Mechanisms of Ataxia Telangiectasia Mutated (ATM) Control in the DNA Damage Response to Oxidative Stress, Epigenetic Regulation, and Persistent Innate Immune Suppression Following Sepsis
Abstract
1. Introduction
2. Physiological Regulation and Response to RONS
2.1. Antioxidant Response to Elevated RONS
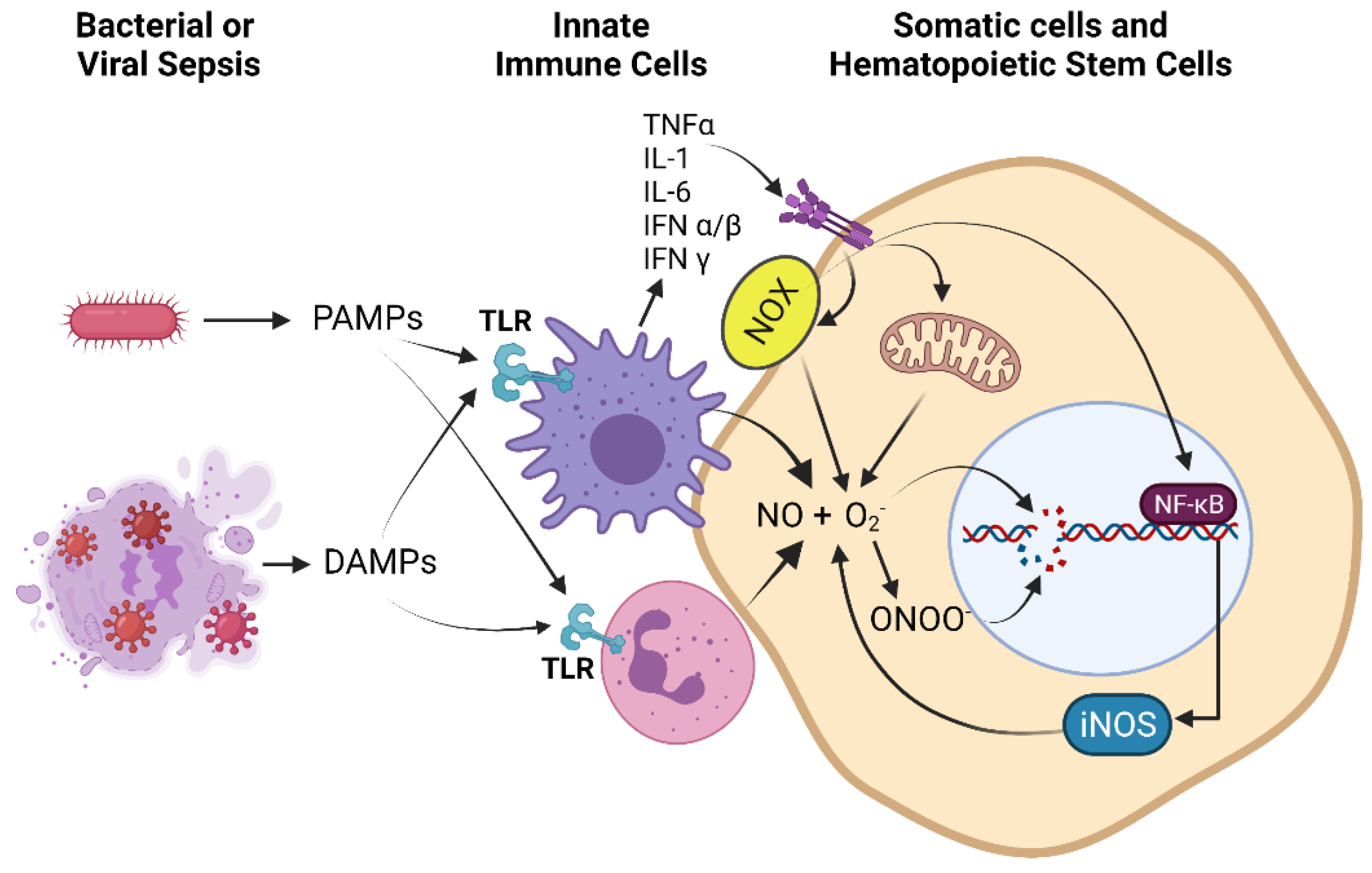
2.2. Sources of ROS and RNS in Pathophysiological States
2.3. Activation of the DNA Damage Response (DDR) by RONS
2.4. Involvement of the DDR in Epigenetic Modifications
2.5. RONS can Directly Participate in Chemical Modification of Chromatin
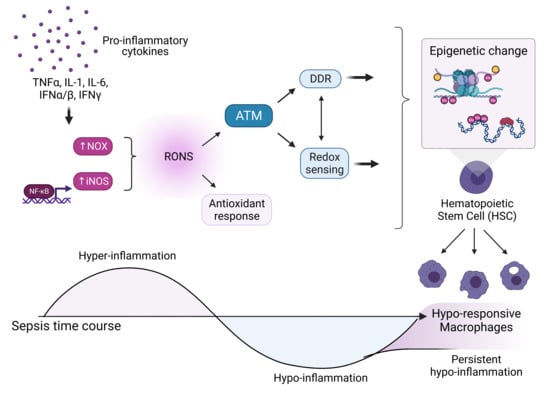
3. Pathological States Associated with High Oxidants Result in Epigenetic Changes to Stem Cells
3.1. Sepsis as a Model for Elevated Oxidative Stress and Epigenetic Modifications
3.2. Evidence for Epigenetic Changes to Innate Immune Cells
3.3. Evidence for Epigenetic Changes to HSCs from Sepsis
3.4. Evidence of DNA Damage from Sepsis
4. The Upregulation of the DDR and the Epigenetic Modifications to Stem Cells May Be Causally Linked
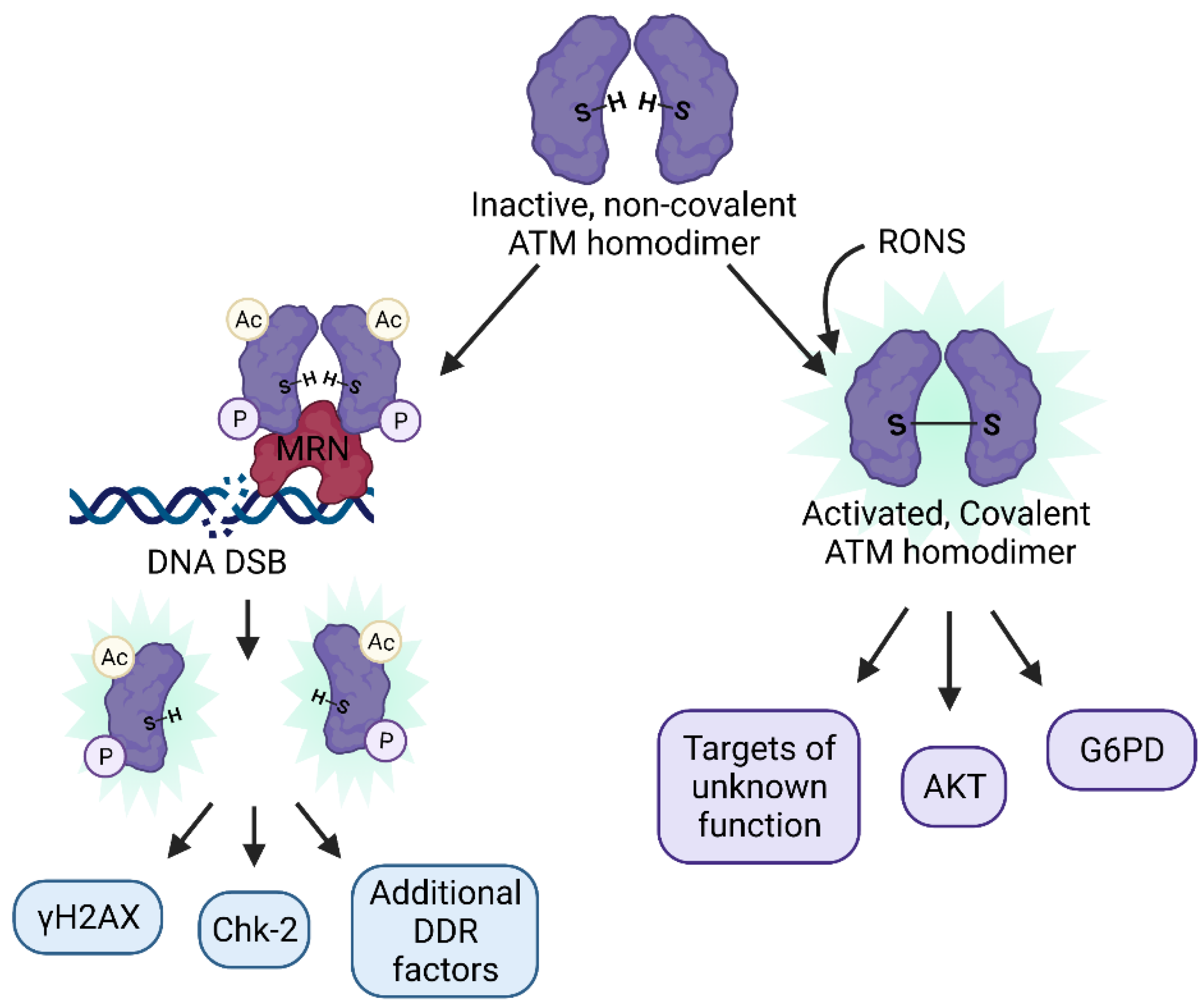
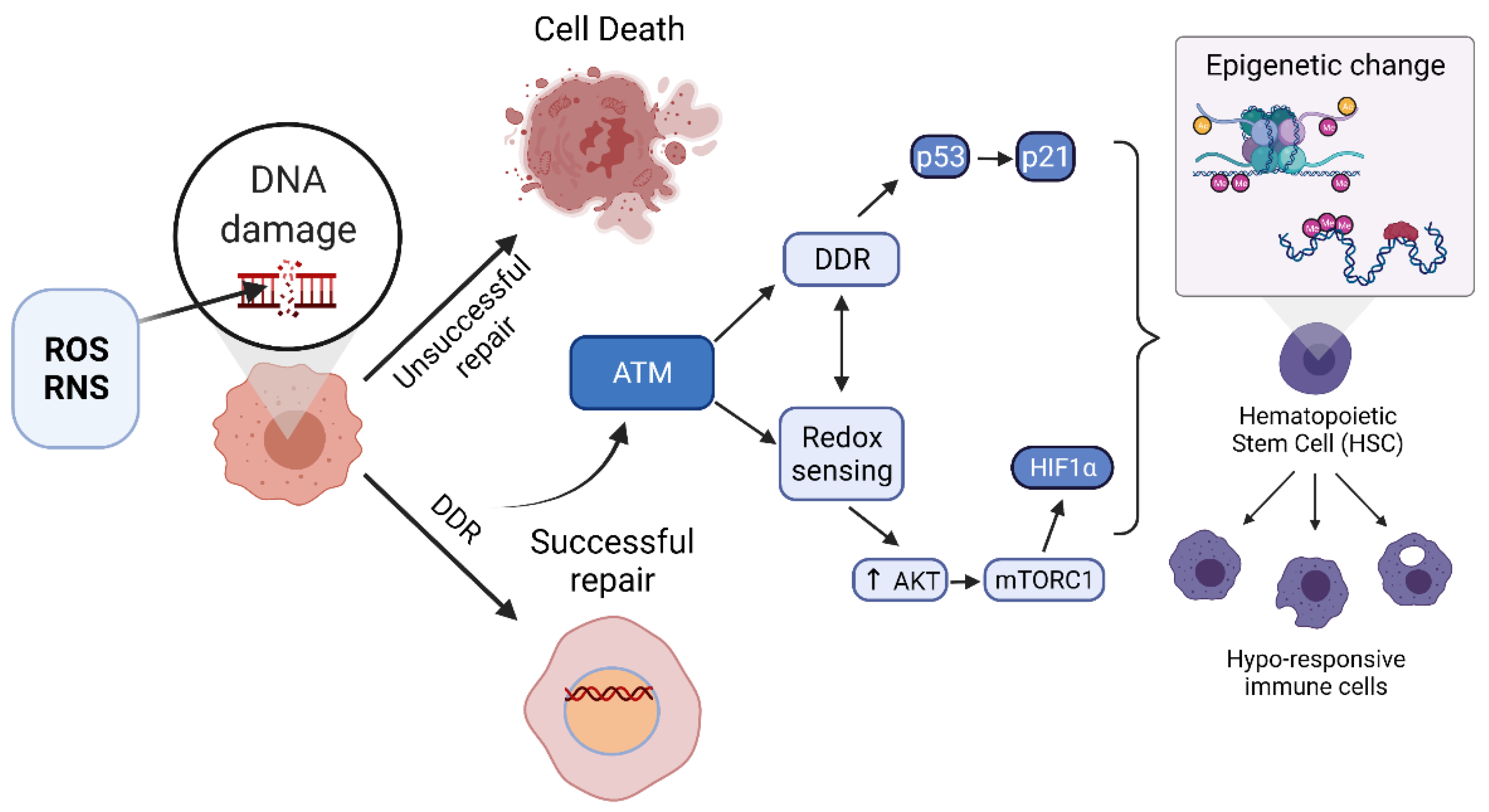
4.1. Dual Roles of ATM as a Master Regulator of the DDR and a Redox Sensor
4.2. ATM as a Regulator of Epigenetic Change
4.3. NFκB as an Epigenetic Modifier
4.4. Epigenetic Regulation of iNOS Gene
4.5. ATM Regulation of Hematopoietic Stem Cells (HSCs)
4.6. ATM as a Coordinator of Epigenetic Change during Sepsis
5. Involvement of Additional DDR Factors in Coordinating Epigenetic Changes
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Buetler, T.M.; Krauskopf, A.; Ruegg, U.T. Role of Superoxide as a Signaling Molecule. Physiology 2004, 19, 120–123. [Google Scholar] [CrossRef] [PubMed]
- D’Autréaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef]
- Moldogazieva, N.T.; Mokhosoev, I.M.; Feldman, N.B.; Lutsenko, S.V. ROS and RNS signalling: Adaptive redox switches through oxidative/nitrosative protein modifications. Free Radic. Res. 2018, 52, 507–543. [Google Scholar] [CrossRef]
- Behl, C. Oxidative stress in Alzheimer’s disease: Implications for prevention and therapy. Subcell. Biochem. 2005, 38, 65–78. [Google Scholar] [PubMed]
- Browne, S.E.; Beal, M.F. Oxidative damage in Huntington’s disease pathogenesis. Antioxid. Redox Signal. 2006, 8, 2061–2073. [Google Scholar] [CrossRef] [PubMed]
- Sugamura, K.; Keaney, J.F., Jr. Reactive oxygen species in cardiovascular disease. Free Radic. Biol. Med. 2011, 51, 978–992. [Google Scholar] [CrossRef]
- Filippin, L.I.; Vercelino, R.; Marroni, N.P.; Xavier, R.M. Redox signalling and the inflammatory response in rheumatoid arthritis. Clin. Exp. Immunol. 2008, 152, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Duong, T.Q. Methylene blue treatment in experimental ischemic stroke: A mini review. Brain Circ. 2016, 2, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Liguori, I.; Russo, G.; Curcio, F.; Bulli, G.; Aran, L.; Della-Morte, D.; Gargiulo, G.; Testa, G.; Cacciatore, F.; Bonaduce, D.; et al. Oxidative stress, aging, and diseases. Clin. Interv. Aging 2018, 13, 757–772. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef] [PubMed]
- Prauchner, C.A. Oxidative stress in sepsis: Pathophysiological implications justifying antioxidant co-therapy. Burns 2017, 43, 471–485. [Google Scholar] [CrossRef]
- Maya-Mendoza, A.; Ostrakova, J.; Kosar, M.; Hall, A.; Duskova, P.; Mistrik, M.; Merchut-Maya, J.M.; Hodny, Z.; Bartkova, J.; Christensen, C.; et al. Myc and Ras oncogenes engage different energy metabolism programs and evoke distinct patterns of oxidative and DNA replication stress. Mol. Oncol. 2015, 9, 601–616. [Google Scholar] [CrossRef]
- Park, M.T.; Kim, M.J.; Suh, Y.; Kim, R.K.; Kim, H.; Lim, E.J.; Yoo, K.C.; Lee, G.H.; Kim, Y.H.; Hwang, S.G.; et al. Novel signaling axis for ROS generation during K-Ras-induced cellular transformation. Cell Death Differ. 2014, 21, 1185–1197. [Google Scholar] [CrossRef]
- Souliotis, V.L.; Vlachogiannis, N.I.; Pappa, M.; Argyriou, A.; Ntouros, P.A.; Sfikakis, P.P. DNA Damage Response and Oxidative Stress in Systemic Autoimmunity. Int. J. Mol. Sci. 2020, 21, 55. [Google Scholar] [CrossRef]
- Florentino, P.T.V.; Mendes, D.; Vitorino, F.N.L.; Martins, D.J.; Cunha, J.P.C.; Mortara, R.A.; Menck, C.F.M. DNA damage and oxidative stress in human cells infected by Trypanosoma cruzi. PLoS Pathog. 2021, 17, e1009502. [Google Scholar] [CrossRef]
