, M.; Freeman, B.A.; Vacek, J. Electrophilic characteristics and aqueous behavior of fatty acid nitroalkenes. Redox Biol. 2021, 38, 101756. [Google Scholar] [CrossRef]
- Franz, J.; Bereau, T.; Pannwitt, S.; Anbazhagan, V.; Lehr, A.; Nubbemeyer, U.; Dietz, U.; Bonn, M.; Weidner, T.; Schneider, D. Nitrated Fatty Acids Modulate the Physical Properties of Model Membranes and the Structure of Transmembrane Proteins. Chemistry 2017, 23, 9690–9697. [Google Scholar] [CrossRef]
- Joyard, J.; Maréchal, E.; Miège, C.; Block, M.A.; Dorne, A.-J.; Douce, R. Structure, distribution and biosynthesis of glycerolipids from higher plant chloroplasts. In Lipids in Photosynthesis: Structure, Function and Genetics; Springer: Dordrecht, The Netherlands, 1998; pp. 21–52. [Google Scholar]
- Nakamura, Y. Plant Phospholipid Diversity: Emerging Functions in Metabolism and Protein-Lipid Interactions. Trends Plant Sci. 2017, 22, 1027–1040. [Google Scholar] [CrossRef]
- Platre, M.P.; Bayle, V.; Armengot, L.; Bareille, J.; Marquès-Bueno, M.D.M.; Creff, A.; Maneta-Peyret, L.; Fiche, J.B.; Nollmann, M.; Miège, C.; et al. Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 2019, 364, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Reszczyńska, E.; Hanaka, A. Lipids Composition in Plant Membranes. Cell Biochem. Biophys. 2020, 78, 401–414. [Google Scholar] [CrossRef]
- Gerth, K.; Lin, F.; Menzel, W.; Krishnamoorthy, P.; Stenzel, I.; Heilmann, M.; Heilmann, I. Guilt by Association: A Phenotype-Based View of the Plant Phosphoinositide Network. Annu. Rev. Plant Biol. 2017, 68, 349–374. [Google Scholar] [CrossRef] [PubMed]
- Noack, L.C.; Jaillais, Y. Functions of Anionic Lipids in Plants. Annu. Rev. Plant Biol. 2020, 71, 71–102. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.-A. 5 Sterol metabolism and functions in higher plants. In Lipid Metabolism and Membrane Biogenesis; Springer: Berlin/Heidelberg, Germany, 2004; pp. 183–211. [Google Scholar]
- Michaelson, L.V.; Napier, J.A.; Molino, D.; Faure, J.D. Plant sphingolipids: Their importance in cellular organization and adaption. Biochim. Biophys. Acta 2016, 1861, 1329–1335. [Google Scholar] [CrossRef] [PubMed]
- Sperling, P.; Franke, S.; Lüthje, S.; Heinz, E. Are glucocerebrosides the predominant sphingolipids in plant plasma membranes? Plant Physiol. Biochem. 2005, 43, 1031–1038. [Google Scholar] [CrossRef]
- Borner, G.H.; Sherrier, D.J.; Weimar, T.; Michaelson, L.V.; Hawkins, N.D.; Macaskill, A.; Napier, J.A.; Beale, M.H.; Lilley, K.S.; Dupree, P. Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 2005, 137, 104–116. [Google Scholar] [CrossRef]
- Lefebvre, B.; Furt, F.; Hartmann, M.A.; Michaelson, L.V.; Carde, J.P.; Sargueil-Boiron, F.; Rossignol, M.; Napier, J.A.; Cullimore, J.; Bessoule, J.J.; et al. Characterization of lipid rafts from Medicago truncatula root plasma membranes: A proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 2007, 144, 402–418. [Google Scholar] [CrossRef] [PubMed]
- Coursol, S.; Fan, L.M.; Le Stunff, H.; Spiegel, S.; Gilroy, S.; Assmann, S.M. Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 2003, 423, 651–654. [Google Scholar] [CrossRef]
- Saucedo-García, M.; Guevara-García, A.; González-Solís, A.; Cruz-García, F.; Vázquez-Santana, S.; Markham, J.E.; Lozano-Rosas, M.G.; Dietrich, C.R.; Ramos-Vega, M.; Cahoon, E.B.; et al. MPK6, sphinganine and the LCB2a gene from serine palmitoyltransferase are required in the signaling pathway that mediates cell death induced by long chain bases in Arabidopsis. New Phytol. 2011, 191, 943–957. [Google Scholar] [CrossRef]
- Xie, L.J.; Chen, Q.F.; Chen, M.X.; Yu, L.J.; Huang, L.; Chen, L.; Wang, F.Z.; Xia, F.N.; Zhu, T.R.; Wu, J.X.; et al. Unsaturation of very-long-chain ceramides protects plant from hypoxia-induced damages by modulating ethylene signaling in Arabidopsis. PLoS Genet. 2015, 11, e1005143. [Google Scholar] [CrossRef]
