RedEfish: Generation of the Polycistronic mScarlet: GSG-T2A: Ttpa Zebrafish Line
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Husbandry and Strains
2.2. Construction of Donor Plasmid and Guide RNA Vectors for Knock-In
2.3. Microinjection
2.4. Mutation Detection and Insertion Mapping
2.5. In Vivo Fluorescence Detection
2.6. Egg Quality and Embryo Morphological Assessment
2.7. Immunolocalization and Western Blot Analysis
2.8. Statistical Methods
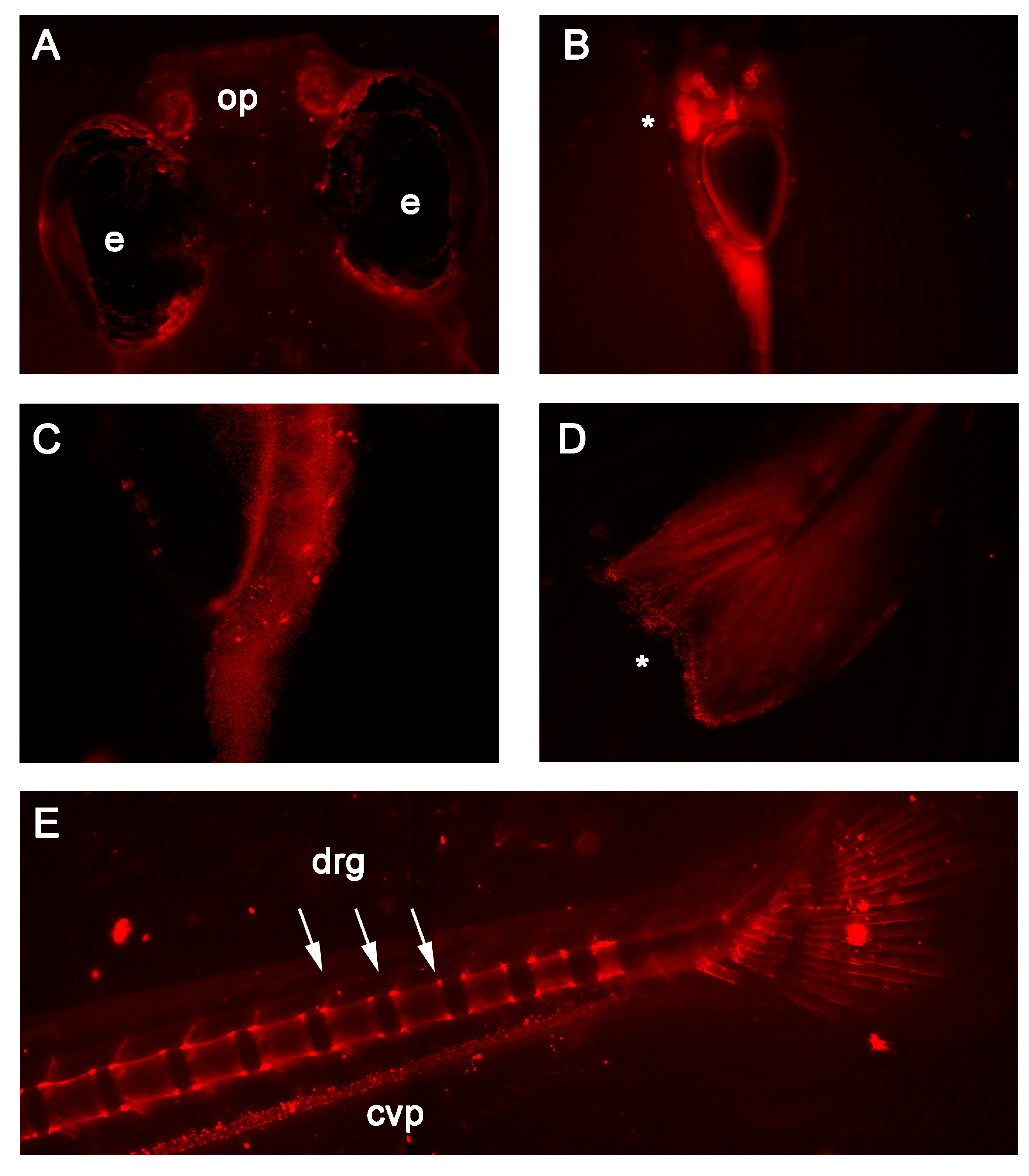
3. Results
Embryo Morphology and Egg Quality Assessment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Evans, H.M.; Bishop, K.S. On the existence of a hitherto unrecognized dietary factor essential for reproduction. Science 1922, 56, 650–651. [Google Scholar] [CrossRef]
- Shah, R.S.; Rajalakshmi, R.; Bhatt, R.V.; Hazra, M.N.; Patel, B.C.; Swamy, N.B.; Patel, T.V. Vitamin E status of the newborn in relation to gestational age, birth weight and maternal vitamin E status. Br. J. Nutr. 1987, 58, 191–198. [Google Scholar] [CrossRef]
- Shamim, A.A.; Schulze, K.; Merrill, R.D.; Kabir, A.; Christian, P.; Shaikh, S.; Wu, L.; Ali, H.; Labrique, A.B.; Mehra, S.; et al. First-trimester plasma tocopherols are associated with risk of miscarriage in rural Bangladesh. Am. J. Clin. Nutr. 2015, 101, 294–301. [Google Scholar] [CrossRef]
- Gagne, A.; Wei, S.Q.; Fraser, W.D.; Julien, P. Absorption, transport, and bioavailability of vitamin e and its role in pregnant women. J. Obstet. Gynaecol. Can. 2009, 31, 210–217. [Google Scholar] [CrossRef]
- Baydas, G.; Karatas, F.; Gursu, M.F.; Bozkurt, H.A.; Ilhan, N.; Yasar, A.; Canatan, H. Antioxidant vitamin levels in term and preterm infants and their relation to maternal vitamin status. Arch. Med. Res. 2002, 33, 276–280. [Google Scholar] [CrossRef]
- Scholl, T.O.; Chen, X.; Sims, M.; Stein, T.P. Vitamin E: Maternal concentrations are associated with fetal growth. Am. J. Clin. Nutr. 2006, 84, 1442–1448. [Google Scholar] [CrossRef]
- Schuelke, M. Ataxia with Vitamin E Deficiency. GeneReviews® [Internet] 2005 May 20 [Updated 2016 Oct 13]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1241/ (accessed on 30 March 2021).
- Di Donato, I.; Bianchi, S.; Federico, A. Ataxia with vitamin E deficiency: Update of molecular diagnosis. Neurol. Sci. 2010, 31, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Ghelfi, M.; Atkinson, J.; Parker, R.; Qian, J.; Carlin, C.; Manor, D. Vitamin E and phosphoinositides regulate the intracellular localization of the hepatic alpha-tocopherol transfer protein. J. Biol. Chem. 2016, 291, 17028–17039. [Google Scholar] [CrossRef]
