Purification, Physicochemical Properties, and Antioxidant Activities of Two Low-Molecular-Weight Polysaccharides from Ganoderma leucocontextum Fruiting Bodies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemical Reagents
2.2. Extraction of Crude Polysaccharide
2.3. Fractionation and Purification
2.4. Components Analysis
2.5. Structural Characteristics
2.5.1. Molecular Weight Detection
2.5.2. Monosaccharide Composition Analysis
2.5.3. Fourier-Transform Infrared Spectroscopy (FT-IR) Analysis
2.5.4. Congo Red Test
2.5.5. X-ray Diffraction (XRD) Analysis
2.5.6. Molecular Surface Morphology Analysis
2.5.7. Methylation Analysis
2.5.8. Nuclear Magnetic Resonance (NMR) Spectroscopy Analysis
2.6. Evaluation for In Vitro Antioxidant Activities
2.6.1. ABTS Radical Cation Decolorization Assay
2.6.2. Hydroxyl Radical Scavenging Assay
2.6.3. Superoxide Anion Scavenging Assay
2.6.4. Ferric Reducing Antioxidant Power Assay
2.6.5. Oxygen Radical Antioxidant Capacity Assay
2.7. Intracellular Antioxidant Activities of GLP-1
2.7.1. Cell Culture
2.7.2. Measurement of Cell Viability and LDH Release
2.7.3. Determination of Intracellular Reactive Oxygen Species
2.7.4. Determination of MDA, GSH, GSSG, CAT, and GSH-Px Levels
2.8. Statistical Analysis
3. Results and Discussion
3.1. Purification, Component Analysis and Molecular Weights of GLP-1 and GLP-2
3.2. Monosaccharide Compositions of GLP-1 and GLP-2
3.3. FT-IR Spectra of GLP-1 and GLP-2
3.4. Chain Conformation and Crystalline Characteristics of GLP-1 and GLP-2
3.5. Morphological Properties of GLP-1 and GLP-2
3.6. Antioxidant Activities of GLP-1 and GLP-2 In Vitro
3.7. Protective Effects of GLP-1 on tBHP-Induced Oxidative Damage in NIH3T3 Cells
3.8. Linkage Features of GLP-1
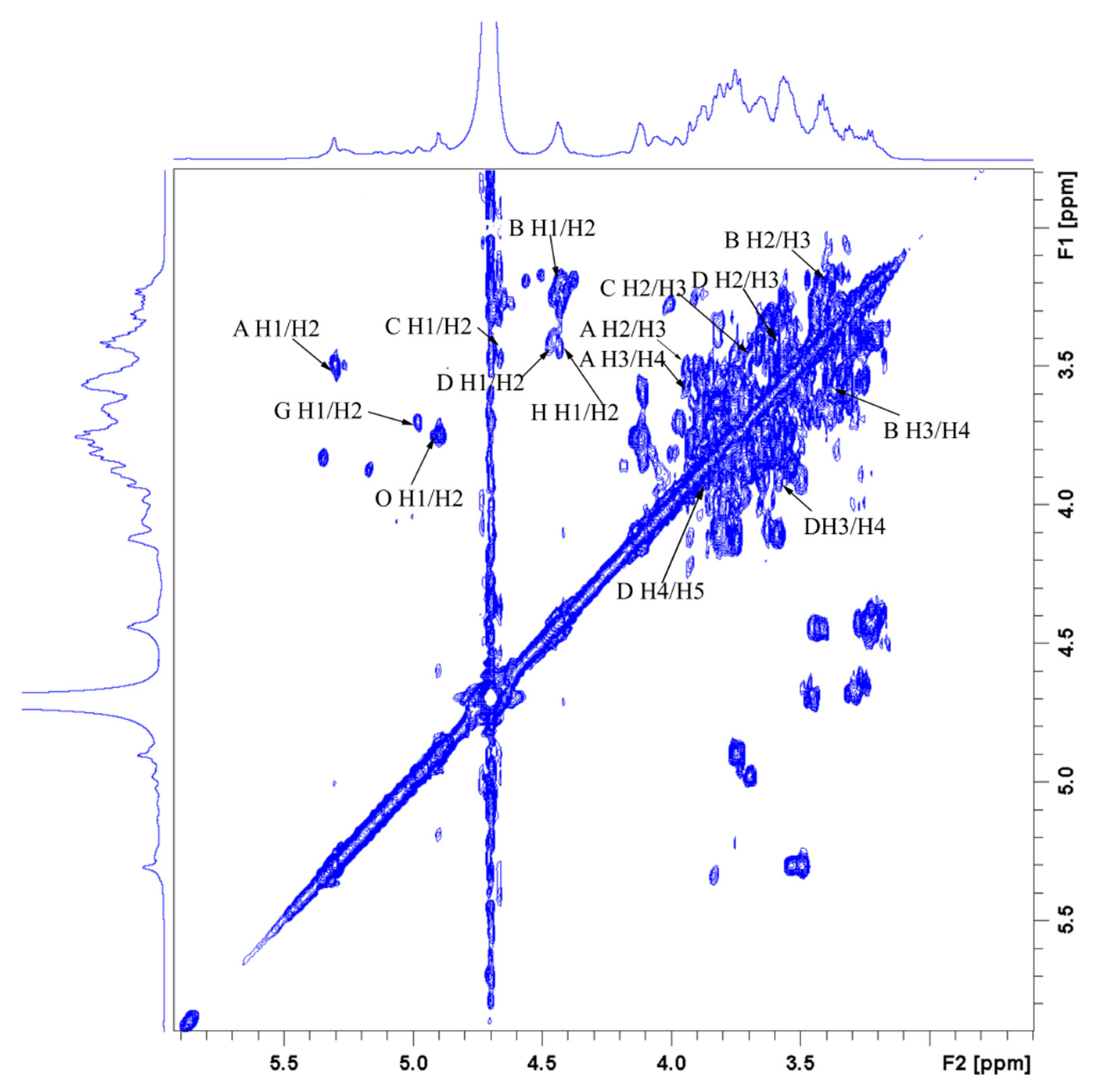
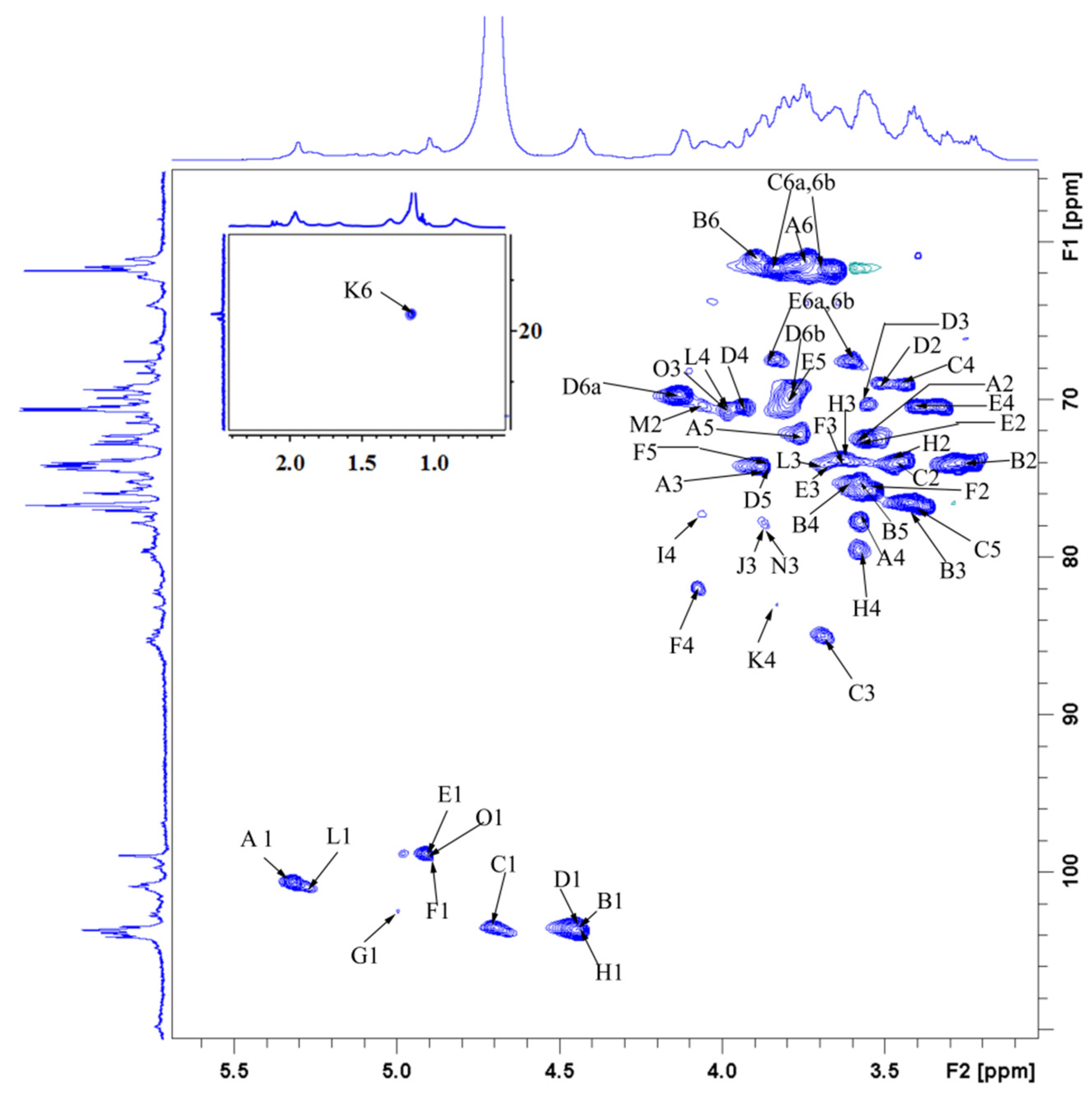
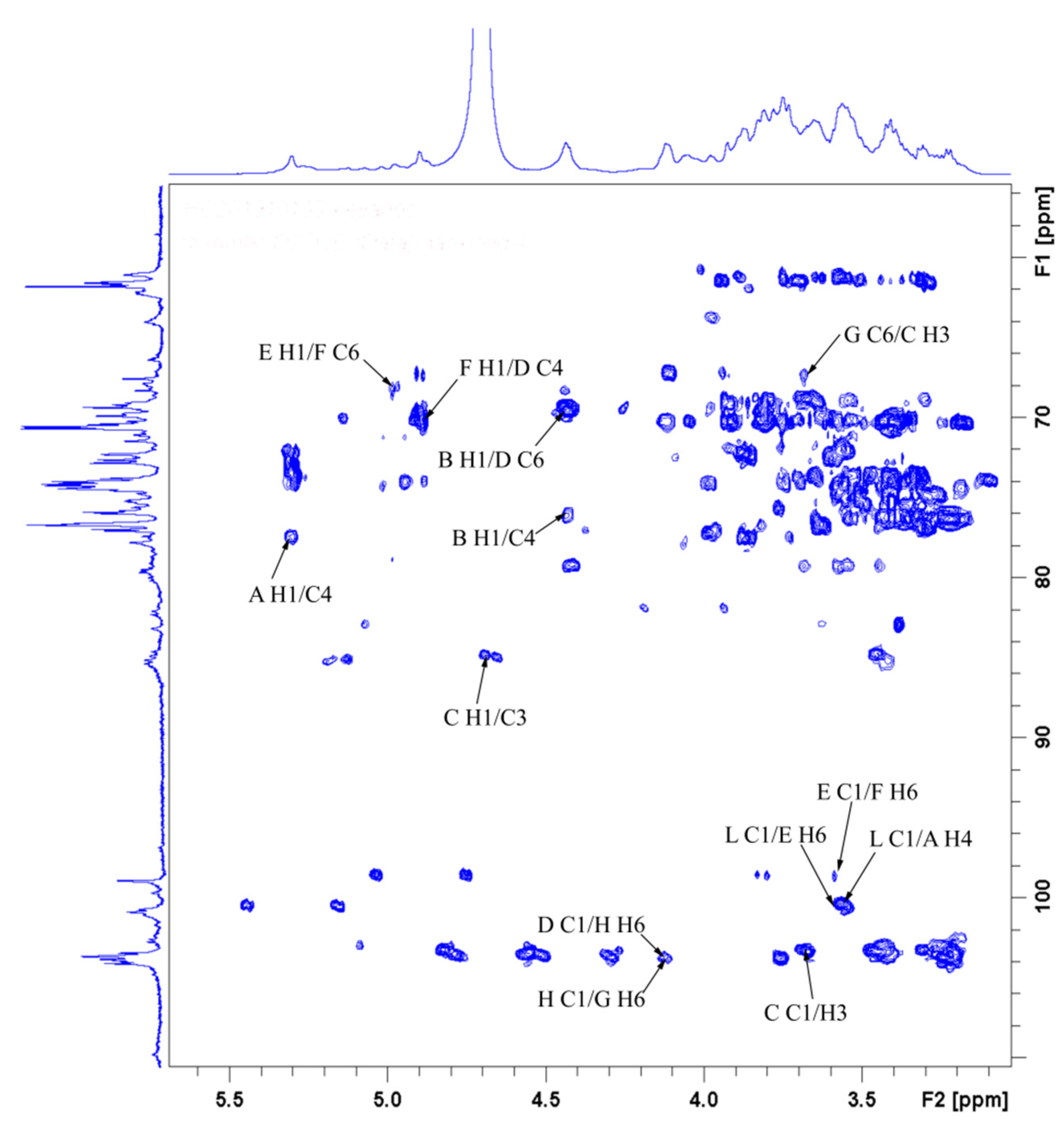
3.9. NMR-Derived Structural Characteristics of GLP-1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dikalov, S.I.; Harrison, D.G. Methods for detection of mitochondrial and cellular reactive oxygen species. Antioxid. Redox Sign. 2014, 20, 372–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.D.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef] [PubMed]
- Rani, V.; Deep, G.; Singh, R.K.; Palle, K.; Yadav, U.C.S. Oxidative stress and metabolic disorders: Pathogenesis and therapeutic strategies. Life Sci. 2016, 148, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Jomova, K.; Rhodes, C.J.; Kuca, K.; Musilek, K. Redox- and non-redox-metal-induced formation of free radicals and their role in human disease. Arch. Toxicol. 2016, 90, 1–37. [Google Scholar] [CrossRef]
- Poprac, P.; Jomova, K.; Simunkova, M.; Kollar, V.; Rhodes, C.J.; Valko, M. Targeting free radicals in oxidative stress-related human diseases. Trends Pharmacol. Sci. 2017, 38, 592–607. [Google Scholar] [CrossRef]