- Moreau, R.A.; Nyström, L.; Whitaker, B.D.; Winkler-Moser, J.K.; Baer, D.J.; Gebauer, S.K.; Hicks, K.B. Phytosterols and their derivatives: Structural diversity, distribution, metabolism, analysis, and health-promoting uses. Prog. Lipid Res. 2018, 70, 35–61. [Google Scholar] [CrossRef] [PubMed]
- Rogowska, A.; Szakiel, A. The role of sterols in plant response to abiotic stress. Phytochem. Rev. 2020, 19, 1525–1538. [Google Scholar] [CrossRef]
- Xu, C.; Shanklin, J. Triacylglycerol Metabolism, Function, and Accumulation in Plant Vegetative Tissues. Annu. Rev. Plant Biol. 2016, 67, 179–206. [Google Scholar] [CrossRef]
- Bates, P.D.; Browse, J. The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J. 2011, 68, 387–399. [Google Scholar] [CrossRef]
- Batthyany, C.; Schopfer, F.J.; Baker, P.R.; Durán, R.; Baker, L.M.; Huang, Y.; Cerveñansky, C.; Branchaud, B.P.; Freeman, B.A. Reversible post-translational modification of proteins by nitrated fatty acids in vivo. J. Biol. Chem. 2006, 281, 20450–20463. [Google Scholar] [CrossRef] [PubMed]
- Sculptoreanu, A.; Kullmann, F.A.; Artim, D.E.; Bazley, F.A.; Schopfer, F.; Woodcock, S.; Freeman, B.A.; de Groat, W.C. Nitro-oleic acid inhibits firing and activates TRPV1- and TRPA1-mediated inward currents in dorsal root ganglion neurons from adult male rats. J. Pharmacol. Exp. Ther. 2010, 333, 883–895. [Google Scholar] [CrossRef]
- Zhang, J.; Villacorta, L.; Chang, L.; Fan, Z.; Hamblin, M.; Zhu, T.; Chen, C.S.; Cole, M.P.; Schopfer, F.J.; Deng, C.X.; et al. Nitro-oleic acid inhibits angiotensin II-induced hypertension. Circ. Res. 2010, 107, 540–548. [Google Scholar] [CrossRef]
- Artim, D.E.; Bazely, F.; Daugherty, S.L.; Sculptoreanu, A.; Koronowski, K.B.; Schopfer, F.J.; Woodcock, S.R.; Freeman, B.A.; de Groat, W.C. Nitro-oleic acid targets transient receptor potential (TRP) channels in capsaicin sensitive afferent nerves of rat urinary bladder. Exp. Neurol. 2011, 232, 90–99. [Google Scholar] [CrossRef]
- Bonacci, G.; Schopfer, F.J.; Batthyany, C.I.; Rudolph, T.K.; Rudolph, V.; Khoo, N.K.; Kelley, E.E.; Freeman, B.A. Electrophilic fatty acids regulate matrix metalloproteinase activity and expression. J. Biol. Chem. 2011, 286, 16074–16081. [Google Scholar] [CrossRef] [PubMed]
- Gil, M.; Graña, M.; Schopfer, F.J.; Wagner, T.; Denicola, A.; Freeman, B.A.; Alzari, P.M.; Batthyány, C.; Durán, R. Inhibition of Mycobacterium tuberculosis PknG by non-catalytic rubredoxin domain specific modification: Reaction of an electrophilic nitro-fatty acid with the Fe-S center. Free Radic. Biol. Med. 2013, 65, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Fazzari, M.; Khoo, N.; Woodcock, S.R.; Li, L.; Freeman, B.A.; Schopfer, F.J. Generation and esterification of electrophilic fatty acid nitroalkenes in triacylglycerides. Free Radic. Biol. Med. 2015, 87, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Fazzari, M.; Khoo, N.K.; Woodcock, S.R.; Jorkasky, D.K.; Li, L.; Schopfer, F.J.; Freeman, B.A. Nitro-fatty acid pharmacokinetics in the adipose tissue compartment. J. Lipid Res. 2017, 58, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Fazzari, M.; Vitturi, D.A.; Woodcock, S.R.; Salvatore, S.R.; Freeman, B.A.; Schopfer, F.J. Electrophilic fatty acid nitroalkenes are systemically transported and distributed upon esterification to complex lipids. J. Lipid Res. 2019, 60, 388–399. [Google Scholar] [CrossRef] [PubMed]
- Lancashire, P.D.; Bleiholder, H.; Boom, T.V.D.; Langelüddeke, P.; Stauss, R.; Weber, E.; Witzenberger, A. A uniform decimal code for growth stages of crops and weeds. Ann. Appl. Biol. 1991, 119, 561–601. [Google Scholar] [CrossRef]