- Kono, N.; Arai, H. α-Tocopherol transfer protein. In Vitamin E; Chemistry and Nutritional Benefits; Niki, E., Williamson, G., Marangoni, A.G., Gerrard, K.A., Eds.; Food Chemistry, Function and Analysis; Royal Society of Chemistry: Cambridge, UK, 2019; Volume 11, pp. 64–74. [Google Scholar]
- Jauniaux, E.; Cindrova-Davies, T.; Johns, J.; Dunster, C.; Hempstock, J.; Kelly, F.J.; Burton, G.J. Distribution and transfer pathways of antioxidant molecules inside the first trimester human gestational sac. J. Clin. Endocrinol. Metab. 2004, 89, 1452–1458. [Google Scholar] [CrossRef] [PubMed]
- Kaempf-Rotzoll, D.E.; Igarashi, K.; Aoki, J.; Jishage, K.; Suzuki, H.; Tamai, H.; Linderkamp, O.; Arai, H. Alpha-tocopherol transfer protein is specifically localized at the implantation site of pregnant mouse uterus. Biol. Reprod. 2002, 67, 599–604. [Google Scholar] [CrossRef]
- Garcia, G.R.; Noyes, P.D.; Tanguay, R.L. Advancements in zebrafish applications for 21st century toxicology. Pharmacol. Ther. 2016, 161, 11–21. [Google Scholar] [CrossRef]
- Driever, W.; Solnica-Krezel, L.; Schier, A.F.; Neuhauss, S.C.; Malicki, J.; Stemple, D.L.; Stainier, D.Y.; Zwartkruis, F.; Abdelilah, S.; Rangini, Z.; et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 1996, 123, 37–46. [Google Scholar] [CrossRef]
- Amsterdam, A.; Nissen, R.M.; Sun, Z.; Swindell, E.C.; Farrington, S.; Hopkins, N. Identification of 315 genes essential for early zebrafish development. Proc. Natl. Acad. Sci. USA 2004, 101, 12792–12797. [Google Scholar] [CrossRef]
- Gonsar, N.; Coughlin, A.; Clay-Wright, J.A.; Borg, B.R.; Kindt, L.M.; Liang, J.O. Temporal and spatial requirements for Nodal-induced anterior mesendoderm and mesoderm in anterior neurulation. Genesis 2016, 54, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Kindt, L.M.; Coughlin, A.R.; Perosino, T.R.; Ersfeld, H.N.; Hampton, M.; Liang, J.O. Identification of transcripts potentially involved in neural tube closure using RNA sequencing. Genesis 2018, 56, e23096. [Google Scholar] [CrossRef]
- Ma, P.; Swartz, M.R.; Kindt, L.M.; Kangas, A.M.; Liang, J.O. Temperature Sensitivity of Neural Tube Defects in Zoep Mutants. Zebrafish 2015, 12, 448–456. [Google Scholar] [CrossRef]
- Lee, M.S.; Bonner, J.R.; Bernard, D.J.; Sanchez, E.L.; Sause, E.T.; Prentice, R.R.; Burgess, S.M.; Brody, L.C. Disruption of the folate pathway in zebrafish causes developmental defects. BMC Dev. Biol. 2012, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Fishman, M.C. Zebrafish—The canonical vertebrate. Science 2001, 294, 1290–1291. [Google Scholar] [CrossRef] [PubMed]
- Kirkwood, J.S.; Lebold, K.M.; Miranda, C.L.; Wright, C.L.; Miller, G.W.; Tanguay, R.L.; Barton, C.L.; Traber, M.G.; Stevens, J.F. Vitamin C deficiency activates the purine nucleotide cycle in zebrafish. J. Biol. Chem. 2012, 287, 3833–3841. [Google Scholar] [CrossRef]