- Zhou, Y.; Ma, W.; Wang, L.; Sun, W.; Li, M.; Zhang, W.; Liu, Y.; Song, X.; Fan, Y. Characterization and antioxidant activity of the oligo-maltose fraction from Polygonum Cillinerve. Carbohyd. Polym. 2019, 226, 115307. [Google Scholar] [CrossRef]
- Yang, W.; Wang, L.; Gong, L.; Lu, Y.; Pan, W.; Wang, Y.; Zhang, W.; Chen, Y. Structural characterization and antioxidant activities of a novel polysaccharide fraction from the fruiting bodies of Craterellus cornucopioides. Int. J. Biol. Macromol. 2018, 117, 473–482. [Google Scholar] [CrossRef]
- Kang, Q.; Chen, S.; Li, S.; Wang, B.; Liu, X.; Hao, L.; Lu, J. Comparison on characterization and antioxidant activity of polysaccharides from Ganoderma lucidum by ultrasound and conventional extraction. Int. J. Biol. Macromol. 2019, 124, 1137–1144. [Google Scholar] [CrossRef]
- Yu, Q.; Nie, S.; Wang, J.; Yin, P.; Li, W.; Xie, M. Polysaccharide from Ganoderma atrum induces tumor necrosis factor-alpha secretion via phosphoinositide 3-kinase/Akt, mitogen-activated protein kinase and nuclear factor-kappa B signaling pathways in RAW264.7 cells. Int. Immunopharmacol. 2012, 14, 362–368. [Google Scholar] [CrossRef]
- Wang, J.; Cao, B.; Zhao, H.; Feng, J. Emerging roles of Ganoderma lucidum in anti-aging. Aging Dis. 2017, 8, 691–707. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Shi, L.; Ding, K. Structure elucidation and anti-tumor activity in vivo of a polysaccharide from spores of Ganoderma lucidum (Fr.) Karst. Int. J. Biol. Macromol. 2019, 141, 693–699. [Google Scholar] [CrossRef]
- Chen, Y.; Xie, M.; Nie, S.; Li, C.; Wang, Y. Purification, composition analysis and antioxidant activity of a polysaccharide from the fruiting bodies of Ganoderma atrum. Food Chem. 2008, 107, 231–241. [Google Scholar] [CrossRef]
- Tseng, Y.; Yang, J.; Mau, J. Antioxidant properties of polysaccharides from Ganoderma tsugae. Food Chem. 2008, 107, 732–738. [Google Scholar] [CrossRef]
- Chen, X.; Chen, Y.; Li, S.; Chen, Y.; Lan, J.; Liu, L. Free radical scavenging of Ganoderma lucidum polysaccharides and its effect on antioxidant enzymes and immunity activities in cervical carcinoma rats. Carbohyd. Polym. 2009, 77, 389–393. [Google Scholar] [CrossRef]
- Yang, Q.; Wang, S.; Xie, Y.; Sun, J.; Wang, J. HPLC analysis of Ganoderma lucidum polysaccharides and its effect on antioxidant enzymes activity and Bax, Bcl-2 expression. Int. J. Biol. Macromol. 2010, 46, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Hu, H.; Deng, W.; Wu, S.; Wang, D.; Tsering, T. Ganoderma leucocontextum, a new member of the G. lucidum complex from southwestern China. Mycoscience 2015, 56, 81–85. [Google Scholar] [CrossRef]