- Boyes, D.C.; Zayed, A.M.; Ascenzi, R.; McCaskill, A.J.; Hoffman, N.E.; Davis, K.R.; Görlach, J. Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 2001, 13, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Leterrier, M.; Barroso, J.B.; Valderrama, R.; Palma, J.M.; Corpas, F.J. NADP-dependent isocitrate dehydrogenase from Arabidopsis roots contributes in the mechanism of defence against the nitro-oxidative stress induced by salinity. Sci. World J. 2012, 2012, 694740. [Google Scholar] [CrossRef]
- Cellier, F.; Conéjéro, G.; Ricaud, L.; Luu, D.T.; Lepetit, M.; Gosti, F.; Casse, F. Characterization of AtCHX17, a member of the cation/H+ exchangers, CHX family, from Arabidopsis thaliana suggests a role in K+ homeostasis. Plant J. 2004, 39, 834–846. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Gutbrod, K.; Peisker, H.; Dörmann, P. Direct Infusion Mass Spectrometry for Complex Lipid Analysis. Methods Mol. Biol. 2021, 2295, 101–115. [Google Scholar] [CrossRef]
- Baker, P.R.; Lin, Y.; Schopfer, F.J.; Woodcock, S.R.; Groeger, A.L.; Batthyany, C.; Sweeney, S.; Long, M.H.; Iles, K.E.; Baker, L.M.; et al. Fatty acid transduction of nitric oxide signaling: Multiple nitrated unsaturated fatty acid derivatives exist in human blood and urine and serve as endogenous peroxisome proliferator-activated receptor ligands. J. Biol. Chem. 2005, 280, 42464–42475. [Google Scholar] [CrossRef]
- Bonacci, G.; Baker, P.R.; Salvatore, S.R.; Shores, D.; Khoo, N.K.; Koenitzer, J.R.; Vitturi, D.A.; Woodcock, S.R.; Golin-Bisello, F.; Cole, M.P.; et al. Conjugated linoleic acid is a preferential substrate for fatty acid nitration. J. Biol. Chem. 2012, 287, 44071–44082. [Google Scholar] [CrossRef]
- Nadtochiy, S.M.; Zhu, Q.M.; Urciuoli, W.; Rafikov, R.; Black, S.M.; Brookes, P.S. Nitroalkenes confer acute cardioprotection via adenine nucleotide translocase 1. J. Biol. Chem. 2012, 287, 3573–3580. [Google Scholar] [CrossRef] [PubMed]
- Bonaventure, G.; Salas, J.J.; Pollard, M.R.; Ohlrogge, J.B. Disruption of the FATB gene in Arabidopsis demonstrates an essential role of saturated fatty acids in plant growth. Plant Cell 2003, 15, 1020–1033. [Google Scholar] [CrossRef] [PubMed]
- Tjellström, H.; Yang, Z.; Allen, D.K.; Ohlrogge, J.B. Rapid kinetic labeling of Arabidopsis cell suspension cultures: Implications for models of lipid export from plastids. Plant Physiol. 2012, 158, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D.; Black, M. Seeds: Physiology of Development and Germination; Springer Science & Business Media: London, UK, 2013. [Google Scholar]
- Harker, M.; Hellyer, A.; Clayton, J.C.; Duvoix, A.; Lanot, A.; Safford, R. Co-ordinate regulation of sterol biosynthesis enzyme activity during accumulation of sterols in developing rape and tobacco seed. Planta 2003, 216, 707–715. [Google Scholar] [CrossRef]
- Albertos, P.; Romero-Puertas, M.C.; Tatematsu, K.; Mateos, I.; Sánchez-Vicente, I.; Nambara, E.; Lorenzo, O. S-nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 2015, 6, 8669. [Google Scholar] [CrossRef]
- Barros, M.; Fleuri, L.; Macedo, G. Seed lipases: Sources, applications and properties-a review. Braz. J. Chem. Eng. 2010, 27, 15–29. [Google Scholar] [CrossRef]
- Quettier, A.L.; Shaw, E.; Eastmond, P.J. SUGAR-DEPENDENT6 encodes a mitochondrial flavin adenine dinucleotide-dependent glycerol-3-p dehydrogenase, which is required for glycerol catabolism and post germinative seedling growth in Arabidopsis. Plant Physiol. 2008, 148, 519–528. [Google Scholar] [CrossRef]
- Graham, I.A. Seed storage oil mobilization. Annu. Rev. Plant Biol. 2008, 59, 115–142. [Google Scholar] [CrossRef]
- Quettier, A.L.; Eastmond, P.J. Storage oil hydrolysis during early seedling growth. Plant Physiol. Biochem. 2009, 47, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Greer, M.S.; Weselake, R.J. Plant phospholipase A: Advances in molecular biology, biochemistry, and cellular function. Biomol. Concepts 2013, 4, 527–532. [Google Scholar] [CrossRef]
