Demyristoylation of the Cytoplasmic Redox Protein Trx-h2 Is Critical for Inducing a Rapid Cold Stress Response in Plants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Plasmid Construction and Plant Transformation
2.3. Detection of the Myristoylation of Trx-h2 In Vivo
2.4. Subcellular Fractionation of Nuclear and Non-Nuclear Proteins
2.5. Co-immunoprecipitation (Co-IP) Assay
2.6. Bimolecular Fluorescence Complementation (BiFC) Assay
2.7. Detection of the Structural Switching of CBF1 In Vivo
2.8. Purification of Trx-h2, Trx-h2(G/A), and Maltose Binding Protein (MBP)-CBF1 Recombinant Proteins
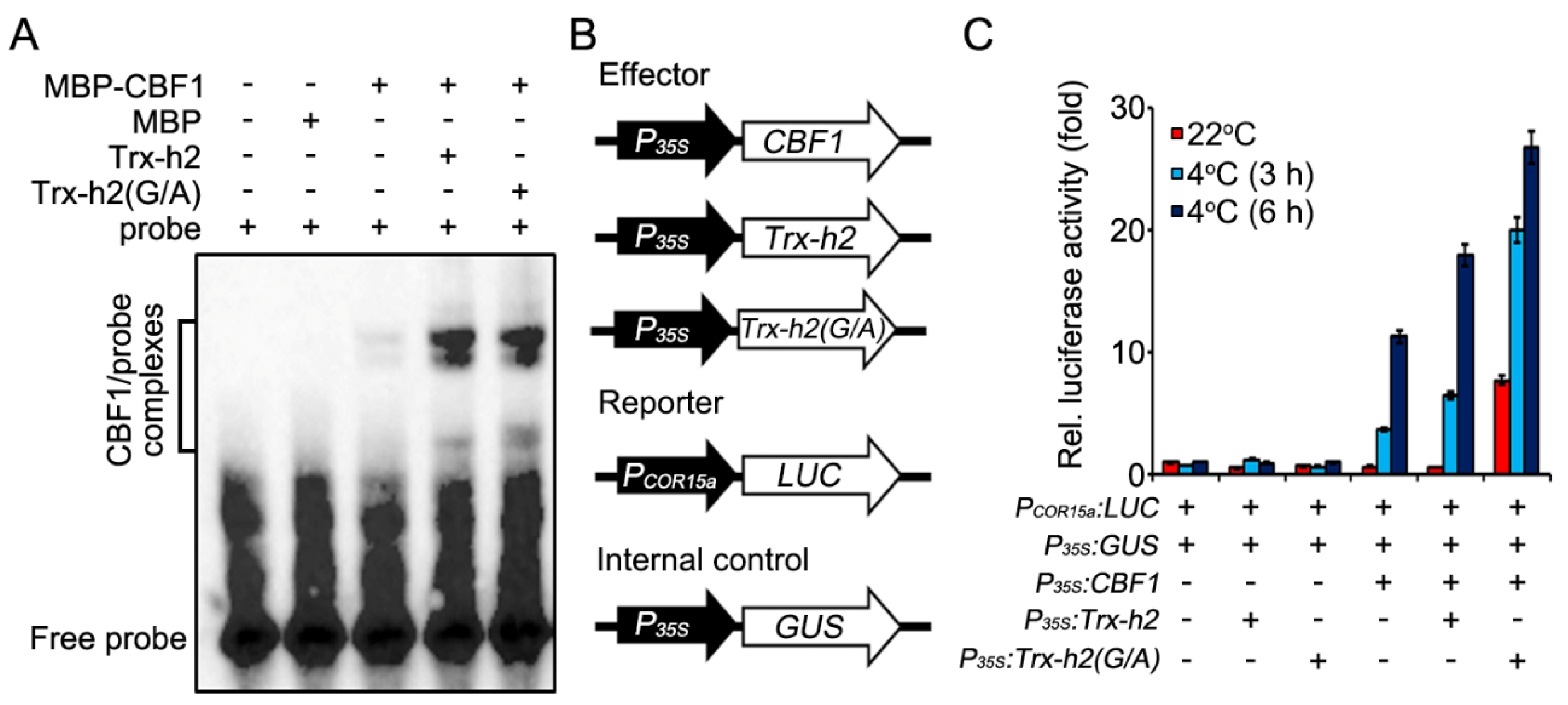
2.9. Electrophoretic Mobility Shift Assay (EMSA)
2.10. CBF1 Transactivation Assay
2.11. RNA Isolation and Quantitative Real-time PCR (qRT-PCR)
2.12. Freezing Tolerance Assay
2.13. Statistical Analysis
3. Results
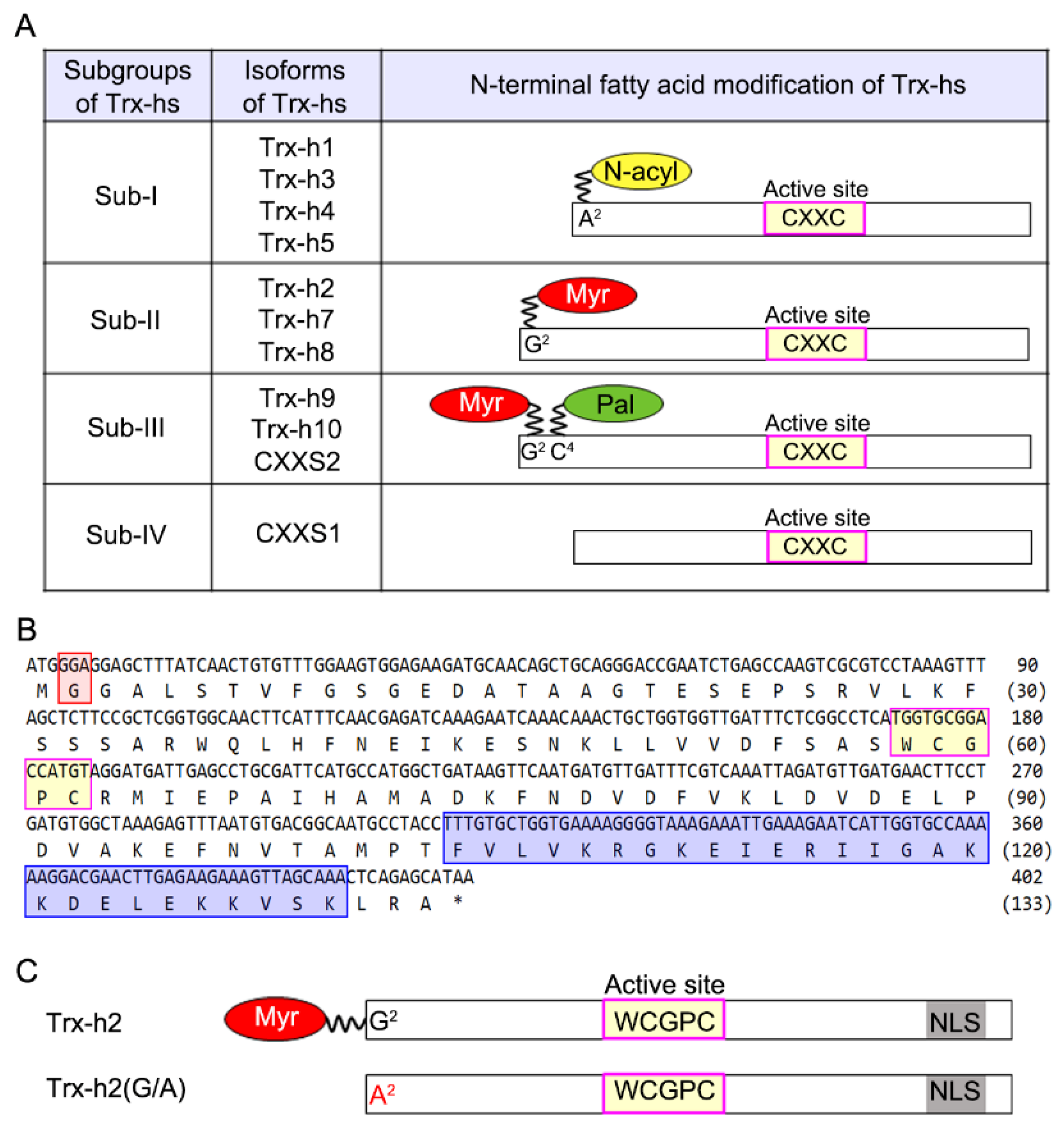
3.1. Myristoylation of Arabidopsis Trx-h2 Is Dependent on the Second Amino Acid of Trx-h2, Gly2
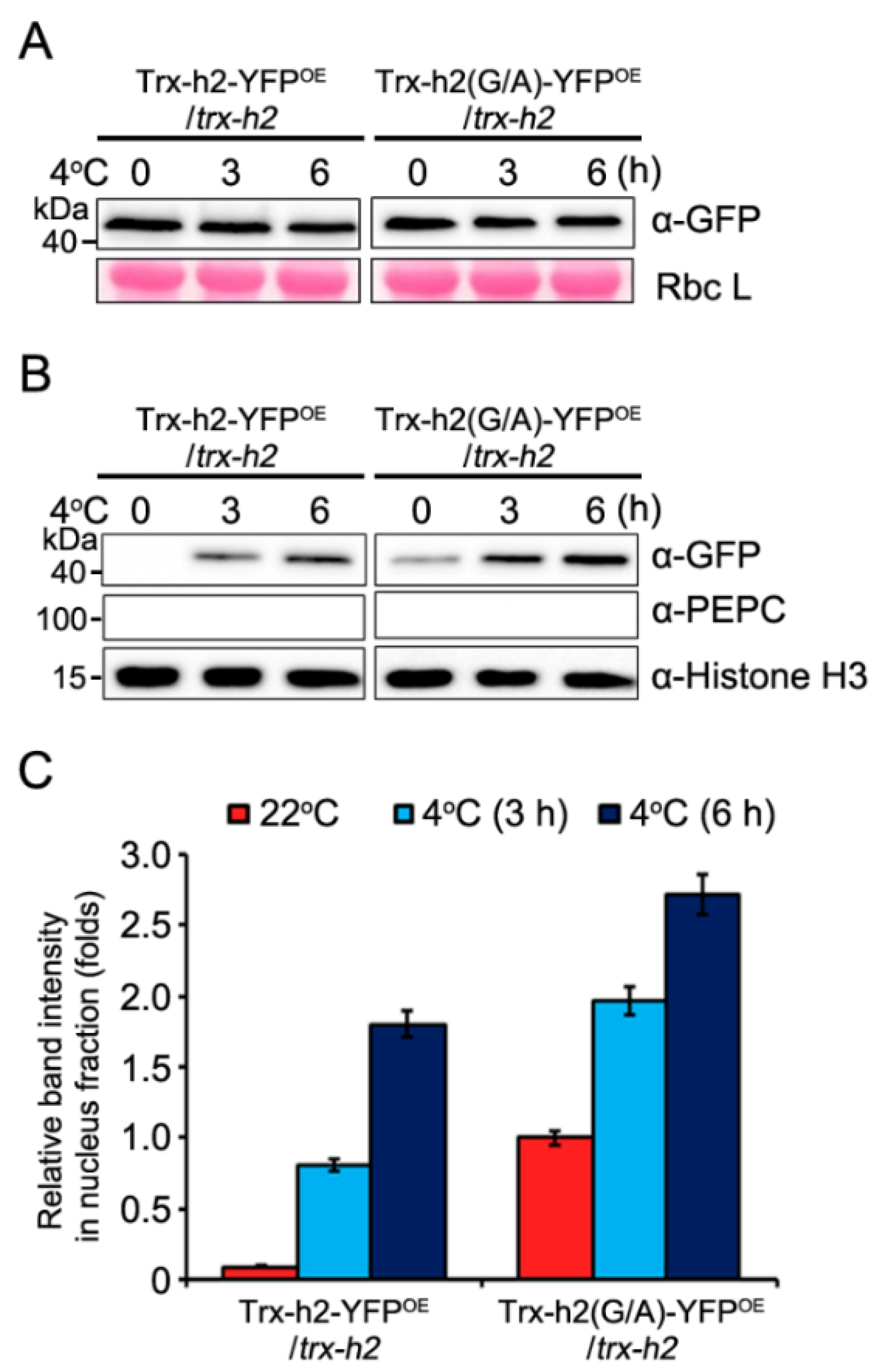
3.2. Kinetics of the Nuclear Translocation of Trx-h2 and Trx-h2(G/A) in Plants
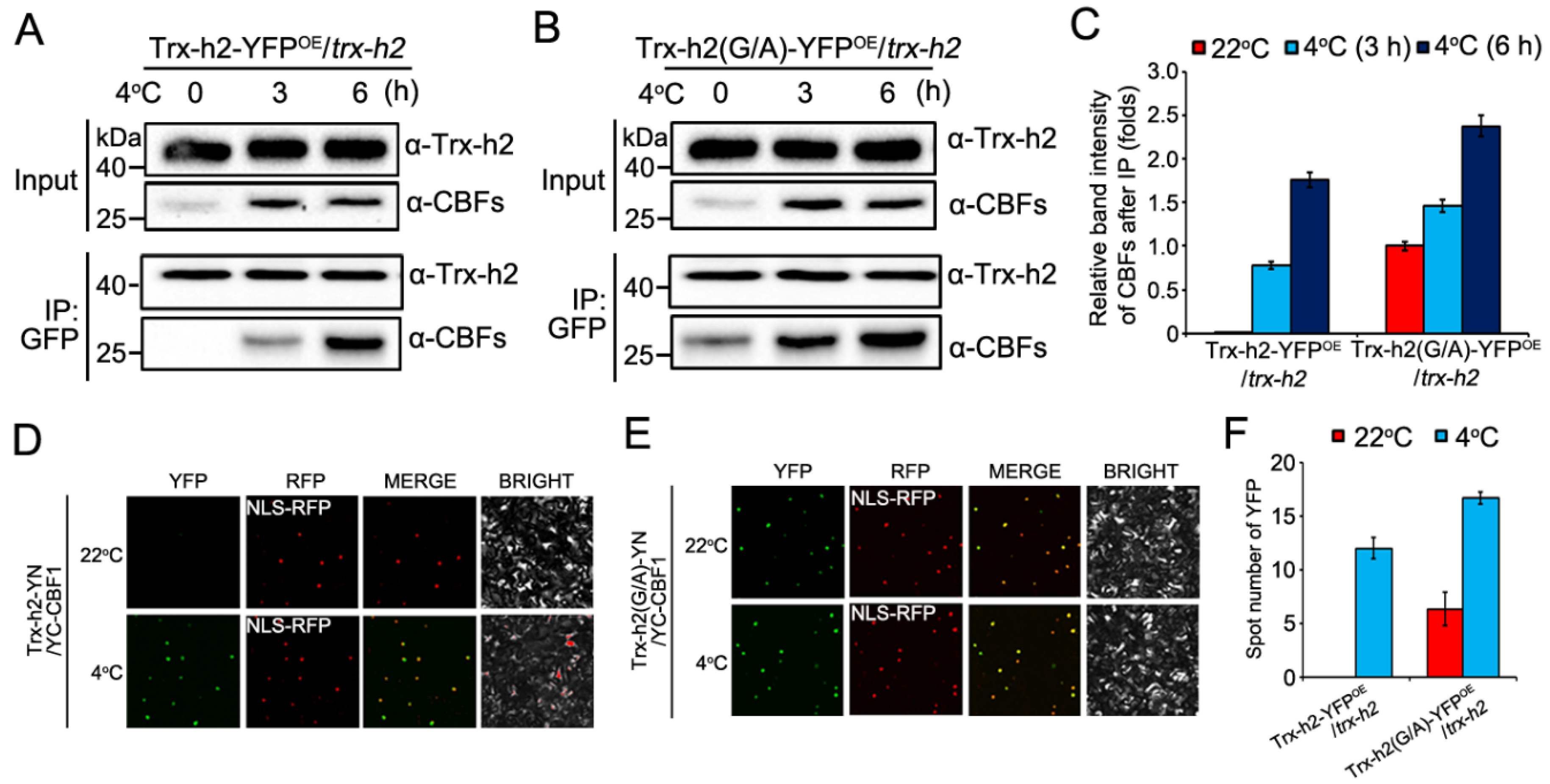
3.3. Effect of Myristoylation on the Interaction of Trx-h2 with CBFs
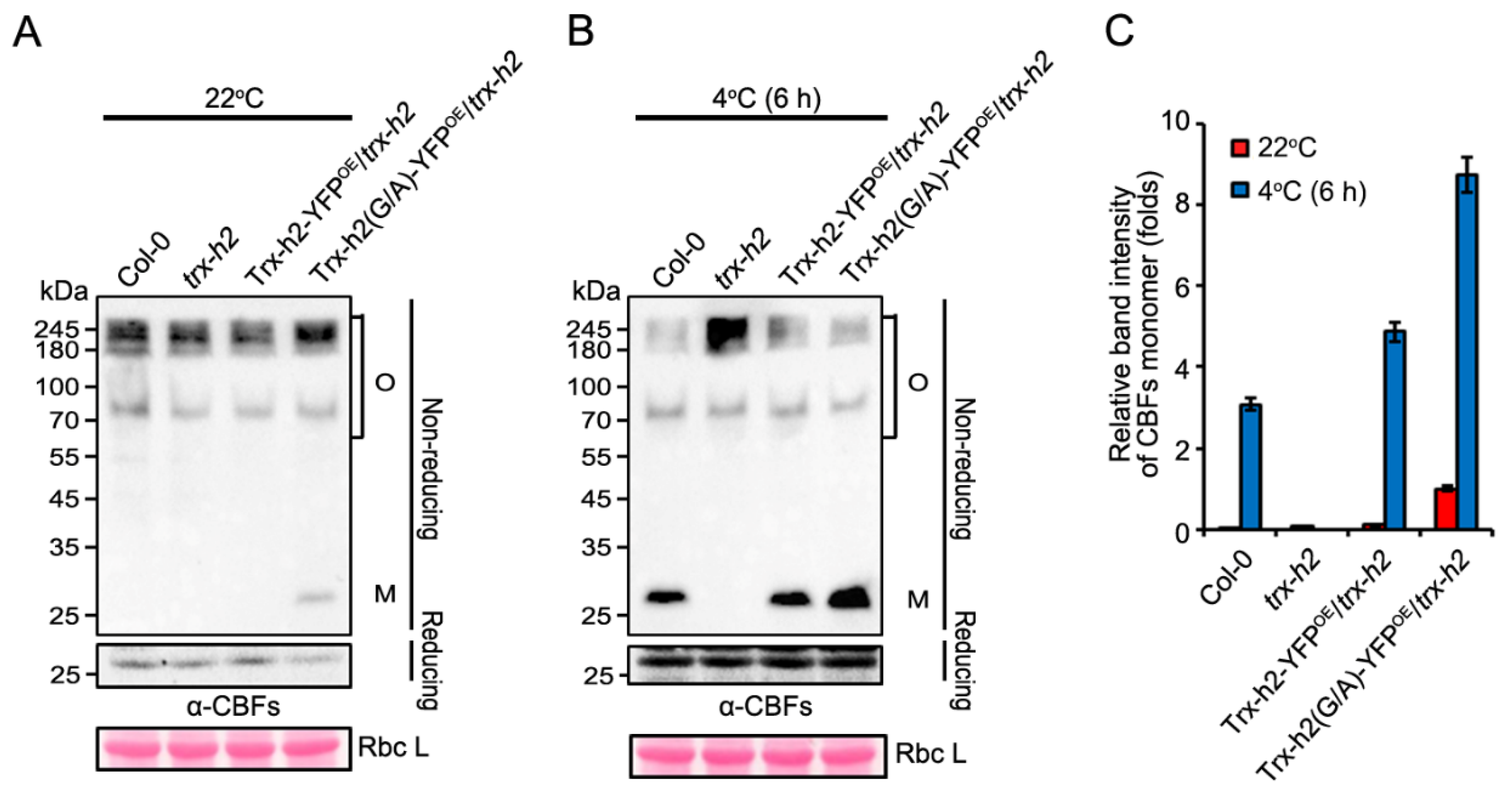
3.4. Trx-h2(G/A) Reduces and Structurally Alters CBF1 at Warm and Cold Temperatures
3.5. Comparison of Trx-h2 and Trx-h2(G/A) for the CBF1 Activation under the Warm and Cold Temperatures
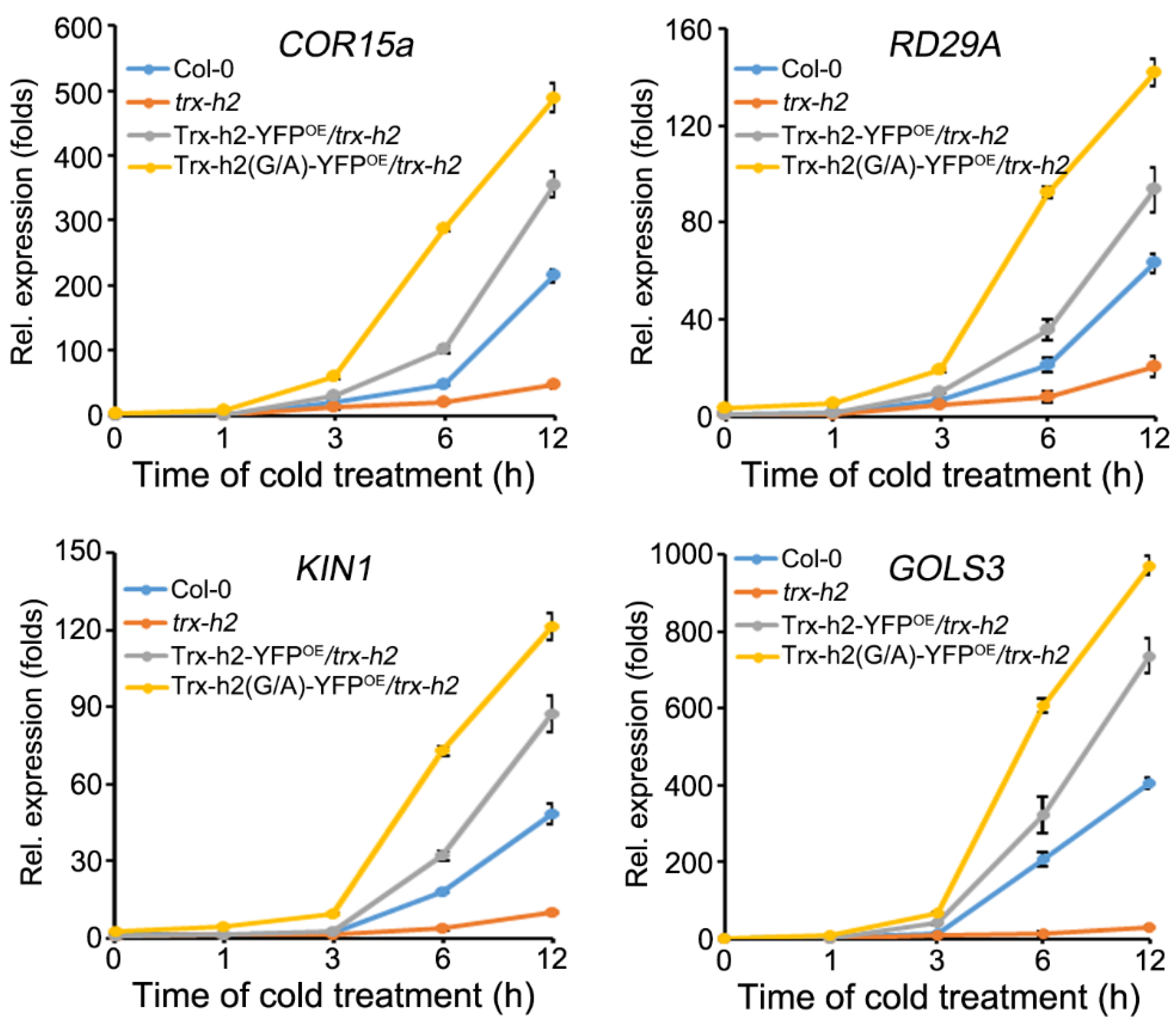
3.6. Comparison of the Ability of Trx-h2 and Trx-h2(G/A) to Activate CORs Expression under Cold Stress
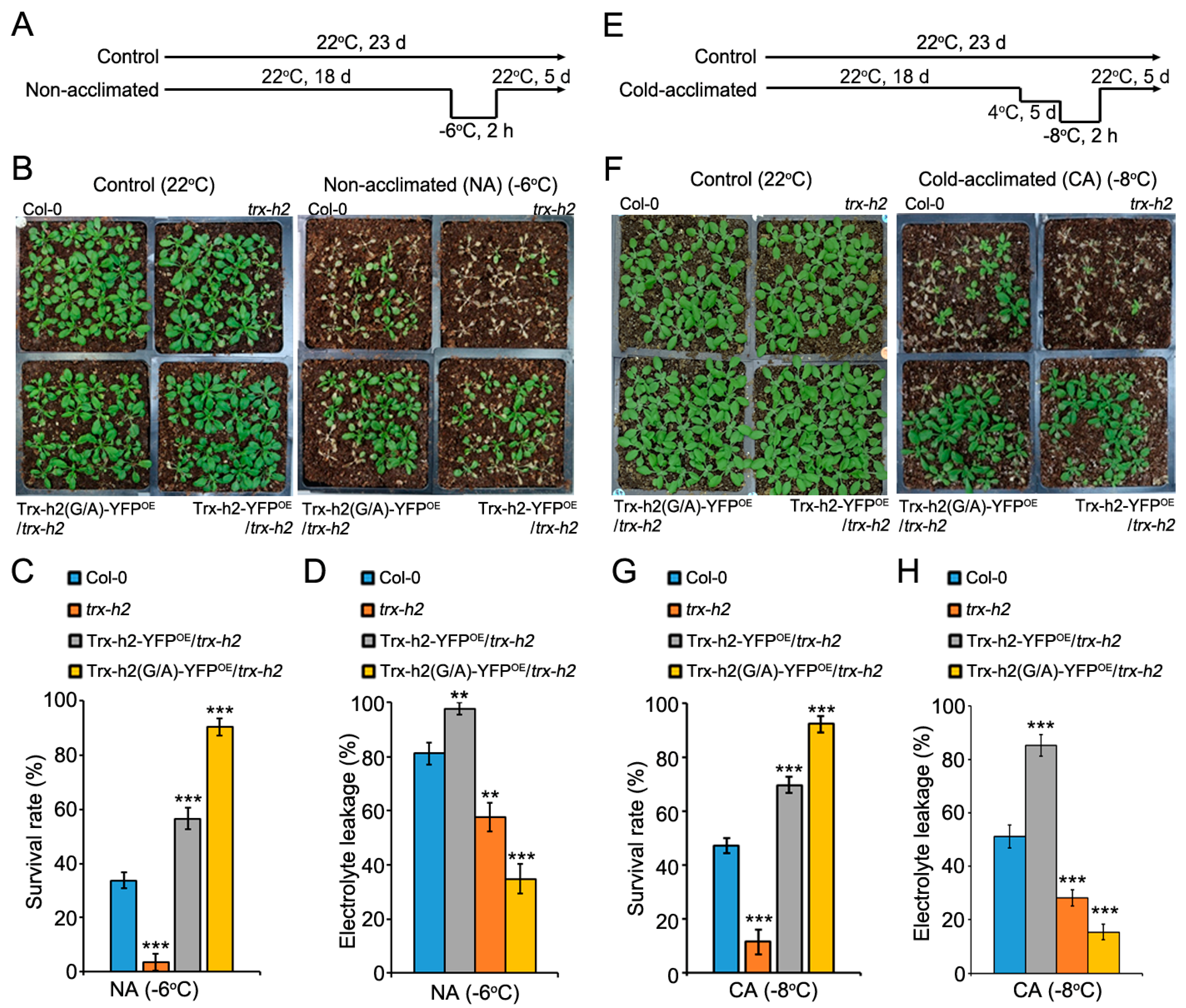