- Wang, K.; Bao, L.; Ma, K.; Zhang, J.; Chen, B.; Han, J.; Ren, J.; Luo, H.; Liu, H. A novel class of alpha-glucosidase and HMG-CoA reductase inhibitors from Ganoderma leucocontextum and the anti-diabetic properties of ganomycin I in KK-A(y) mice. Eur. J. Med. Chem. 2017, 127, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xie, Y.; Peng, J.; Hu, H.; Wu, Q.; Yang, B.B. Ganoderiol F purified from Ganoderma leucocontextum retards cell cycle progression by inhibiting CDK4/CDK6. Cell Cycle 2019, 18, 3030–3043. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, J.; Ren, J.; Wang, W.; Xiong, W.; Zhang, Y.; Bao, L.; Liu, H. Triterpenes and meroterpenes with neuroprotective effects from Ganoderma leucocontextum. Chem. Biodivers. 2018, 15, e17005675. [Google Scholar] [CrossRef]
- Gao, X.; Qi, J.; Ho, C.; Li, B.; Mu, J.; Zhang, Y.; Hu, H.; Mo, W.; Chen, Z.; Xie, Y. Structural characterization and immunomodulatory activity of a water-soluble polysaccharide from Ganoderma leucocontextum fruiting bodies. Carbohyd. Polym. 2020, 249, 116874. [Google Scholar] [CrossRef]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Bradford, M.M. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Niu, Y.; Shang, P.; Chen, L.; Zhang, H.; Gong, L.; Zhang, X.; Yu, W.; Xu, Y.; Wang, Q.; Yu, L.L. Characterization of a novel alkali-soluble heteropolysaccharide from tetraploid Gynostemma pentaphyllum Makino and its potential anti-inflammatory and antioxidant properties. J. Agric. Food Chem. 2014, 62, 3783–3790. [Google Scholar] [CrossRef]
- Chen, W.; Zhu, X.; Ma, J.; Zhang, M.; Wu, H. Structural elucidation of a novel pectin-polysaccharide from the petal of Saussurea laniceps and the mechanism of its anti-HBV activity. Carbohyd. Polym. 2019, 223, 115077. [Google Scholar] [CrossRef] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, L.; Rui, X.; Li, W.; Chen, X.; Jiang, M.; Dong, M. Enhancement of the antioxidant capacity of soy whey by fermentation with Lactobacillus plantarum B1-6. J. Funct. Foods 2015, 12, 33–44. [Google Scholar] [CrossRef]
- Li, W.; Ji, J.; Chen, X.; Jiang, M.; Rui, X.; Dong, M. Structural elucidation and antioxidant activities of exopolysaccharides from Lactobacillus helveticus MB2-1. Carbohyd. Polym. 2014, 102, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Benzie, I.; Szeto, Y.T. Total antioxidant capacity of teas by the ferric reducing/antioxidant power assay. J. Agric. Food Chem. 1999, 47, 633–636. [Google Scholar] [CrossRef]
- Davalos, A.; Gomez-Cordoves, C.; Bartolome, B. Extending applicability of the oxygen radical absorbance capacity (ORAC-fluorescein) assay. J. Agric. Food Chem. 2004, 52, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ma, Z.; Zhang, L.; Fang, Y.; Jiang, F.; Phillips, G.O. Structure and chain conformation of water-soluble heteropolysaccharides from Ganoderma lucidum. Carbohyd. Polym. 2011, 86, 844–851. [Google Scholar] [CrossRef]