- Geyik, S.; Altunısık, E.; Neyal, A.M.; Taysi, S. Oxidative stress and DNA damage in patients with migraine. J. Headache Pain 2016, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Bahar, I.; Elay, G.; Başkol, G.; Sungur, M.; Donmez-Altuntas, H. Increased DNA damage and increased apoptosis and necrosis in patients with severe sepsis and septic shock. J. Crit. Care 2018, 43, 271–275. [Google Scholar] [CrossRef]
- Lee, S.R.; Kwon, K.S.; Kim, S.R.; Rhee, S.G. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 1998, 273, 15366–15372. [Google Scholar] [CrossRef]
- Salmeen, A.; Andersen, J.N.; Myers, M.P.; Meng, T.C.; Hinks, J.A.; Tonks, N.K.; Barford, D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature 2003, 423, 769–773. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, M.; Yu, Z.X.; Ferrans, V.J.; Irani, K.; Finkel, T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science 1995, 270, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Bae, Y.S.; Kang, S.W.; Seo, M.S.; Baines, I.C.; Tekle, E.; Chock, P.B.; Rhee, S.G. Epidermal growth factor (EGF)-induced generation of hydrogen peroxide. Role in EGF receptor-mediated tyrosine phosphorylation. J. Biol. Chem. 1997, 272, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Finkel, T. Signal transduction by reactive oxygen species. J. Cell Biol. 2011, 194, 7–15. [Google Scholar] [CrossRef]
- Niethammer, P.; Grabher, C.; Look, A.T.; Mitchison, T.J. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 2009, 459, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Kepp, O.; Kroemer, G. Mitochondria: Master regulators of danger signalling. Nat. Rev. Mol. Cell Biol. 2012, 13, 780–788. [Google Scholar] [CrossRef]
- Tuteja, N.; Chandra, M.; Tuteja, R.; Misra, M.K. Nitric Oxide as a Unique Bioactive Signaling Messenger in Physiology and Pathophysiology. J. Biomed. Biotechnol. 2004, 2004, 227–237. [Google Scholar] [CrossRef]
- Deshmukh, P.; Unni, S.; Krishnappa, G.; Padmanabhan, B. The Keap1-Nrf2 pathway: Promising therapeutic target to counteract ROS-mediated damage in cancers and neurodegenerative diseases. Biophys. Rev. 2017, 9, 41–56. [Google Scholar] [CrossRef]
- Nguyen, T.; Nioi, P.; Pickett, C.B. The Nrf2-Antioxidant Response Element Signaling Pathway and Its Activation by Oxidative Stress. J. Biol. Chem. 2009, 284, 13291–13295. [Google Scholar] [CrossRef]
- Santos Sánchez, N.; Salas-Coronado, R.; Villanueva, C.; Hernandez-Carlos, B. Antioxidant Compounds and Their Antioxidant Mechanism. In Antioxidants; IntechOpen Limited: London, UK, 2019; pp. 1–28. [Google Scholar] [CrossRef]
- Brown, D.I.; Griendling, K.K. Nox proteins in signal transduction. Free Radic. Biol. Med. 2009, 47, 1239–1253. [Google Scholar] [CrossRef]
- Bedard, K.; Krause, K.-H. The NOX Family of ROS-Generating NADPH Oxidases: Physiology and Pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef]
- Van Houten, B.; Santa-Gonzalez, G.A.; Camargo, M. DNA repair after oxidative stress: Current challenges. Curr. Opin. Toxicol. 2018, 7, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.; Landi, R.; Pezone, A.; Morano, A.; Zuchegna, C.; Romano, A.; Muller, M.T.; Gottesman, M.E.; Porcellini, A.; Avvedimento, E.V. DNA damage and Repair Modify DNA methylation and Chromatin Domain of the Targeted Locus: Mechanism of allele methylation polymorphism. Sci. Rep. 2016, 6, 33222. [Google Scholar] [CrossRef]
- Ronco, C.; Martin, A.R.; Demange, L.; Benhida, R. ATM, ATR, CHK1, CHK2 and WEE1 inhibitors in cancer and cancer stem cells. Medchemcomm 2016, 8, 295–319. [Google Scholar] [CrossRef]
- Paull, T.T. Mechanisms of ATM Activation. Annu. Rev. Biochem. 2015, 84, 711–738. [Google Scholar] [CrossRef]
- Bakkenist, C.J.; Kastan, M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003, 421, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Paull, T.T. ATM Activation by DNA Double-Strand Breaks through the Mre11-Rad50-Nbs1 Complex. Science 2005, 308, 551–554. [Google Scholar] [CrossRef]
- Pellegrini, M.; Celeste, A.; Difilippantonio, S.; Guo, R.; Wang, W.; Feigenbaum, L.; Nussenzweig, A. Autophosphorylation at serine 1987 is dispensable for murine Atm activation in vivo. Nature 2006, 443, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Daniel, J.A.; Pellegrini, M.; Lee, J.-H.; Paull, T.T.; Feigenbaum, L.; Nussenzweig, A. Multiple autophosphorylation sites are dispensable for murine ATM activation in vivo. J. Cell Biol. 2008, 183, 777–783. [Google Scholar] [CrossRef]