3.7. Comparison of the Freezing Tolerance of Plants Expressing Trx-h2 and Trx-h2(G/A)
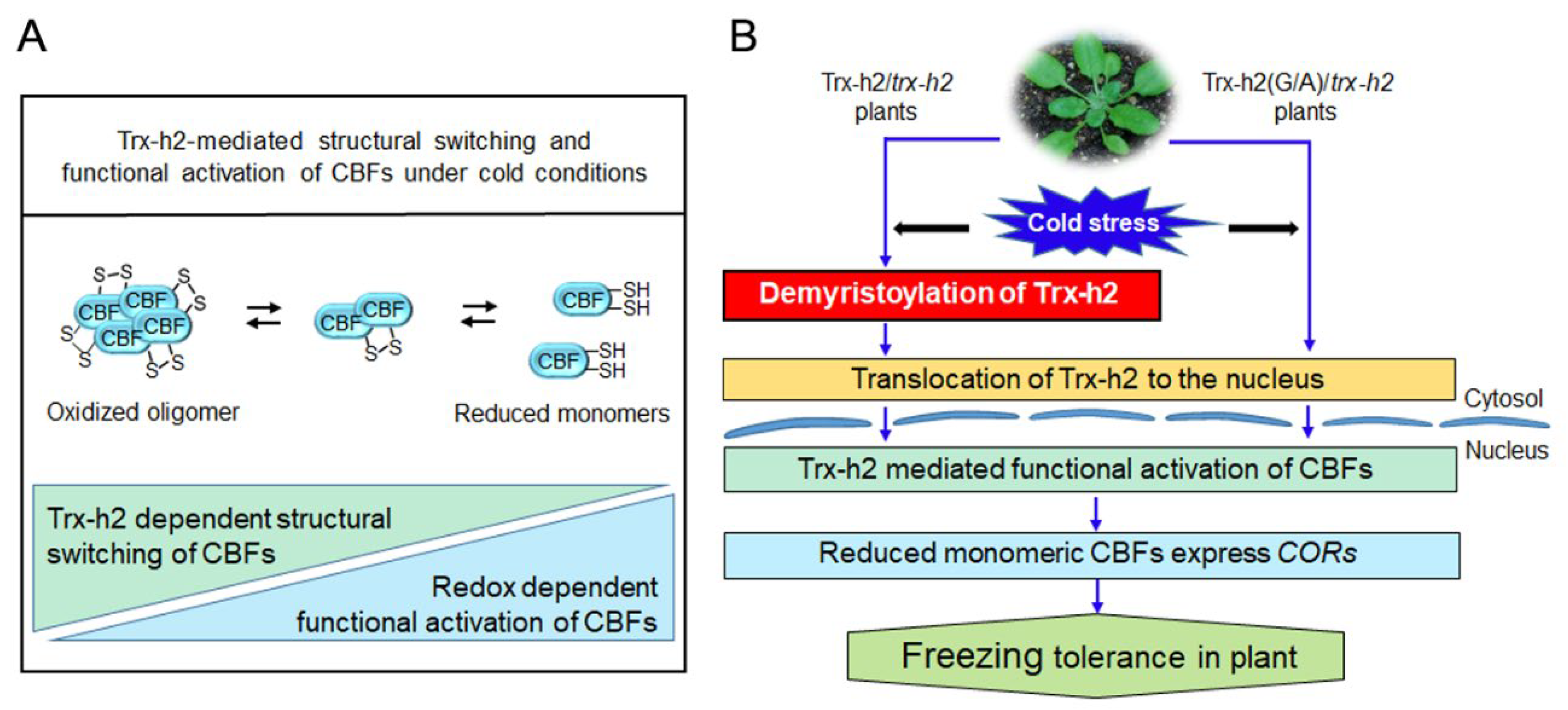
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006, 11, 15–19. [Google Scholar] [CrossRef]
- Apel, K.; Hirt, H. Reactive Oxygen Species: Metabolism, Oxidative Stress, and Signal Transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.; Panda, P.; Sahoo, L.; Panda, S.K. Reactive oxygen species signaling in plants under abiotic stress. Plant Signal. Behav. 2013, 8, e23681. [Google Scholar] [CrossRef]
- Waszczak, C.; Carmody, M.; Kangasjärvi, J. Reactive Oxygen Species in Plant Signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [PubMed]
- Zandalinas, S.I.; Fichman, Y.; Devireddy, A.R.; Sengupta, S.; Azad, R.K.; Mittler, R. Systemic signaling during abiotic stress combination in plants. Proc. Natl. Acad. Sci. USA 2020, 117, 13810–13820. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox Signaling in Plants. Antioxid. Redox Signal. 2013, 18, 2087–2090. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Suzuki, N.; Miller, G.; Tognetti, V.B.; Vandepoele, K.; Gollery, M.; Shulaev, V.; Van Breusegem, F. ROS signaling: The new wave? Trends Plant Sci. 2011, 16, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Ullah, F.; Zhou, D.-X.; Yi, M.; Zhao, Y. Mechanisms of ROS Regulation of Plant Development and Stress Responses. Front. Plant Sci. 2019, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Castro, B.; Citterico, M.; Kimura, S.; Stevens, D.M.; Wrzaczek, M.; Coaker, G. Stress-Induced Reactive Oxygen Species Compartmentalization, Perception and Signalling. Nat. Plants 2021, 7, 403–412. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Lee, E.S.; Kang, C.H.; Park, J.H.; Lee, S.Y.; Lee, M.E.S.; Park, M.J.H. Physiological Significance of Plant Peroxiredoxins and the Structure-Related and Multifunctional Biochemistry of Peroxiredoxin 1. Antioxid. Redox Signal. 2018, 28, 625–639. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.H.; Lee, K.O.; Chi, Y.H.; Jung, B.G.; Park, S.K.; Park, J.H.; Lee, J.R.; Lee, S.S.; Moon, J.C.; Yun, J.W.; et al. Two Enzymes in One: Two Yeast Peroxiredoxins Display Oxidative Stress-Dependent Switching from a Peroxidase to a Molecular Chaperone Function. Cell 2004, 117, 625–635. [Google Scholar] [CrossRef]
- Chae, H.B.; Kim, M.G.; Kang, C.H.; Park, J.H.; Lee, E.S.; Lee, S.-U.; Chi, Y.H.; Paeng, S.K.; Bin Bae, S.; Wi, S.D.; et al. Redox sensor QSOX1 regulates plant immunity by targeting GSNOR to modulate ROS generation. Mol. Plant 2021. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Ren, H.; Liu, R.; Li, B.; Wu, T.; Sun, F.; Liu, H.; Wang, X.; Dong, H. An h-Type Thioredoxin Functions in Tobacco Defense Responses to Two Species of Viruses and an Abiotic Oxidative Stress. Mol. Plant Microbe Interact. 2010, 23, 1470–1485. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-C.; Ren, J.-P.; Cho, M.-J.; Zhou, S.-M.; Kim, Y.-B.; Guo, H.-X.; Wong, J.H.; Niu, H.-B.; Kim, H.-K.; Morigasaki, S.; et al. The Level of Expression of Thioredoxin is Linked to Fundamental Properties and Applications of Wheat Seeds. Mol. Plant 2009, 2, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Wong, J.H.; Feldman, L.J.; Lemaux, P.G.; Buchanan, B.B. A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc. Natl. Acad. Sci. USA 2010, 107, 3900–3905. [Google Scholar] [CrossRef]