- Lebold, K.M.; Lohr, C.V.; Barton, C.L.; Miller, G.W.; Labut, E.M.; Tanguay, R.L.; Traber, M.G. Chronic vitamin E deficiency promotes vitamin C deficiency in zebrafish leading to degenerative myopathy and impaired swimming behavior. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2013, 157, 382–389. [Google Scholar] [CrossRef]
- Lebold, K.M.; Kirkwood, J.S.; Taylor, A.W.; Choi, J.; Barton, C.L.; Miller, G.W.; La Du, J.; Jump, D.B.; Stevens, J.F.; Tanguay, R.L.; et al. Novel liquid chromatography-mass spectrometry method shows that vitamin E deficiency depletes arachidonic and docosahexaenoic acids in zebrafish (Danio rerio) embryos. Redox Biol. 2013, 2, 105–113. [Google Scholar] [CrossRef] [PubMed]
- McDougall, M.; Choi, J.; Kim, H.K.; Bobe, G.; Stevens, J.F.; Cadenas, E.; Tanguay, R.; Traber, M.G. Lethal dysregulation of energy metabolism during embryonic vitamin E deficiency. Free Radic. Biol. Med. 2017, 104, 324–332. [Google Scholar] [CrossRef]
- McDougall, M.; Choi, J.; Kim, H.K.; Bobe, G.; Stevens, J.F.; Cadenas, E.; Tanguay, R.; Traber, M.G. Lipid quantitation and metabolomics data from vitamin E-deficient and -sufficient zebrafish embryos from 0 to 120 hours-post-fertilization. Data Brief 2017, 11, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Head, B.; Leonard, S.W.; Choi, J.; Tanguay, R.L.; Traber, M.G. Vitamin E deficiency dysregulates thiols, amino acids and related molecules during zebrafish embryogenesis. Redox Biol. 2021, 38, 101784. [Google Scholar] [CrossRef] [PubMed]
- Head, B.; Ramsey, S.A.; Kioussi, C.; Tanguay, R.L.; Traber, M.G. Vitamin E Deficiency Disrupts Gene Expression Networks during Zebrafish Development. Nutrients 2021, 13, 468. [Google Scholar] [CrossRef] [PubMed]
- Head, B.; La Du, J.; Tanguay, R.L.; Kioussi, C.; Traber, M.G. Vitamin E is necessary for zebrafish nervous system development. Sci. Rep. 2020, 10, 15028. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.W.; Ulatowski, L.; Labut, E.M.; Lebold, K.M.; Manor, D.; Atkinson, J.; Barton, C.L.; Tanguay, R.L.; Traber, M.G. The alpha-tocopherol transfer protein is essential for vertebrate embryogenesis. PLoS ONE 2012, 7, e47402. [Google Scholar] [CrossRef]
- Bindels, D.S.; Haarbosch, L.; van Weeren, L.; Postma, M.; Wiese, K.E.; Mastop, M.; Aumonier, S.; Gotthard, G.; Royant, A.; Hink, M.A.; et al. mScarlet: A bright monomeric red fluorescent protein for cellular imaging. Nat. Methods 2017, 14, 53–56. [Google Scholar] [CrossRef]
- Velez, M.R. Generation of New Transgenic Zebrafish Lines for Studying Neuronal Circuits Underlying Behavior in Zebrafish. Master in Molecular Genetics and Biomedicine, Universidade Nova de Lisboa, RUN—Repositorio Universidade Nova. Available online: http://hdl.handle.net/10362/58095 (accessed on 18 May 2021).