- Dupré, A.; Boyer-Chatenet, L.; Gautier, J. Two-step activation of ATM by DNA and the Mre11–Rad50–Nbs1 complex. Nat. Struct. Mol. Biol. 2006, 13, 451–457. [Google Scholar] [CrossRef]
- Kozlov, S.V.; Graham, M.E.; Peng, C.; Chen, P.; Robinson, P.J.; Lavin, M.F. Involvement of novel autophosphorylation sites in ATM activation. EMBO J. 2006, 25, 3504–3514. [Google Scholar] [CrossRef] [PubMed]
- Weintz, G.; Olsen, J.V.; Frühauf, K.; Niedzielska, M.; Amit, I.; Jantsch, J.; Mages, J.; Frech, C.; Dölken, L.; Mann, M.; et al. The phosphoproteome of toll-like receptor-activated macrophages. Mol. Syst. Biol. 2010, 6, 371. [Google Scholar] [CrossRef]
- Neves-Costa, A.; Moita, L.F. Modulation of inflammation and disease tolerance by DNA damage response pathways. FEBS J. 2017, 284, 680–698. [Google Scholar] [CrossRef] [PubMed]
- Beyfuss, K.; Hood, D.A. A systematic review of p53 regulation of oxidative stress in skeletal muscle. Redox Rep. 2018, 23, 100–117. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Siddik, Z.H.; Huang, Z.; Wang, R.; Koomen, J.; Kobayashi, R.; Khokhar, A.R.; Kuang, J. Induction of p21 by p53 following DNA damage inhibits both Cdk4 and Cdk2 activities. Oncogene 2005, 24, 2929–2943. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, C.; Bernstein, H. The Role of DNA Repair and the Epigenetic Markers Left after Repair in Neurologic Functions, Including Memory and Learning. In DNA Repair-An Update; Mognato, M., Ed.; 2019; Available online: https://www.intechopen.com/chapters/65132 (accessed on 16 June 2021).
- Rossetto, D.; Truman, A.W.; Kron, S.J.; Côté, J. Epigenetic Modifications in Double-Strand Break DNA Damage Signaling and Repair. Clin. Cancer Res. 2010, 16, 4543–4552. [Google Scholar] [CrossRef]
- Kietzmann, T.; Petry, A.; Shvetsova, A.; Gerhold, J.M.; Görlach, A. The epigenetic landscape related to reactive oxygen species formation in the cardiovascular system. Br. J. Pharm. 2017, 174, 1533–1554. [Google Scholar] [CrossRef]
- Afanas’ev, I. New nucleophilic mechanisms of ros-dependent epigenetic modifications: Comparison of aging and cancer. Aging Dis. 2014, 5, 52–62. [Google Scholar] [CrossRef]
- Afanas’ev, I. Mechanisms of superoxide signaling in epigenetic processes: Relation to aging and cancer. Aging Dis. 2015, 6, 216–227. [Google Scholar] [CrossRef]
- Khan, M.A.; Alam, K.; Dixit, K.; Rizvi, M.M. Role of peroxynitrite induced structural changes on H2B histone by physicochemical method. Int. J. Biol. Macromol. 2016, 82, 31–38. [Google Scholar] [CrossRef]
- García-Giménez, J.L.; Òlaso, G.; Hake, S.B.; Bönisch, C.; Wiedemann, S.M.; Markovic, J.; Dasí, F.; Gimeno, A.; Pérez-Quilis, C.; Palacios, O.; et al. Histone h3 glutathionylation in proliferating mammalian cells destabilizes nucleosomal structure. Antioxid. Redox Signal. 2013, 19, 1305–1320. [Google Scholar] [CrossRef]
- Galligan, J.J.; Rose, K.L.; Beavers, W.N.; Hill, S.; Tallman, K.A.; Tansey, W.P.; Marnett, L.J. Stable histone adduction by 4-oxo-2-nonenal: A potential link between oxidative stress and epigenetics. J. Am. Chem Soc. 2014, 136, 11864–11866. [Google Scholar] [CrossRef]
- Xiong, F.; Xiao, D.; Zhang, L. Norepinephrine causes epigenetic repression of PKCε gene in rodent hearts by activating Nox1-dependent reactive oxygen species production. FASEB J. 2012, 26, 2753–2763. [Google Scholar] [CrossRef]
- Patterson, A.J.; Xiao, D.; Xiong, F.; Dixon, B.; Zhang, L. Hypoxia-derived oxidative stress mediates epigenetic repression of PKCε gene in foetal rat hearts. Cardiovasc. Res. 2012, 93, 302–310. [Google Scholar] [CrossRef]
- Nanduri, J.; Makarenko, V.; Reddy, V.D.; Yuan, G.; Pawar, A.; Wang, N.; Khan, S.A.; Zhang, X.; Kinsman, B.; Peng, Y.-J.; et al. Epigenetic regulation of hypoxic sensing disrupts cardiorespiratory homeostasis. Proc. Natl. Acad. Sci. USA 2012, 109, 2515–2520. [Google Scholar] [CrossRef] [PubMed]