- Park, S.K.; Jung, Y.J.; Lee, J.R.; Lee, Y.M.; Jang, H.H.; Lee, S.S.; Park, J.H.; Kim, S.Y.; Moon, J.C.; Lee, S.Y.; et al. Heat-Shock and Redox-Dependent Functional Switching of an h-Type Arabidopsis Thioredoxin from a Disulfide Reductase to a Molecular Chaperone. Plant Physiol. 2009, 150, 552–561. [Google Scholar] [CrossRef]
- Tada, Y.; Spoel, S.; Pajerowska-Mukhtar, K.; Mou, Z.; Song, J.; Wang, C.; Zuo, J.; Dong, X. Plant Immunity Requires Conformational Charges of NPR1 via S-Nitrosylation and Thioredoxins. Science 2008, 321, 952–956. [Google Scholar] [CrossRef]
- Hemsley, P.A. The importance of lipid modified proteins in plants. N. Phytol. 2014, 205, 476–489. [Google Scholar] [CrossRef]
- Friso, G.; Van Wijk, K.J. Update: Post-Translational Protein Modifications in Plant Metabolism. Plant Physiol. 2015, 169, 1469–1487. [Google Scholar] [CrossRef]
- He, Z.; Huang, T.; Ao, K.; Yan, X.; Huang, Y. Sumoylation, Phosphorylation, and Acetylation Fine-Tune the Turnover of Plant Immunity Components Mediated by Ubiquitination. Front. Plant Sci. 2017, 8, 1682. [Google Scholar] [CrossRef]
- Park, J.H.; Kang, C.H.; Nawkar, G.M.; Lee, E.S.; Paeng, S.K.; Chae, H.B.; Chi, Y.H.; Kim, W.Y.; Yun, D.-J.; Lee, S.Y. EMR, a cytosolic-abundant ring finger E3 ligase, mediates ER-associated protein degradation in Arabidopsis. N. Phytol. 2018, 220, 163–177. [Google Scholar] [CrossRef]