- Wang, M.; Chen, G.; Chen, D.; Ye, H.; Sun, Y.; Zeng, X.; Liu, Z. Purified fraction of polysaccharides from Fuzhuan brick tea modulates the composition and metabolism of gut microbiota in anaerobic fermentation in vitro. Int. J. Biol. Macromol. 2019, 140, 858–870. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Wu, J.; Huang, M.; Zhao, M.; Sun, W.; Sun, X.; Zheng, F. Structural characterization and immuno-stimulating activities of a novel polysaccharide from Huangshui, a byproduct of Chinese Baijiu. Food Res. Int. 2020, 136, 109493. [Google Scholar] [CrossRef]
- Zhou, W.; Zhao, Y.; Yan, Y.; Mi, J.; Lu, L.; Luo, Q.; Li, X.; Zeng, X.; Cao, Y. Antioxidant and immunomodulatory activities in vitro of polysaccharides from bee collected pollen of Chinese wolfberry. Int. J. Biol. Macromol. 2020, 163, 190–199. [Google Scholar] [CrossRef]
- Liu, Z.; Jiao, Y.; Lu, H.; Shu, X.; Chen, Q. Chemical characterization, antioxidant properties and anticancer activity of exopolysaccharides from Floccularia luteovirens. Carbohyd. Polym. 2020, 229, 115432. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, J.; Huang, X.; Nie, S. Structural characteristics and rheological properties of high viscous glucan from fruit body of Dictyophora rubrovolvata. Food Hydrocoll. 2020, 101, 105514. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, M.; Wen, C.; Zhou, J.; Gu, J.; Duan, Y.; Zhang, H.; Ren, X.; Ma, H. Structural characterization and immunostimulatory activity of a novel polysaccharide isolated with subcritical water from Sagittaria sagittifolia L. Int. J. Biol. Macromol. 2019, 133, 11–20. [Google Scholar] [CrossRef]
- Cheng, Y.; Xiao, X.; Li, X.; Song, D.; Lu, Z.; Wang, F.; Wang, Y. Characterization, antioxidant property and cytoprotection of exopolysaccharide-capped elemental selenium particles synthesized by Bacillus paralicheniformis SR14. Carbohyd. Polym. 2017, 178, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Kpodo, F.M.; Agbenorhevi, J.K.; Alba, K.; Bingham, R.J.; Oduro, I.N.; Morris, G.A.; Kontogiorgos, V. Pectin isolation and characterization from six okra genotypes. Food Hydrocoll. 2017, 72, 323–330. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Cheung, P.C.K. Structure and immunomodulatory activity of microparticulate mushroom sclerotial beta-glucan prepared from Polyporus rhinoceros. J. Agric. Food Chem. 2019, 67, 9070–9078. [Google Scholar] [CrossRef]
- Gu, J.; Zhang, H.; Wen, C.; Zhang, J.; He, Y.; Ma, H.; Duan, Y. Purification, characterization, antioxidant and immunological activity of polysaccharide from Sagittaria sagittifolia L. Food Res. Int. 2020, 136, 109345. [Google Scholar] [CrossRef]