- Coulter, J.B.; O’Driscoll, C.M.; Bressler, J.P. Hydroquinone increases 5-hydroxymethylcytosine formation through ten eleven translocation 1 (TET1) 5-methylcytosine dioxygenase. J. Biol. Chem. 2013, 288, 28792–28800. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Yang, Y.; Wang, X.; Chong, Z.; Yin, R.; Song, S.H.; Zhao, C.; Li, C.; Huang, H.; Sun, B.F.; et al. Redox-active quinones induces genome-wide DNA methylation changes by an iron-mediated and Tet-dependent mechanism. Nucleic Acids Res. 2014, 42, 1593–1605. [Google Scholar] [CrossRef]
- Miao, Z.; He, Y.; Xin, N.; Sun, M.; Chen, L.; Lin, L.; Li, J.; Kong, J.; Jin, P.; Xu, X. Altering 5-hydroxymethylcytosine modification impacts ischemic brain injury. Hum. Mol. Genet. 2015, 24, 5855–5866. [Google Scholar] [CrossRef]
- Mariani, C.J.; Vasanthakumar, A.; Madzo, J.; Yesilkanal, A.; Bhagat, T.; Yu, Y.; Bhattacharyya, S.; Wenger, R.H.; Cohn, S.L.; Nanduri, J.; et al. TET1-mediated hydroxymethylation facilitates hypoxic gene induction in neuroblastoma. Cell Rep. 2014, 7, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, H.M.; Wang, W.; Sen, S.; Destefano Shields, C.; Lee, S.S.; Zhang, Y.W.; Clements, E.G.; Cai, Y.; Van Neste, L.; Easwaran, H.; et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG Islands. Cancer Cell 2011, 20, 606–619. [Google Scholar] [CrossRef]
- Chervona, Y.; Costa, M. The control of histone methylation and gene expression by oxidative stress, hypoxia, and metals. Free Radic. Biol. Med. 2012, 53, 1041–1047. [Google Scholar] [CrossRef]
- Niu, Y.; DesMarais, T.L.; Tong, Z.; Yao, Y.; Costa, M. Oxidative stress alters global histone modification and DNA methylation. Free Radic. Biol. Med. 2015, 82, 22–28. [Google Scholar] [CrossRef]
- Huang, M.; Cai, S.; Su, J. The Pathogenesis of Sepsis and Potential Therapeutic Targets. Int. J. Mol. Sci. 2019, 20, 5376. [Google Scholar] [CrossRef]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Cross, D.; Drury, R.; Hill, J.; Pollard, A.J. Epigenetics in Sepsis: Understanding Its Role in Endothelial Dysfunction, Immunosuppression, and Potential Therapeutics. Front. Immunol. 2019, 10, 1363. [Google Scholar] [CrossRef]
- Boomer, J.S.; Green, J.M.; Hotchkiss, R.S. The changing immune system in sepsis: Is individualized immuno-modulatory therapy the answer? Virulence 2014, 5, 45–56. [Google Scholar] [CrossRef]
- Arts, R.J.W.; Gresnigt, M.S.; Joosten, L.A.B.; Netea, M.G. Cellular metabolism of myeloid cells in sepsis. J. Leukoc. Biol. 2017, 101, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.-C.; Scicluna, B.P.; Arts, R.J.W.; Gresnigt, M.S.; Lachmandas, E.; Giamarellos-Bourboulis, E.J.; Kox, M.; Manjeri, G.R.; Wagenaars, J.A.L.; Cremer, O.L.; et al. Broad defects in the energy metabolism of leukocytes underlie immunoparalysis in sepsis. Nat. Immunol. 2016, 17, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, M.D.; Moore, F.A. Persistent inflammatory, immunosuppressed, catabolic syndrome (PICS): A new phenotype of multiple organ failure. J. Adv. Nutr. Hum. Metab. 2015, 1, e784. [Google Scholar]
- Wang, X.; Buechler, N.L.; Woodruff, A.G.; Long, D.L.; Zabalawi, M.; Yoza, B.K.; McCall, C.E.; Vachharajani, V. Sirtuins and Immuno-Metabolism of Sepsis. Int. J. Mol. Sci. 2018, 19, 2738. [Google Scholar] [CrossRef]
- Guo, R.F.; Ward, P.A. Role of oxidants in lung injury during sepsis. Antioxid. Redox Signal. 2007, 9, 1991–2002. [Google Scholar] [CrossRef]
- Bomans, K.; Schenz, J.; Sztwiertnia, I.; Schaack, D.; Weigand, M.A.; Uhle, F. Sepsis Induces a Long-Lasting State of Trained Immunity in Bone Marrow Monocytes. Front. Immunol. 2018, 9, 2685. [Google Scholar] [CrossRef]
- Foster, S.L.; Hargreaves, D.C.; Medzhitov, R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature 2007, 447, 972–978. [Google Scholar] [CrossRef]
- Yao, Y.; Xu, X.-H.; Jin, L. Macrophage Polarization in Physiological and Pathological Pregnancy. Front. Immunol. 2019, 10, 792. [Google Scholar] [CrossRef]
- Butcher, S.K.; O’Carroll, C.E.; Wells, C.A.; Carmody, R.J. Toll-Like Receptors Drive Specific Patterns of Tolerance and Training on Restimulation of Macrophages. Front. Immunol. 2018, 9, 933. [Google Scholar] [CrossRef]
- Fernández-Morera, J.L.; Calvanese, V.; Rodríguez-Rodero, S.; Menéndez-Torre, E.; Fraga, M.F. Epigenetic regulation of the immune system in health and disease. Tissue Antigens 2010, 76, 431–439. [Google Scholar] [CrossRef]
- Busslinger, M.; Tarakhovsky, A. Epigenetic control of immunity. Cold Spring Harb. Perspect. Biol. 2014, 6, a019307. [Google Scholar] [CrossRef]