- Sandoval, G. Lipases and Phospholipases; Springer: New York, NY, USA, 2018. [Google Scholar]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The Defective in Anther Dehiscience gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef] [PubMed]
- Lee, O.R.; Kim, S.J.; Kim, H.J.; Hong, J.K.; Ryu, S.B.; Lee, S.H.; Ganguly, A.; Cho, H.T. Phospholipase A(2) is required for PIN-FORMED protein trafficking to the plasma membrane in the Arabidopsis root. Plant Cell 2010, 22, 1812–1825. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, J.H.; Lyu, J.I.; Woo, H.R.; Lim, P.O. New insights into the regulation of leaf senescence in Arabidopsis. J. Exp. Bot. 2018, 69, 787–799. [Google Scholar] [CrossRef]
- Hong, Y.; Zhao, J.; Guo, L.; Kim, S.C.; Deng, X.; Wang, G.; Zhang, G.; Li, M.; Wang, X. Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 2016, 62, 55–74. [Google Scholar] [CrossRef]
- MacDonald, G.E.; Lada, R.R.; Caldwell, C.D.; Udenigwe, C.; MacDonald, M.T. Potential roles of fatty acids and lipids in postharvest needle abscission physiology. Am. J. Plant Sci. 2019, 10, 1069–1089. [Google Scholar] [CrossRef]
- Pokotylo, I.; Kravets, V.; Martinec, J.; Ruelland, E. The phosphatidic acid paradox: Too many actions for one molecule class? Lessons from plants. Prog. Lipid Res. 2018, 71, 43–53. [Google Scholar] [CrossRef]
- Geisler, A.C.; Rudolph, T.K. Nitroalkylation--a redox sensitive signaling pathway. Biochim. Biophys. Acta 2012, 1820, 777–784. [Google Scholar] [CrossRef]
- Matsoukas, I.G. Florigens and antiflorigens: A molecular genetic understanding. Essays Biochem. 2015, 58, 133–149. [Google Scholar] [CrossRef]
- Nakamura, Y.; Andrés, F.; Kanehara, K.; Liu, Y.C.; Dörmann, P.; Coupland, G. Arabidopsis florigen FT binds to diurnally oscillating phospholipids that accelerate flowering. Nat. Commun. 2014, 5, 3553. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y. Membrane Lipid Oscillation: An Emerging System of Molecular Dynamics in the Plant Membrane. Plant Cell Physiol. 2018, 59, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Jaillais, Y.; Parcy, F. Lipid-mediated regulation of flowering time. Science 2021, 373, 1086–1087. [Google Scholar] [CrossRef] [PubMed]
- Yamaoka, Y.; Yu, Y.; Mizoi, J.; Fujiki, Y.; Saito, K.; Nishijima, M.; Lee, Y.; Nishida, I. PHOSPHATIDYLSERINE SYNTHASE1 is required for microspore development in Arabidopsis thaliana. Plant J. 2011, 67, 648–661. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Yin, H.; Gao, P.; Hu, X.; Yang, J.; Liu, Z.; Fu, X.; Luo, D. Phosphatidylserine synthase 1 is required for inflorescence meristem and organ development in Arabidopsis. J. Integr. Plant Biol. 2013, 55, 682–695. [Google Scholar] [CrossRef]
- Kaup, M.T.; Froese, C.D.; Thompson, J.E. A role for diacylglycerol acyltransferase during leaf senescence. Plant Physiol. 2002, 129, 1616–1626. [Google Scholar] [CrossRef]
- Lin, W.; Oliver, D.J. Role of triacylglycerols in leaves. Plant Sci. 2008, 175, 233–237. [Google Scholar] [CrossRef]
- Wewer, V.; Dombrink, I.; vom Dorp, K.; Dörmann, P. Quantification of sterol lipids in plants by quadrupole time-of-flight mass spectrometry. J. Lipid Res. 2011, 52, 1039–1054. [Google Scholar] [CrossRef]
- Li, L.; Zhao, J.; Zhao, Y.; Lu, X.; Zhou, Z.; Zhao, C.; Xu, G. Comprehensive investigation of tobacco leaves during natural early senescence via multi-platform metabolomics analyses. Sci. Rep. 2016, 6, 37976. [Google Scholar] [CrossRef]
- Troncoso-Ponce, M.A.; Cao, X.; Yang, Z.; Ohlrogge, J.B. Lipid turnover during senescence. Plant Sci. 2013, 205–206, 13–19. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).