- Karve, T.M.; Cheema, A.K. Small Changes Huge Impact: The Role of Protein Posttranslational Modifications in Cellular Homeostasis and Disease. J. Amino Acids 2011, 2011, 1–13. [Google Scholar] [CrossRef]
- Kim, M.-Y.; Bae, J.-S.; Kim, T.-H.; Park, J.-M.; Ahn, Y.H. Role of Transcription Factor Modifications in the Pathogenesis of Insulin Resistance. Exp. Diabetes Res. 2011, 2012, 1–16. [Google Scholar] [CrossRef]
- Martin, D.D.; Beauchamp, E.; Berthiaume, L.G. Post-Translational Myristoylation: Fat Matters in Cellular Life and Death. Biochimie 2011, 93, 18–31. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, X.; Chen, X.; Aramsangtienchai, P.; Tong, Z.; Lin, H. Protein Lipidation: Occurrence, Mechanisms, Biological Functions, and Enabling Technologies. Chem. Rev. 2017, 118, 919–988. [Google Scholar] [CrossRef]
- Lee, E.S.; Park, J.H.; Wi, S.D.; Kang, C.H.; Chi, Y.H.; Chae, H.B.; Paeng, S.K.; Ji, M.G.; Kim, W.-Y.; Kim, M.G.; et al. Redox-dependent structural switch and CBF activation confer freezing tolerance in plants. Nat. Plants 2021, 7, 914–922. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A. Floral dip: A simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Nawkar, G.M.; Kang, C.H.; Maibam, P.; Park, J.H.; Jung, Y.J.; Chae, H.B.; Chi, Y.H.; Jung, I.J.; Kim, W.Y.; Yun, D.-J.; et al. HY5, a positive regulator of light signaling, negatively controls the unfolded protein response inArabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 2084–2089. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Xia, C.; Gao, L.; Hao, C.; Zhao, G.; Jia, J.; Kong, X. A Novel Wheat C-bZIP Gene, TabZIP14-B, Participates in Salt and Freezing Tolerance in Transgenic Plants. Front. Plant Sci. 2017, 8, 710. [Google Scholar] [CrossRef]