- Rozi, P.; Abuduwaili, A.; Mutailifu, P.; Gao, Y.; Rakhmanberdieva, R.; Aisa, H.A.; Yili, A. Sequential extraction, characterization and antioxidant activity of polysaccharides from Fritillaria pallidiflora Schrenk. Int. J. Biol. Macromol. 2019, 131, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Chen, W.; Zhang, W.; Zhang, H. Adulteration identification of some fungal polysaccharides with SEM, XRD, IR and optical rotation: A primary approach. Carbohyd. Polym. 2009, 78, 620–625. [Google Scholar] [CrossRef]
- Kong, L.; Yu, L.; Feng, T.; Yin, X.; Liu, T.; Dong, L. Physicochemical characterization of the polysaccharide from Bletilla striata: Effect of drying method. Carbohyd. Polym. 2015, 125, 1–8. [Google Scholar] [CrossRef]
- Marszalek, P.E.; Dufrene, Y.F. Stretching single polysaccharides and proteins using atomic force microscopy. Chem. Soc. Rev. 2012, 41, 3523–3534. [Google Scholar] [CrossRef]
- Deng, Y.; Li, M.; Chen, L.; Chen, X.; Lu, J.; Zhao, J.; Li, S. Chemical characterization and immunomodulatory activity of acetylated polysaccharides from Dendrobium devonianum. Carbohyd. Polym. 2018, 180, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, J.; Li, Q.; Zhang, Q.; You, R.; Cheng, Y.; Luo, L.; Zhang, Y. Structural differences and conformational characterization of five bioactive polysaccharides from Lentinus edodes. Food Res. Int. 2014, 62, 223–232. [Google Scholar] [CrossRef]
- Giannotti, M.I.; Rinaudo, M.; Vancso, G.J. Force spectroscopy of hyaluronan by atomic force microscopy: From hydrogen-bonded networks toward single-chain behavior. Biomacromolecules 2007, 8, 2648–2652. [Google Scholar] [CrossRef]
- Li, J.; Yuan, P.; Wang, X.; Aipire, A.; Li, M.; Yang, J.; Tao, H.; Ying, T.; Fu, C.; Wei, X.; et al. Purification, characterization and bioactivities of polysaccharides from Pleurotus ferulae. Food Funct. 2017, 8, 1905–1914. [Google Scholar] [CrossRef]
- Lopez-Alarcon, C.; Denicola, A. Evaluating the antioxidant capacity of natural products: A review on chemical and cellular-based assays. Anal. Chim. Acta 2013, 763, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Shahidi, F.; Ambigaipalan, P. Phenolics and polyphenolics in foods, beverages and spices: Antioxidant activity and health effects - A review. J. Funct. Foods 2015, 18, 820–897. [Google Scholar] [CrossRef]
- Liu, W.; Wang, H.; Pang, X.; Yao, W.; Gao, X. Characterization and antioxidant activity of two low-molecular-weight polysaccharides purified from the fruiting bodies of Ganoderma lucidum. Int. J. Biol. Macromol. 2010, 46, 451–457. [Google Scholar] [CrossRef]
- Shang, H.; Zhou, H.; Duan, M.; Li, R.; Wu, H.; Lou, Y. Extraction condition optimization and effects of drying methods on physicochemical properties and antioxidant activities of polysaccharides from comfrey (Symphytum officinale L.) root. Int. J. Biol. Macromol. 2018, 112, 889–899. [Google Scholar] [CrossRef]