- Seeley, J.J.; Baker, R.G.; Mohamed, G.; Bruns, T.; Hayden, M.S.; Deshmukh, S.D.; Freedberg, D.E.; Ghosh, S. Induction of innate immune memory via microRNA targeting of chromatin remodelling factors. Nature 2018, 559, 114–119. [Google Scholar] [CrossRef]
- Beltrán-García, J.; Osca-Verdegal, R.; Romá-Mateo, C.; Carbonell, N.; Ferreres, J.; Rodríguez, M.; Mulet, S.; García-López, E.; Pallardó, F.V.; García-Giménez, J.L. Epigenetic biomarkers for human sepsis and septic shock: Insights from immunosuppression. Epigenomics 2020, 12, 617–646. [Google Scholar] [CrossRef]
- Davis, F.M.; Gallagher, K.A. Epigenetic Mechanisms in Monocytes/Macrophages Regulate Inflammation in Cardiometabolic and Vascular Disease. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 623–634. [Google Scholar] [CrossRef]
- Roy, D.G.; Chen, J.; Mamane, V.; Ma, E.H.; Muhire, B.M.; Sheldon, R.D.; Shorstova, T.; Koning, R.; Johnson, R.M.; Esaulova, E.; et al. Methionine Metabolism Shapes T Helper Cell Responses through Regulation of Epigenetic Reprogramming. Cell Metab. 2020, 31, 250–266.e9. [Google Scholar] [CrossRef]
- Weiterer, S.; Uhle, F.; Lichtenstern, C.; Siegler, B.H.; Bhuju, S.; Jarek, M.; Bartkuhn, M.; Weigand, M.A. Sepsis induces specific changes in histone modification patterns in human monocytes. PLoS ONE 2015, 10, e0121748. [Google Scholar] [CrossRef]
- Chan, G.C.; Fish, J.E.; Mawji, I.A.; Leung, D.D.; Rachlis, A.C.; Marsden, P.A. Epigenetic basis for the transcriptional hyporesponsiveness of the human inducible nitric oxide synthase gene in vascular endothelial cells. J. Immunol. 2005, 175, 3846–3861. [Google Scholar] [CrossRef]
- de Andrés, M.C.; Imagawa, K.; Hashimoto, K.; Gonzalez, A.; Roach, H.I.; Goldring, M.B.; Oreffo, R.O.C. Loss of methylation in CpG sites in the NF-κB enhancer elements of inducible nitric oxide synthase is responsible for gene induction in human articular chondrocytes. Arthritis Rheum. 2013, 65, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Skirecki, T.; Zielińska-Borkowska, U.; Kawiak, J.; Hoser, G. Sepsis modulates the human hematopoietic stem cell compartment in peripheral blood and bone marrow. Crit. Care 2014, 18, P33. [Google Scholar] [CrossRef]
- Wang, P.; Wang, J.; Li, Y.-H.; Wang, L.; Shang, H.-C.; Wang, J.-X. Phenotypical Changes of Hematopoietic Stem and Progenitor Cells in Sepsis Patients: Correlation with Immune Status? Front. Pharmacol. 2021, 11, 640203. [Google Scholar] [CrossRef]
- Skirecki, T.; Mikaszewska-Sokolewicz, M.; Godlewska, M.; Dołęgowska, B.; Czubak, J.; Hoser, G.; Kawiak, J.; Zielińska-Borkowska, U. Mobilization of Stem and Progenitor Cells in Septic Shock Patients. Sci. Rep. 2019, 9, 3289. [Google Scholar] [CrossRef]
- Pinheiro da Silva, F.; Machado, M.C.C. Septic Shock and the Aging Process: A Molecular Comparison. Front. Immunol. 2017, 8, 1389. [Google Scholar] [CrossRef]
- Zhang, H.; Rodriguez, S.; Wang, L.; Wang, S.; Serezani, H.; Kapur, R.; Cardoso, A.A.; Carlesso, N. Sepsis Induces Hematopoietic Stem Cell Exhaustion and Myelosuppression through Distinct Contributions of TRIF and MYD88. Stem Cell Rep. 2016, 6, 940–956. [Google Scholar] [CrossRef]
- Rodriguez, S.; Chora, A.; Goumnerov, B.; Mumaw, C.; Goebel, W.S.; Fernandez, L.; Baydoun, H.; HogenEsch, H.; Dombkowski, D.M.; Karlewicz, C.A.; et al. Dysfunctional expansion of hematopoietic stem cells and block of myeloid differentiation in lethal sepsis. Blood 2009, 114, 4064–4076. [Google Scholar] [CrossRef]
- Seyfried, A.N.; Maloney, J.M.; MacNamara, K.C. Macrophages Orchestrate Hematopoietic Programs and Regulate HSC Function during Inflammatory Stress. Front. Immunol. 2020, 11, 1499. [Google Scholar] [CrossRef]
- Nastasi, C.; Mannarino, L.; D’Incalci, M. DNA Damage Response and Immune Defense. Int. J. Mol. Sci. 2020, 21, 7504. [Google Scholar] [CrossRef]
- Morales, A.J.; Carrero, J.A.; Hung, P.J.; Tubbs, A.T.; Andrews, J.M.; Edelson, B.T.; Calderon, B.; Innes, C.L.; Paules, R.S.; Payton, J.E.; et al. A type I IFN-dependent DNA damage response regulates the genetic program and inflammasome activation in macrophages. eLife 2017, 6, e24655. [Google Scholar] [CrossRef]
- Kamsler, A. Increased oxidative stress in ataxia telangiectasia evidenced by alterations in redox state of brains from Atm-deficient mice. Cancer Res. 2001, 61, 1849–1854. [Google Scholar]