- Antinucci, P.; Hindges, R. A crystal-clear zebrafish for in vivo imaging. Sci. Rep. 2016, 6, 29490. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Anderson, R.; Stainier, D.Y.; Spradling, A.C.; Halpern, M.E. Transcriptional silencing and reactivation in transgenic zebrafish. Genetics 2009, 182, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Aksoy, Y.A.; Nguyen, D.T.; Chow, S.; Chung, R.S.; Guillemin, G.J.; Cole, N.J.; Hesselson, D. Chemical reprogramming enhances homology-directed genome editing in zebrafish embryos. Commun. Biol. 2019, 2, 198. [Google Scholar] [CrossRef]
- Truong, L.; Reif, D.M.; St Mary, L.; Geier, M.C.; Truong, H.D.; Tanguay, R.L. Multidimensional in vivo hazard assessment using zebrafish. Toxicol. Sci. 2014, 137, 212–233. [Google Scholar] [CrossRef] [PubMed]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 2000, 29, 52–54. [Google Scholar] [CrossRef] [PubMed]
- Jardine, D.; Litvak, M.K. Direct yolk sac volume manipulation of zebrafish embryos and the relationship between offspring size and yolk sac volume. J. Fish Biol. 2003, 63, 388–397. [Google Scholar] [CrossRef]
- Uusi-Heikkila, S.; Wolter, C.; Meinelt, T.; Arlinghaus, R. Size-dependent reproductive success of wild zebrafish Danio rerio in the laboratory. J. Fish Biol. 2010, 77, 552–569. [Google Scholar] [CrossRef]
- Truong, L.; Harper, S.L.; Tanguay, R.L. Evaluation of embryotoxicity using the zebrafish model. Methods Mol. Biol. 2011, 691, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Podda, M.; Weber, C.; Traber, M.G.; Packer, L. Simultaneous determination of tissue tocopherols, tocotrienols, ubiquinols, and ubiquinones. J. Lipid Res. 1996, 37, 893–901. [Google Scholar] [CrossRef]
- Axton, E.R.; Beaver, L.M.; St Mary, L.; Truong, L.; Logan, C.R.; Spagnoli, S.; Prater, M.C.; Keller, R.M.; Garcia-Jaramillo, M.; Ehrlicher, S.E.; et al. Treatment with nitrate, but not nitrite, lowers the oxygen cost of exercise and decreases glycolytic intermediates while increasing fatty acid metabolites in exercised zebrafish. J. Nutr. 2019, 149, 2120–2132. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, S.R.; Li, L.H.; Park, H.J.; Park, J.H.; Lee, K.Y.; Kim, M.K.; Shin, B.A.; Choi, S.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE 2011, 6, e18556. [Google Scholar] [CrossRef]
- Kono, N.; Arai, H. Intracellular transport of fat-soluble vitamins A and E. Traffic 2015, 16, 19–34. [Google Scholar] [CrossRef]
- Panagabko, C.; Morley, S.; Hernandez, M.; Cassolato, P.; Gordon, H.; Parsons, R.; Manor, D.; Atkinson, J. Ligand specificity in the CRAL-TRIO protein family. Biochemistry 2003, 42, 6467–6474. [Google Scholar] [CrossRef] [PubMed]
- Arita, M.; Nomura, K.; Arai, H.; Inoue, K. Alpha-tocopherol transfer protein stimulates the secretion of alpha-tocopherol from a cultured liver cell line through a brefeldin A-insensitive pathway. Proc. Natl. Acad. Sci. USA 1997, 94, 12437–12441. [Google Scholar] [CrossRef] [PubMed]
- Terasawa, Y.; Ladha, Z.; Leonard, S.W.; Morrow, J.D.; Newland, D.; Sanan, D.; Packer, L.; Traber, M.G.; Farese, R.V., Jr. Increased atherosclerosis in hyperlipidemic mice deficient in alpha -tocopherol transfer protein and vitamin E. Proc. Natl. Acad. Sci. USA 2000, 97, 13830–13834. [Google Scholar] [CrossRef]