- Cai, L.; Zou, S.; Liang, D.; Luan, L. Structural characterization, antioxidant and hepatoprotective activities of polysaccharides from Sophorae tonkinensis Radix. Carbohyd. Polym. 2018, 184, 354–365. [Google Scholar] [CrossRef]
- Huang, S.; Ding, S.; Fan, L. Antioxidant activities of five polysaccharides from Inonotus obliquus. Int. J. Biol. Macromol. 2012, 50, 1183–1187. [Google Scholar] [CrossRef]
- Song, H.; Zhang, Q.; Zhang, Z.; Wang, J. In vitro antioxidant activity of polysaccharides extracted from Bryopsis plumose. Carbohyd. Polym. 2010, 80, 1057–1061. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, W.; Wen, P.; Shen, M.; Li, H.; Ren, Y.; Xiao, Y.; Song, Q.; Chen, Y.; Yu, Q.; et al. Two water-soluble polysaccharides from mung bean skin: Physicochemical characterization, antioxidant and antibacterial activities. Food Hydrocoll. 2020, 100, 105412. [Google Scholar] [CrossRef]
- Wang, Z.; Yi, K.; Lin, Q.; Yang, L.; Chen, X.; Chen, H.; Liu, Y.; Wei, D. Free radical sensors based on inner-cutting graphene field-effect transistors. Nat. Commun. 2019, 10, 1544. [Google Scholar] [CrossRef] [Green Version]
- Poljsak, B.; Suput, D.; Milisav, I. Achieving the balance between ROS and antioxidants: When to use the synthetic antioxidants. Oxid. Med. Cell. Longev. 2013, 2013, 956792. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Caro, G.; Sarria, B.; Madrona, A.; Espartero, J.L.; Goya, L.; Bravo, L.; Mateos, R. Alkyl hydroxytyrosyl ethers show protective effects against oxidative stress in HepG2 cells. J. Agric. Food Chem. 2011, 59, 5964–5976. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, L.; Wang, R.; Luo, X.; Li, Y.; Chen, Z. Protective effects of rice dreg protein hydrolysates against hydrogen peroxide-induced oxidative stress in HepG-2 cells. Food Funct. 2016, 7, 1429–1437. [Google Scholar] [CrossRef] [PubMed]
- Lue, J.; Lin, P.H.; Yao, Q.; Chen, C. Chemical and molecular mechanisms of antioxidants: Experimental approaches and model systems. J. Cell. Mol. Med. 2010, 14, 840–860. [Google Scholar] [CrossRef] [PubMed]
- Brigelius-Flohe, R.; Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 3289–3303. [Google Scholar] [CrossRef]
- Li, Q.; Feng, Y.; He, W.; Wang, L.; Wang, R.; Dong, L.; Wang, C. Post-screening characterisation and in vivo evaluation of an anti-inflammatory polysaccharide fraction from Eucommia ulmoides. Carbohyd. Polym. 2017, 169, 304–314. [Google Scholar] [CrossRef]
- Zhan, Q.; Wang, Q.; Lin, R.; He, P.; Lai, F.; Zhang, M.; Wu, H. Structural characterization and immunomodulatory activity of a novel acid polysaccharide isolated from the pulp of Rosa laevigata Michx fruit. Int. J. Biol. Macromol. 2020, 145, 1080–1090. [Google Scholar] [CrossRef]