- Quick, K.L.; Dugan, L.L. Superoxide stress identifies neurons at risk in a model of ataxia-telangiectasia. Ann. Neurol. 2001, 49, 627–635. [Google Scholar] [CrossRef]
- Ito, K.; Hirao, A.; Arai, F.; Matsuoka, S.; Takubo, K.; Hamaguchi, I.; Nomiyama, K.; Hosokawa, K.; Sakurada, K.; Nakagata, N.; et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature 2004, 431, 997–1002. [Google Scholar] [CrossRef]
- Guo, Z.; Kozlov, S.; Lavin, M.F.; Person, M.D.; Paull, T.T. ATM Activation by Oxidative Stress. Science 2010, 330, 517–521. [Google Scholar] [CrossRef]
- Ditch, S.; Paull, T.T. The ATM protein kinase and cellular redox signaling: Beyond the DNA damage response. Trends Biochem. Sci. 2012, 37, 15–22. [Google Scholar] [CrossRef]
- Zhang, Y.; Lee, J.-H.; Paull, T.T.; Gehrke, S.; D’Alessandro, A.; Dou, Q.; Gladyshev, V.N.; Schroeder, E.A.; Steyl, S.K.; Christian, B.E.; et al. Mitochondrial redox sensing by the kinase ATM maintains cellular antioxidant capacity. Sci. Signal. 2018, 11, eaaq0702. [Google Scholar] [CrossRef]
- Chessa, L.; Petrinelli, P.; Antonelli, A.; Fiorlli, M.; Elli, R.; Marcucci, L.; Federico, A.; Gandini, E. Heterogeneity in ataxia-telangiectasia: Classical phenotype associated with intermediate cellular radiosensitivity. Am. J. Med. Genet. 1992, 42, 741–746. [Google Scholar] [CrossRef]
- Gilad, S. Genotype-Phenotype Relationships in Ataxia-Telangiectasia and Variants. Am. J. Hum. Genet. 1998, 62, 551–561. [Google Scholar] [CrossRef]
- Toyoshima, M. Ataxia-telangiectasia without immunodeficiency: Novel point mutations within and adjacent to the phosphatidylinositol 3-kinase-like domain. Am. J. Med. Genet. 1998, 75, 141–144. [Google Scholar] [CrossRef]
- Bencokova, Z.; Kaufmann, M.R.; Pires, I.M.; Lecane, P.S.; Giaccia, A.J.; Hammond, E.M. ATM Activation and Signaling under Hypoxic Conditions. Mol. Cell. Biol. 2009, 29, 526–537. [Google Scholar] [CrossRef]
- Valvezan, A.J.; Manning, B.D. Molecular logic of mTORC1 signalling as a metabolic rheostat. Nat. Metab. 2019, 1, 321–333. [Google Scholar] [CrossRef]
- Khalil, H.; Tummala, H.; Zhelev, N. ATM in focus: A damage sensor and cancer target. BioDiscovery 2012, 1, e8936. [Google Scholar] [CrossRef]
- Cam, H.; Easton, J.B.; High, A.; Houghton, P.J. mTORC1 Signaling under Hypoxic Conditions Is Controlled by ATM-Dependent Phosphorylation of HIF-1α. Mol. Cell 2010, 40, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Mimura, I.; Tanaka, T.; Nangaku, M. Novel therapeutic strategy with hypoxia-inducible factors via reversible epigenetic regulation mechanisms in progressive tubulointerstitial fibrosis. Semin Nephrol. 2013, 33, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Laribee, R.N. Transcriptional and Epigenetic Regulation by the Mechanistic Target of Rapamycin Complex 1 Pathway. J. Mol. Biol. 2018, 430, 4874–4890. [Google Scholar] [CrossRef]
- Rohde, J.R.; Cardenas, M.E. The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 2003, 23, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.K.; Bertram, P.G.; Ai, W.; Drenan, R.; Zheng, X.F.S. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 2003, 22, 6045–6056. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, L.; Lu, A.; Han, Y.; Colangelo, D.; Bukata, C.; Scibetta, A.; Yousefzadeh, M.J.; Li, X.; Gurkar, A.U.; et al. ATM is a key driver of NF-κB-dependent DNA-damage-induced senescence, stem cell dysfunction and aging. Aging 2020, 12, 4688–4710. [Google Scholar] [CrossRef]
- Unniyampurath, U.; Crisci, A.; Krishnan, M.N. Loss of Function Genetic Screen Identifies ATM Kinase as a Positive Regulator of TLR3-Mediated NF-κB Activation. iScience 2020, 23, 101356. [Google Scholar] [CrossRef] [PubMed]
- Nakshatri, H.; Appaiah, H.N.; Anjanappa, M.; Gilley, D.; Tanaka, H.; Badve, S.; Crooks, P.A.; Mathews, W.; Sweeney, C.; Bhat-Nakshatri, P. NF-κB-dependent and -independent epigenetic modulation using the novel anti-cancer agent DMAPT. Cell Death Dis. 2015, 6, e1608. [Google Scholar] [CrossRef]
- Stender, J.D.; Pascual, G.; Liu, W.; Kaikkonen, M.U.; Do, K.; Spann, N.J.; Boutros, M.; Perrimon, N.; Rosenfeld, M.G.; Glass, C.K. Control of Proinflammatory Gene Programs by Regulated Trimethylation and Demethylation of Histone H4K20. Mol. Cell 2012, 48, 28–38. [Google Scholar] [CrossRef]