- Kang, C.H.; Lee, Y.M.; Park, J.H.; Nawkar, G.M.; Oh, H.T.; Kim, M.G.; Lee, S.I.; Kim, W.Y.; Yun, D.; Lee, S.Y. Ribosomal P3 protein AtP3B ofArabidopsisacts as both protein and RNA chaperone to increase tolerance of heat and cold stresses. Plant Cell Environ. 2016, 39, 1631–1642. [Google Scholar] [CrossRef]
- Frottin, F.; Martinez, A.; Peynot, P.; Mitra, S.; Holz, R.C.; Giglione, C.; Meinnel, T. The Proteomics of N-terminal Methionine Cleavage. Mol. Cell. Proteom. 2006, 5, 2336–2349. [Google Scholar] [CrossRef]
- Martinez, A.; Traverso, J.A.; Valot, B.; Ferro, M.; Espagne, C.; Ephritikhine, G.; Zivy, M.; Giglione, C.; Meinnel, T. Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics 2008, 8, 2809–2831. [Google Scholar] [CrossRef]
- Traverso, J.A.; Micalella, C.; Martinez, A.; Brown, S.C.; Satiat-Jeunemaitre, B.; Meinnel, T.; Giglione, C. Roles of N-Terminal Fatty Acid Acylations in Membrane Compartment Partitioning: Arabidopsis h-Type Thioredoxins as a Case Study. Plant Cell 2013, 25, 1056–1077. [Google Scholar] [CrossRef]
- Hantschel, O.; Nagar, B.; Guettler, S.; Kretzschmar, J.; Dorey, K.; Kuriyan, J.; Superti-Furga, G. A Myristoyl/Phosphotyrosine Switch Regulates c-Abl. Cell 2003, 112, 845–857. [Google Scholar] [CrossRef]
- Ding, Y.; Lv, J.; Shi, Y.; Gao, J.; Hua, J.; Song, C.; Gong, Z.; Yang, S. EGR 2 phosphatase regulates OST 1 kinase activity and freezing tolerance in Arabidopsis. EMBO J. 2018, 38, e99819. [Google Scholar] [CrossRef]