- Leonard, S.W.; Terasawa, Y.; Farese, R.V., Jr.; Traber, M.G. Incorporation of deuterated RRR- or all-rac-alpha-tocopherol in plasma and tissues of alpha-tocopherol transfer protein–null mice. Am. J. Clin. Nutr. 2002, 75, 555–560. [Google Scholar] [CrossRef]
- Traber, M.G.; Siddens, L.K.; Leonard, S.W.; Schock, B.; Gohil, K.; Krueger, S.K.; Cross, C.E.; Williams, D.E. Alpha-tocopherol modulates Cyp3a expression, increases gamma-CEHC production, and limits tissue gamma-tocopherol accumulation in mice fed high gamma-tocopherol diets. Free Radic. Biol. Med. 2005, 38, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, C.B.; Law, R.D. Cell lineage of zebrafish blastomeres. II. Formation of the yolk syncytial layer. Dev. Biol. 1985, 108, 86–93. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Law, R.D. Cell lineage of zebrafish blastomeres. I. Cleavage pattern and cytoplasmic bridges between cells. Dev. Biol. 1985, 108, 78–85. [Google Scholar] [CrossRef]
- Miyares, R.L.; de Rezende, V.B.; Farber, S.A. Zebrafish yolk lipid processing: A tractable tool for the study of vertebrate lipid transport and metabolism. Dis. Model Mech. 2014, 7, 915–927. [Google Scholar] [CrossRef]
- Quinlivan, V.H.; Farber, S.A. Lipid Uptake, Metabolism, and Transport in the Larval Zebrafish. Front. Endocrinol. (Lausanne) 2017, 8, 319. [Google Scholar] [CrossRef]
- Wilson, M.H.; Rajan, S.; Danoff, A.; White, R.J.; Hensley, M.R.; Quinlivan, V.H.; Recacha, R.; Thierer, J.H.; Tan, F.J.; Busch-Nentwich, E.M.; et al. A point mutation decouples the lipid transfer activities of microsomal triglyceride transfer protein. PLoS Genet 2020, 16, e1008941. [Google Scholar] [CrossRef]
- Schlegel, A.; Stainier, D.Y. Microsomal triglyceride transfer protein is required for yolk lipid utilization and absorption of dietary lipids in zebrafish larvae. Biochemistry 2006, 45, 15179–15187. [Google Scholar] [CrossRef]
- Marza, E.; Barthe, C.; Andre, M.; Villeneuve, L.; Helou, C.; Babin, P.J. Developmental expression and nutritional regulation of a zebrafish gene homologous to mammalian microsomal triglyceride transfer protein large subunit. Dev. Dyn. 2005, 232, 506–518. [Google Scholar] [CrossRef]
- Raabe, M.; Kim, E.; Veniant, M.; Nielsen, L.B.; Young, S.G. Using genetically engineered mice to understand apolipoprotein-B deficiency syndromes in humans. Proc. Assoc. Am. Physicians 1998, 110, 521–530. [Google Scholar] [PubMed]
- Raabe, M.; Flynn, L.M.; Zlot, C.H.; Wong, J.S.; Veniant, M.M.; Hamilton, R.L.; Young, S.G. Knockout of the abetalipoproteinemia gene in mice: Reduced lipoprotein secretion in heterozygotes and embryonic lethality in homozygotes. Proc. Natl. Acad. Sci. USA 1998, 95, 8686–8691. [Google Scholar] [CrossRef]
- Rader, D.J.; Brewer, H.B., Jr. Abetalipoproteinemia. New insights into lipoprotein assembly and vitamin E metabolism from a rare genetic disease. JAMA 1993, 270, 865–869. [Google Scholar] [CrossRef]