- De Santa, F.; Totaro, M.G.; Prosperini, E.; Notarbartolo, S.; Testa, G.; Natoli, G. The Histone H3 Lysine-27 Demethylase Jmjd3 Links Inflammation to Inhibition of Polycomb-Mediated Gene Silencing. Cell 2007, 130, 1083–1094. [Google Scholar] [CrossRef]
- Ge, R.; Wang, Z.; Zeng, Q.; Xu, X.; Olumi, A.F. F-box protein 10, an NF-κB-dependent anti-apoptotic protein, regulates TRAIL-induced apoptosis through modulating c-Fos/c-FLIP pathway. Cell Death Differ. 2011, 18, 1184–1195. [Google Scholar] [CrossRef]
- Vento-Tormo, R.; Rodríguez-Ubreva, J.; Lisio, L.D.; Islam, A.B.; Urquiza, J.M.; Hernando, H.; López-Bigas, N.; Shannon-Lowe, C.; Martínez, N.; Montes-Moreno, S.; et al. NF-κB directly mediates epigenetic deregulation of common microRNAs in Epstein-Barr virus-mediated transformation of B-cells and in lymphomas. Nucleic Acids Res. 2014, 42, 11025–11039. [Google Scholar] [CrossRef]
- O’Gorman, A.; Colleran, A.; Ryan, A.; Mann, J.; Egan, L.J. Regulation of NF-κB responses by epigenetic suppression of IκBα expression in HCT116 intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G96–G105. [Google Scholar] [CrossRef] [PubMed]
- Gregory, D.J.; Zhang, Y.; Kobzik, L.; Fedulov, A.V. Specific transcriptional enhancement of inducible nitric oxide synthase by targeted promoter demethylation. Epigenetics 2013, 8, 1205–1212. [Google Scholar] [CrossRef] [PubMed]
- Handy, D.; Loscalzo, J. Epigenetics and the Regulation of Nitric Oxide. In Nitrite and Nitrate in Human Health and Disease, 2nd ed.; Bryan, N.S., Loscalzo, J., Eds.; Humana Press/Springer Nature: Gewerbestrasse, Switzerland, 2017; pp. 33–52. [Google Scholar] [CrossRef]
- Hotokezaka, Y.; Katayama, I.; Nakamura, T. ATM-associated signalling triggers the unfolded protein response and cell death in response to stress. Commun. Biol. 2020, 3, 378. [Google Scholar] [CrossRef]
- Valentin-Vega, Y.A.; Kastan, M.B. A new role for ATM. Autophagy 2012, 8, 840–841. [Google Scholar] [CrossRef][Green Version]
- Maryanovich, M.; Oberkovitz, G.; Niv, H.; Vorobiyov, L.; Zaltsman, Y.; Brenner, O.; Lapidot, T.; Jung, S.; Gross, A. The ATM-BID pathway regulates quiescence and survival of haematopoietic stem cells. Nat. Cell Biol. 2012, 14, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Imai, Y.; Yamakoshi, K.; Kuninaka, S.; Ohtani, N.; Yoshimoto, S.; Hori, S.; Tachibana, M.; Anderton, E.; Takeuchi, T.; et al. DNA damage signaling triggers degradation of histone methyltransferases through APC/C(Cdh1) in senescent cells. Mol. Cell 2012, 45, 123–131. [Google Scholar] [CrossRef]
- Pazolli, E.; Alspach, E.; Milczarek, A.; Prior, J.; Piwnica-Worms, D.; Stewart, S.A. Chromatin remodeling underlies the senescence-associated secretory phenotype of tumor stromal fibroblasts that supports cancer progression. Cancer Res. 2012, 72, 2251–2261. [Google Scholar] [CrossRef]
- Li, Z.; Kim, J.H.; Kim, J.; Lyu, J.I.; Zhang, Y.; Guo, H.; Nam, H.G.; Woo, H.R. ATM suppresses leaf senescence triggered by DNA double-strand break through epigenetic control of senescence-associated genes in Arabidopsis. New Phytol. 2020, 227, 473–484. [Google Scholar] [CrossRef]
- Ahmad, A.; Vieira, J.d.C.; de Mello, A.H.; de Lima, T.M.; Ariga, S.K.; Barbeiro, D.F.; Barbeiro, H.V.; Szczesny, B.; Törö, G.; Druzhyna, N.; et al. The PARP inhibitor olaparib exerts beneficial effects in mice subjected to cecal ligature and puncture and in cells subjected to oxidative stress without impairing DNA integrity: A potential opportunity for repurposing a clinically used oncological drug for the experimental therapy of sepsis. Pharmacol. Res. 2019, 145, 104263. [Google Scholar] [PubMed]
- Figueiredo, N.; Chora, A.; Raquel, H.; Pejanovic, N.; Pereira, P.; Hartleben, B.; Neves-Costa, A.; Moita, C.; Pedroso, D.; Pinto, A.; et al. Anthracyclines Induce DNA Damage Response-Mediated Protection against Severe Sepsis. Immunity 2013, 39, 874–884. [Google Scholar] [CrossRef]
- Aguilar-Quesada, R.; Muñoz-Gámez, J.A.; Martín-Oliva, D.; Peralta, A.; Valenzuela, M.T.; Matínez-Romero, R.; Quiles-Pérez, R.; Murcia, J.M.-d.; de Murcia, G.; de Almodóvar, M.R.; et al. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol. Biol. 2007, 8, 29. [Google Scholar] [CrossRef]