- Ishitani, M.; Liu, J.; Halfter, U.; Kim, C.-S.; Shi, W.; Zhu, J.-K. SOS3 Function in Plant Salt Tolerance Requires N-Myristoylation and Calcium Binding. Plant Cell 2000, 12, 1667–1677. [Google Scholar] [CrossRef]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Park, J.; Lim, C.J.; Shen, M.; Park, H.J.; Cha, J.-Y.; Iniesto, E.; Rubio, V.; Mengiste, T.; Zhu, J.-K.; Bressan, R.A.; et al. Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proc. Natl. Acad. Sci. USA 2018, 115, E5400–E5409. [Google Scholar] [CrossRef]
- Belda-Palazon, B.; Julian, J.; Coego, A.; Wu, Q.; Zhang, X.; Batistic, O.; AlQuraishi, S.A.; Kudla, J.; An, C.; Rodriguez, P.L. ABA inhibits myristoylation and induces shuttling of the RGLG 1 E3 ligase to promote nuclear degradation of PP 2 CA. Plant J. 2019, 98, 813–825. [Google Scholar] [CrossRef]
- Resh, M.D. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2006, 2, 584–590. [Google Scholar] [CrossRef]
- Burnaevskiy, N.; Fox, T.G.; Plymire, D.A.; Ertelt, J.M.; Weigele, B.A.; Selyunin, A.S.; Way, S.S.; Patrie, S.M.; Alto, N.M. Proteolytic Elimination of N-Myristoyl Modifications by the Shigella Virulence Factor IpaJ. Nat. Cell Biol. 2013, 496, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Burnaevskiy, N.; Peng, T.; Reddick, L.E.; Hang, H.C.; Alto, N.M. Myristoylome Profiling Reveals a Concerted Mechanism of ARF GTPase Deacylation by the Bacterial Protease IpaJ. Mol. Cell 2015, 58, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Hashiguchi, A.; Komatsu, S. Impact of Post-Translational Modifications of Crop Proteins under Abiotic Stress. Proteomes 2016, 4, 42. [Google Scholar] [CrossRef]