- Ru, Y.; Chen, X.; Wang, J.; Guo, L.; Lin, Z.; Peng, X.; Qiu, B. Polysaccharides from Tetrastigma hemsleyanum Diels et Gilg: Extraction optimization, structural characterizations, antioxidant and antihyperlipidemic activities in hyperlipidemic mice. Int. J. Biol. Macromol. 2019, 125, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gu, F.; Cai, C.; Hu, M.; Fan, L.; Hao, J.; Yu, G. Purification, structural characterization, and immunomodulatory activity of the polysaccharides from Ganoderma lucidum. Int. J. Biol. Macromol. 2020, 143, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, G.; Lai, F.; Wu, H. Structural characterization and immunomodulatory activity of a novel polysaccharide from Lepidium meyenii. J. Agric. Food Chem. 2016, 64, 1921–1931. [Google Scholar] [CrossRef] [PubMed]
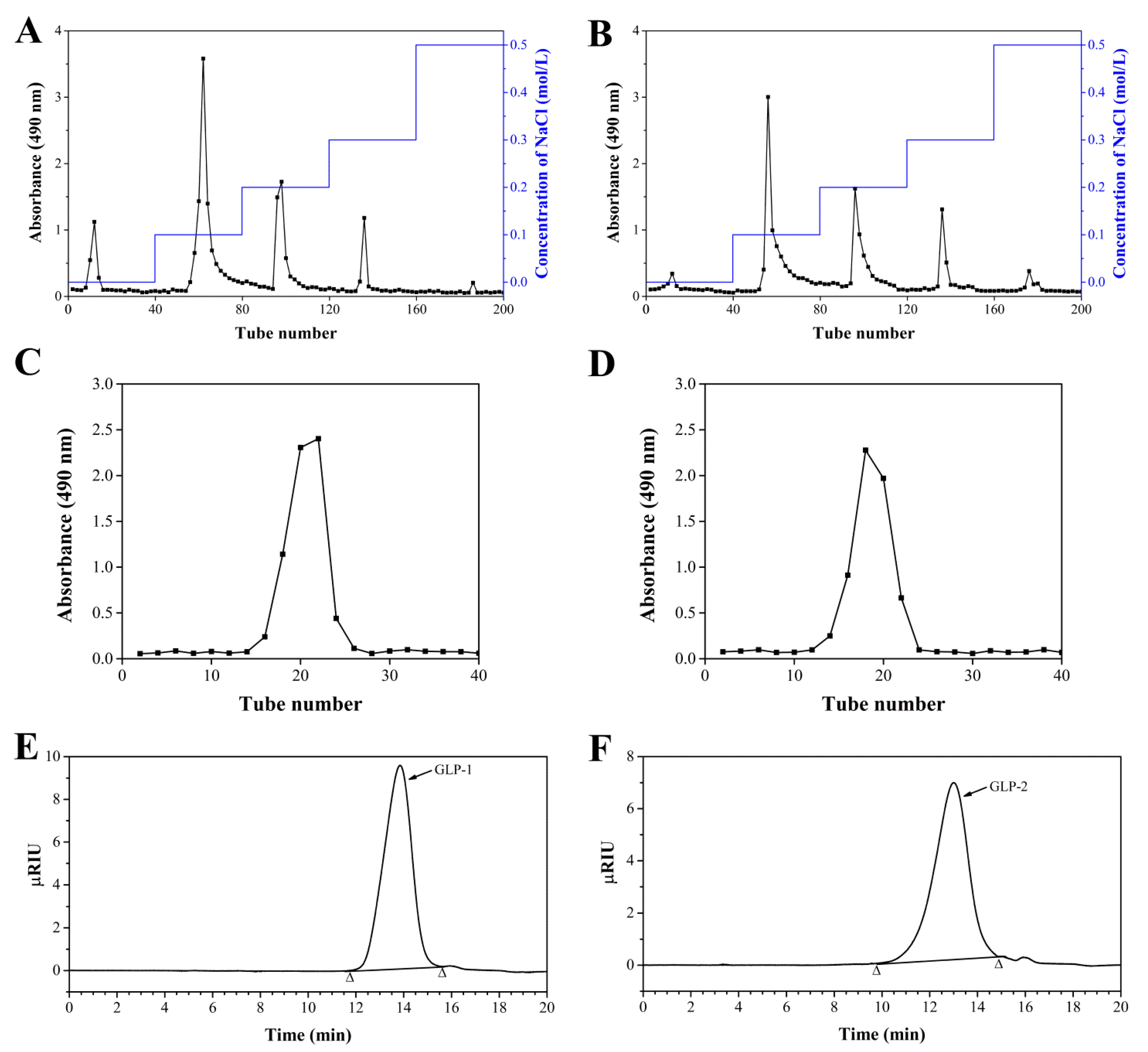



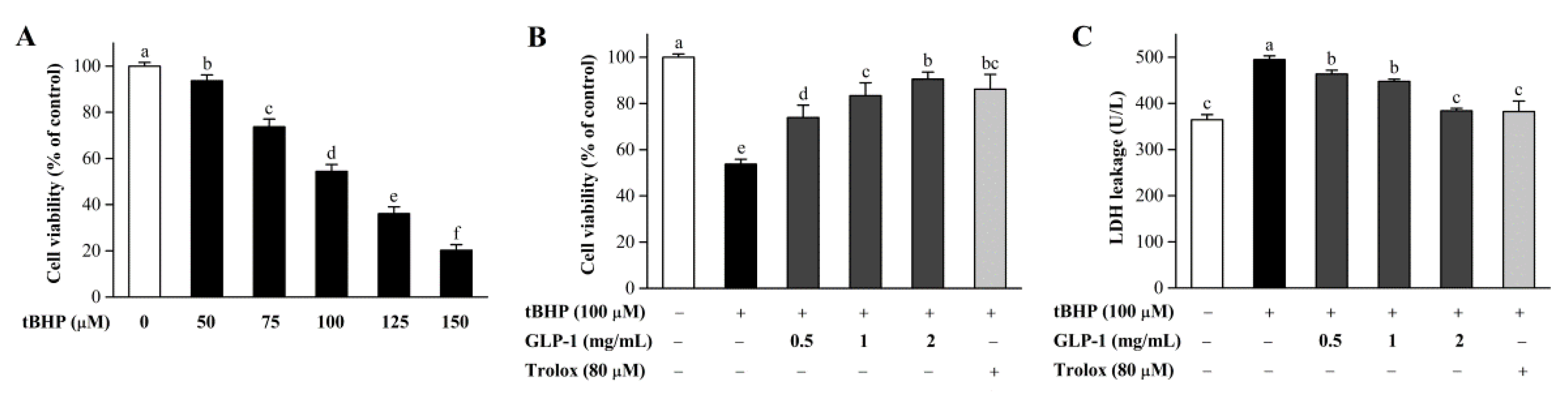
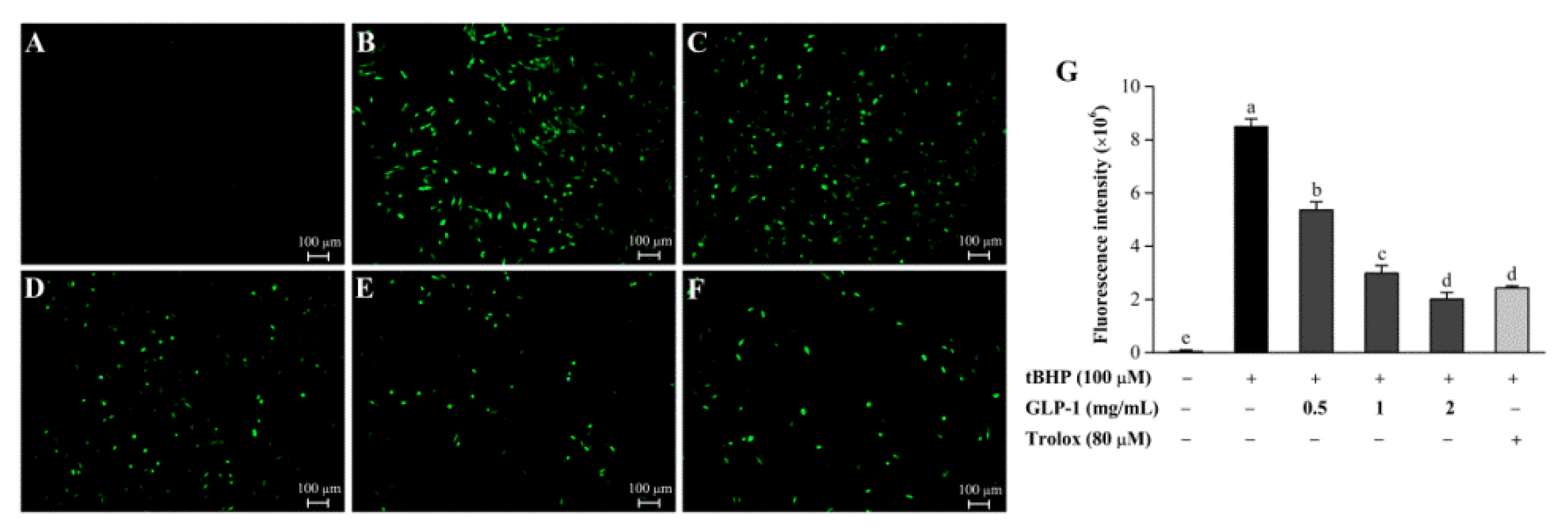

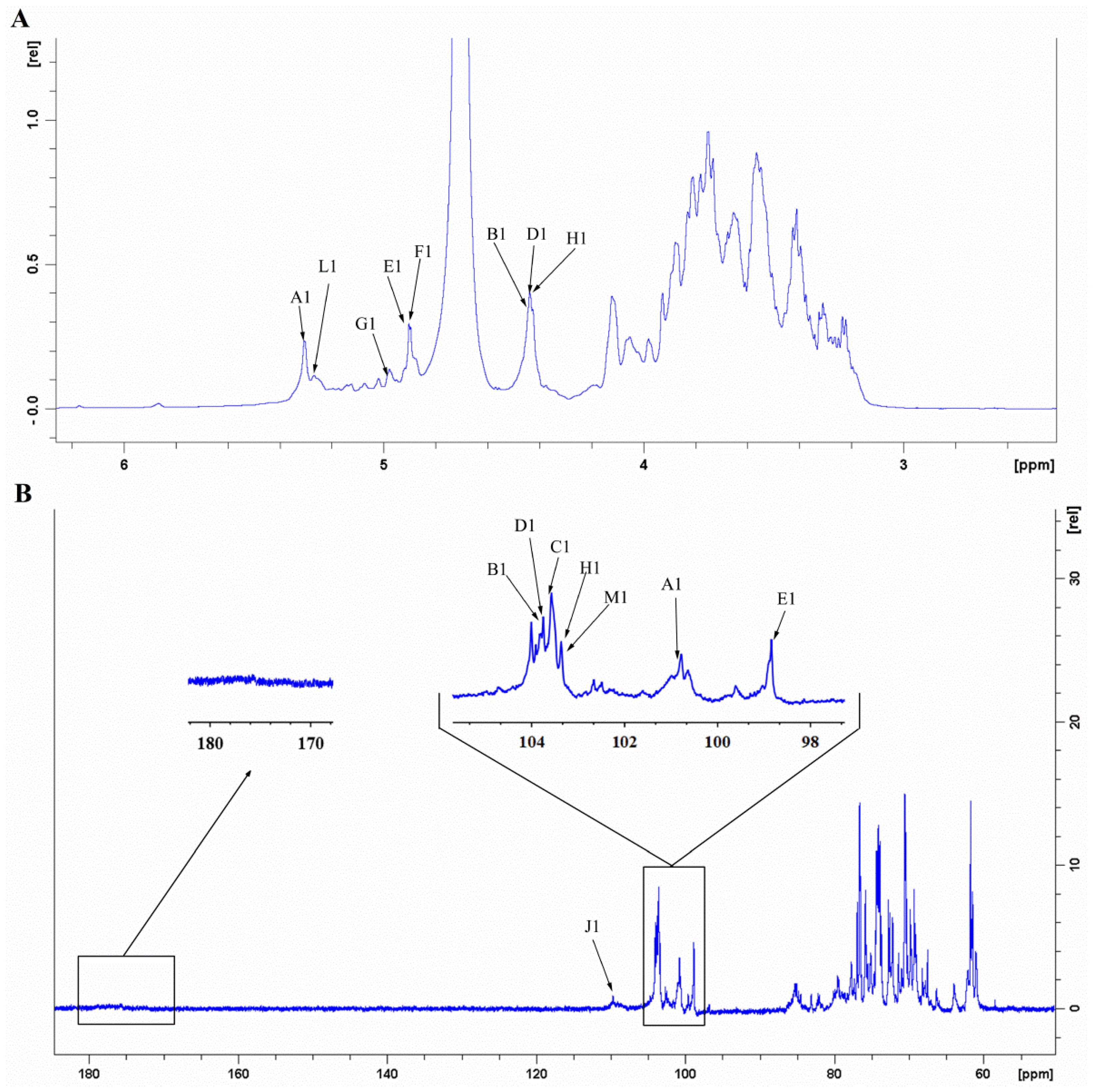

| Sample | GLP-1 | GLP-2 |
|---|---|---|
| Carbohydrate (wt%) | 73.36 ± 1.35 | 72.45 ± 0.49 |
| Protein (wt%) | 0.03 ± 0.01 | 0.08 ± 0.01 |
| Mw (kDa) | 6.31 | 14.07 |
| Mn (kDa) | 5.22 | 10.06 |
| Mw/Mn | 1.21 | 1.40 |
| Methylated Sugar | Linkage Pattern | Molar Ratios | Rt | Mass Fragments (m/z) |
|---|---|---|---|---|
| 2,3,5-Me3-Araf | T-Araf-(1→ | 3.28 | 9.35 | 43,71,87,101,117,129,145,161 |
| 2,3,4-Me3-Arap | T-Arap-(1→ | 1.09 | 10.78 | 43,71,87,101,117,129,131,161 |
| 2,3,4-Me3-Fucp | T-Fucp-(1→ | 0.88 | 11.83 | 43,59,72,89,101,115,117,131,175 |
| 2-Me1-Rhap | →3,4)-Rhap-(1→ | 1.49 | 13.00 | 43,87,99,113,117,129,141,159,173 |
| 2,3-Me2-Xylp | →4)-Xylp-(1→ | 3.25 | 14.69 | 43,71,87,99,101,117,129,161,189 |
| 2,3,4,6-Me4-Glcp | T-Glcp-(1→ | 11.29 | 16.23 | 43,71,87,101,117,129,145,161,205 |
| 2,3,4,6-Me4-Manp | T-Manp-(1→ | 1.46 | 16.45 | 43,71,87,101,117,129,145,161,205 |
| 2,4,6-Me3-Glcp | →3)-Glcp-(1→ | 12.75 | 20.84 | 43,71,85,87,99,101,117,129,161 |
| 2,3,6-Me3-Glcp | →4)-Glcp-(1→ | 25.89 | 21.48 | 43,87,99,101,113,117,129,131,161,173,233 |
| 2,3,4-Me3-Glcp | →6)-Glcp-(1→ | 10.14 | 22.40 | 43,87,99,101,117,129,161,189,233 |
| 2,3,4-Me3-Galp | →6)-Galp-(1→ | 11.97 | 24.35 | 43,87,99,101,117,129,161,189,233 |
| 2,3-Me2-Galp | →4,6)-Galp-(1→ | 3.73 | 27.29 | 43,71,85,87,99,101,117,127,159,161,201 |
| 2,3-Me2-Glcp | →4,6)-Glcp-(1→ | 8.69 | 27.63 | 43,71,85,87,99,101,117,127,159,161,201 |
| 3,4-Me2-Galp | →2,6)-Galp-(1→ | 4.09 | 29.22 | 43,71,87,99,129,189 |
| Glycosyl Residues | H1 | H2 | H3 | H4 | H5 | H6a | H6b |
|---|---|---|---|---|---|---|---|
| C1 | C2 | C3 | C4 | C5 | C6 | ||
| →4)-α-D-Glcp-(1→ | 5.32 | 3.55 | 3.88 | 3.58 | 3.76 | 3.77 | ns |
| A | 100.66 | 72.53 | 74.25 | 77.69 | 72.22 | 61.43 | |
| →4)-β-D-Glcp-(1→ | 4.45 | 3.24 | 3.42 | 3.58 | 3.54 | 3.90 | |
| B | 103.63 | 73.93 | 76.44 | 75.19 | 75.81 | 60.96 | |
| →3)-β-D-Glcp-(1→ | 4.70 | 3.45 | 3.69 | 3.43 | 3.42 | 3.83 | 3.66 |
| C | 103.48 | 73.93 | 85.03 | 69.09 | 76.44 | 61.74 | |