- Chandrasekaran, A.; Idelchik, M.; Melendez, J.A. Redox control of senescence and age-related disease. Redox Biol. 2017, 11, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Mijit, M.; Caracciolo, V.; Melillo, A.; Amicarelli, F.; Giordano, A. Role of p53 in the Regulation of Cellular Senescence. Biomolecules 2020, 10, 420. [Google Scholar] [CrossRef]
- Jung, S.H.; Hwang, H.J.; Kang, D.; Park, H.A.; Lee, H.C.; Jeong, D.; Lee, K.; Park, H.J.; Ko, Y.G.; Lee, J.S. mTOR kinase leads to PTEN-loss-induced cellular senescence by phosphorylating p53. Oncogene 2019, 38, 1639–1650. [Google Scholar] [CrossRef]
- Bartkova, J.; Rezaei, N.; Liontos, M.; Karakaidos, P.; Kletsas, D.; Issaeva, N.; Vassiliou, L.V.; Kolettas, E.; Niforou, K.; Zoumpourlis, V.C.; et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006, 444, 633–637. [Google Scholar] [CrossRef]
- Kruse, J.P.; Gu, W. Modes of p53 regulation. Cell 2009, 137, 609–622. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Fumagalli, M.; Cicalese, A.; Piccinin, S.; Gasparini, P.; Luise, C.; Schurra, C.; Garre, M.; Nuciforo, P.G.; Bensimon, A.; et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006, 444, 638–642. [Google Scholar] [CrossRef]
- Nacarelli, T.; Liu, P.; Zhang, R. Epigenetic Basis of Cellular Senescence and Its Implications in Aging. Genes 2017, 8, 343. [Google Scholar] [CrossRef]
- Narita, M.; Nũnez, S.; Heard, E.; Narita, M.; Lin, A.W.; Hearn, S.A.; Spector, D.L.; Hannon, G.J.; Lowe, S.W. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 2003, 113, 703–716. [Google Scholar] [CrossRef]
- Kovatcheva, M.; Liao, W.; Klein, M.E.; Robine, N.; Geiger, H.; Crago, A.M.; Dickson, M.A.; Tap, W.D.; Singer, S.; Koff, A. ATRX is a regulator of therapy induced senescence in human cells. Nat. Commun. 2017, 8, 386. [Google Scholar] [CrossRef] [PubMed]
- Swanson, E.C.; Manning, B.; Zhang, H.; Lawrence, J.B. Higher-order unfolding of satellite heterochromatin is a consistent and early event in cell senescence. J. Cell Biol. 2013, 203, 929–942. [Google Scholar] [CrossRef] [PubMed]
- Hock, R.; Furusawa, T.; Ueda, T.; Bustin, M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007, 17, 72–79. [Google Scholar] [CrossRef]
- Funayama, R.; Saito, M.; Tanobe, H.; Ishikawa, F. Loss of linker histone H1 in cellular senescence. J. Cell Biol. 2006, 175, 869–880. [Google Scholar] [CrossRef]
- Tasdemir, N.; Banito, A.; Roe, J.S.; Alonso-Curbelo, D.; Camiolo, M.; Tschaharganeh, D.F.; Huang, C.H.; Aksoy, O.; Bolden, J.E.; Chen, C.C.; et al. BRD4 Connects Enhancer Remodeling to Senescence Immune Surveillance. Cancer Discov. 2016, 6, 612–629. [Google Scholar] [CrossRef]
- Hayakawa, T.; Iwai, M.; Aoki, S.; Takimoto, K.; Maruyama, M.; Maruyama, W.; Motoyama, N. SIRT1 suppresses the senescence-associated secretory phenotype through epigenetic gene regulation. PLoS ONE 2015, 10, e0116480. [Google Scholar] [CrossRef]
- Wang, Y.; Schulte, B.A.; LaRue, A.C.; Ogawa, M.; Zhou, D. Total body irradiation selectively induces murine hematopoietic stem cell senescence. Blood 2006, 107, 358–366. [Google Scholar] [CrossRef]
- Wang, Y.; Schulte, B.A.; Zhou, D. Hematopoietic stem cell senescence and long-term bone marrow injury. Cell Cycle 2006, 5, 35–38. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huff, L.A.; Yan, S.; Clemens, M.G. Mechanisms of Ataxia Telangiectasia Mutated (ATM) Control in the DNA Damage Response to Oxidative Stress, Epigenetic Regulation, and Persistent Innate Immune Suppression Following Sepsis. Antioxidants 2021, 10, 1146. https://doi.org/10.3390/antiox10071146
Huff LA, Yan S, Clemens MG. Mechanisms of Ataxia Telangiectasia Mutated (ATM) Control in the DNA Damage Response to Oxidative Stress, Epigenetic Regulation, and Persistent Innate Immune Suppression Following Sepsis. Antioxidants. 2021; 10(7):1146. https://doi.org/10.3390/antiox10071146
Chicago/Turabian StyleHuff, Laura A., Shan Yan, and Mark G. Clemens. 2021. "Mechanisms of Ataxia Telangiectasia Mutated (ATM) Control in the DNA Damage Response to Oxidative Stress, Epigenetic Regulation, and Persistent Innate Immune Suppression Following Sepsis" Antioxidants 10, no. 7: 1146. https://doi.org/10.3390/antiox10071146
APA StyleHuff, L. A., Yan, S., & Clemens, M. G. (2021). Mechanisms of Ataxia Telangiectasia Mutated (ATM) Control in the DNA Damage Response to Oxidative Stress, Epigenetic Regulation, and Persistent Innate Immune Suppression Following Sepsis. Antioxidants, 10(7), 1146. https://doi.org/10.3390/antiox10071146