- Su, M.-G.; Weng, J.T.-Y.; Hsu, J.B.-K.; Huang, K.-Y.; Chi, Y.-H.; Lee, T.-Y. Investigation and identification of functional post-translational modification sites associated with drug binding and protein-protein interactions. BMC Syst. Biol. 2017, 11, 69–80. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, E.S.; Park, J.H.; Wi, S.D.; Chae, H.B.; Paeng, S.K.; Bae, S.B.; Phan, K.A.T.; Kim, M.G.; Kwak, S.-S.; Kim, W.-Y.; et al. Demyristoylation of the Cytoplasmic Redox Protein Trx-h2 Is Critical for Inducing a Rapid Cold Stress Response in Plants. Antioxidants 2021, 10, 1287. https://doi.org/10.3390/antiox10081287
Lee ES, Park JH, Wi SD, Chae HB, Paeng SK, Bae SB, Phan KAT, Kim MG, Kwak S-S, Kim W-Y, et al. Demyristoylation of the Cytoplasmic Redox Protein Trx-h2 Is Critical for Inducing a Rapid Cold Stress Response in Plants. Antioxidants. 2021; 10(8):1287. https://doi.org/10.3390/antiox10081287
Chicago/Turabian StyleLee, Eun Seon, Joung Hun Park, Seong Dong Wi, Ho Byoung Chae, Seol Ki Paeng, Su Bin Bae, Kieu Anh Thi Phan, Min Gab Kim, Sang-Soo Kwak, Woe-Yeon Kim, and et al. 2021. "Demyristoylation of the Cytoplasmic Redox Protein Trx-h2 Is Critical for Inducing a Rapid Cold Stress Response in Plants" Antioxidants 10, no. 8: 1287. https://doi.org/10.3390/antiox10081287
APA StyleLee, E. S., Park, J. H., Wi, S. D., Chae, H. B., Paeng, S. K., Bae, S. B., Phan, K. A. T., Kim, M. G., Kwak, S.-S., Kim, W.-Y., Yun, D.-J., & Lee, S. Y. (2021). Demyristoylation of the Cytoplasmic Redox Protein Trx-h2 Is Critical for Inducing a Rapid Cold Stress Response in Plants. Antioxidants, 10(8), 1287. https://doi.org/10.3390/antiox10081287