| →6)-β-D-Galp-(1→ | 4.45 | 3.43 | 3.55 | 3.94 | 3.88 | 4.14 | 3.78 |
| D | 103.63 | 69.09 | 70.18 | 70.34 | 74.25 | 69.71 | |
| →6)-α-D-Glcp-(1→ | 4.91 | 3.55 | 3.69 | 3.39 | 3.78 | 3.83 | 3.60 |
| E | 98.79 | 72.53 | 74.09 | 70.34 | 69.71 | 67.37 | |
| →4,6)-α-D-Glcp-(1→ | 4.89 | 3.49 | 3.65 | 4.08 | 3.85 | 4.20 | 3.58 |
| F | 99.87 | 75.73 | 74.02 | 81.91 | 74.60 | 68.31 | |
| →2,6)-β-D-Galp-(1→ | 5.00 | 3.73 | ns | ns | ns | 3.91 | 4.12 |
| G | 102.38 | 78.94 | ns | ns | ns | 66.12 | |
| →4,6)-β-D-Galp-(1→ | 4.44 | 3.45 | 3.67 | 3.59 | ns | 3.90 | 4.13 |
| H | 103.36 | 73.76 | 73.75 | 79.71 | ns | 67.06 | |
| →4)-β-L-Xylp-(1→ | 5.08 | 3.63 | 3.99 | 4.07 | 3.65 | 4.03 | |
| I | 102.50 | 73.50 | 74.50 | 77.20 | 63.80 | ||
| α-L-Araf-(1→ | 5.20 | 4.27 | 3.87 | 4.14 | 3.75 | 3.66 | |
| J | 108.95 | 82.65 | 77.97 | 85.24 | 62.59 | ||
| →3,4)-α-L-Rhap-(1→ | 4.96 | 3.91 | 3.69 | 3.83 | 3.86 | 1.15 | |
| K | 103.16 | 72.37 | 79.09 | 83.00 | 75.81 | 16.57 | |
| α-D-Glcp-1→ | 5.26 | 3.60 | 3.70 | 3.95 | 3.99 | 3.60 | 3.82 |
| L | 101.33 | 71.94 | 74.03 | 70.61 | 69.25 | 62.30 | |
| α-D-Manp-1→ | 5.03 | 4.06 | 3.62 | 3.51 | 3.76 | 3.83 | 3.66 |
| M | 103.32 | 70.34 | 70.50 | 68.93 | 76.28 | 61.74 | |
| α-L-Arap-(1→ | 5.14 | 4.22 | 3.87 | 4.04 | 3.29 | 3.74 | |
| N | 109.57 | 85.35 | 77.84 | 85.19 | 63.93 | ||
| α-L-Fucp-1→ | 4.89 | 3.75 | 3.99 | 3.67 | 4.11 | 1.15 | |
| O | 98.79 | 73.26 | 70.65 | 76.28 | 68.15 | 16.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Qi, J.; Ho, C.-T.; Li, B.; Xie, Y.; Chen, S.; Hu, H.; Chen, Z.; Wu, Q. Purification, Physicochemical Properties, and Antioxidant Activities of Two Low-Molecular-Weight Polysaccharides from Ganoderma leucocontextum Fruiting Bodies. Antioxidants 2021, 10, 1145. https://doi.org/10.3390/antiox10071145
Gao X, Qi J, Ho C-T, Li B, Xie Y, Chen S, Hu H, Chen Z, Wu Q. Purification, Physicochemical Properties, and Antioxidant Activities of Two Low-Molecular-Weight Polysaccharides from Ganoderma leucocontextum Fruiting Bodies. Antioxidants. 2021; 10(7):1145. https://doi.org/10.3390/antiox10071145
Chicago/Turabian StyleGao, Xiong, Jiayi Qi, Chi-Tang Ho, Bin Li, Yizhen Xie, Shaodan Chen, Huiping Hu, Zhongzheng Chen, and Qingping Wu. 2021. "Purification, Physicochemical Properties, and Antioxidant Activities of Two Low-Molecular-Weight Polysaccharides from Ganoderma leucocontextum Fruiting Bodies" Antioxidants 10, no. 7: 1145. https://doi.org/10.3390/antiox10071145
APA StyleGao, X., Qi, J., Ho, C.-T., Li, B., Xie, Y., Chen, S., Hu, H., Chen, Z., & Wu, Q. (2021). Purification, Physicochemical Properties, and Antioxidant Activities of Two Low-Molecular-Weight Polysaccharides from Ganoderma leucocontextum Fruiting Bodies. Antioxidants, 10(7), 1145. https://doi.org/10.3390/antiox10071145