- Louwette, S.; Regal, L.; Wittevrongel, C.; Thys, C.; Vandeweeghde, G.; Decuyper, E.; Leemans, P.; De Vos, R.; Van Geet, C.; Jaeken, J.; et al. NPC1 defect results in abnormal platelet formation and function: Studies in Niemann-Pick disease type C1 patients and zebrafish. Hum. Mol. Genet 2013, 22, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, T.; Kikuchi, Y.; Kuroiwa, A.; Takeda, H.; Stainier, D.Y. The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development 2006, 133, 4063–4072. [Google Scholar] [CrossRef]
- Tseng, W.C.; Loeb, H.E.; Pei, W.; Tsai-Morris, C.H.; Xu, L.; Cluzeau, C.V.; Wassif, C.A.; Feldman, B.; Burgess, S.M.; Pavan, W.J.; et al. Modeling Niemann-Pick disease type C1 in zebrafish: A robust platform for in vivo screening of candidate therapeutic compounds. Dis. Model Mech. 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Ulatowski, L.; Parker, R.; Davidson, C.; Yanjanin, N.; Kelley, T.J.; Corey, D.; Atkinson, J.; Porter, F.; Arai, H.; Walkley, S.U.; et al. Altered vitamin E status in Niemann-Pick type C disease. J. Lipid Res. 2011, 52, 1400–1410. [Google Scholar] [CrossRef] [PubMed]
- Yevenes, L.F.; Klein, A.; Castro, J.F.; Marin, T.; Leal, N.; Leighton, F.; Alvarez, A.R.; Zanlungo, S. Lysosomal vitamin E accumulation in Niemann-Pick type C disease. Biochim. Biophys. Acta 2012, 1822, 150–160. [Google Scholar] [CrossRef]
- Narushima, K.; Takada, T.; Yamanashi, Y.; Suzuki, H. Niemann-pick C1-like 1 mediates alpha-tocopherol transport. Mol. Pharmacol. 2008, 74, 42–49. [Google Scholar] [CrossRef]
- Reboul, E.; Soayfane, Z.; Goncalves, A.; Cantiello, M.; Bott, R.; Nauze, M.; Terce, F.; Collet, X.; Comera, C. Respective contributions of intestinal Niemann-Pick C1-like 1 and scavenger receptor class B type I to cholesterol and tocopherol uptake: In vivo v. in vitro studies. Br. J. Nutr. 2012, 107, 1296–1304. [Google Scholar] [CrossRef]
- Shi, X.; Teo, L.S.; Pan, X.; Chong, S.W.; Kraut, R.; Korzh, V.; Wohland, T. Probing events with single molecule sensitivity in zebrafish and Drosophila embryos by fluorescence correlation spectroscopy. Dev. Dyn. 2009, 238, 3156–3167. [Google Scholar] [CrossRef] [PubMed]
- Fechner, H.; Schlame, M.; Guthmann, F.; Stevens, P.A.; Rustow, B. alpha- and delta-tocopherol induce expression of hepatic alpha-tocopherol-transfer-protein mRNA. Biochem. J. 1998, 331 Pt 2, 577–581. [Google Scholar] [CrossRef]
- Thakur, V.; Morley, S.; Manor, D. Hepatic alpha-tocopherol transfer protein: Ligand-induced protection from proteasomal degradation. Biochemistry 2010, 49, 9339–9344. [Google Scholar] [CrossRef]
- Qian, J.; Morley, S.; Wilson, K.; Nava, P.; Atkinson, J.; Manor, D. Intracellular trafficking of vitamin E in hepatocytes: The role of tocopherol transfer protein. J. Lipid Res. 2005, 46, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Taormina, M.J.; Hay, E.A.; Parthasarathy, R. Passive and Active Microrheology of the Intestinal Fluid of the Larval Zebrafish. Biophys. J. 2017, 113, 957–965. [Google Scholar] [CrossRef] [PubMed]
- Whipps, C.M.; Moss, L.G.; Sisk, D.M.; Murray, K.N.; Tobin, D.M.; Moss, J.B. Detection of autofluorescent Mycobacterium chelonae in living zebrafish. Zebrafish 2014, 11, 76–82. [Google Scholar] [CrossRef]
- Fukui, K.; Nakamura, K.; Shirai, M.; Hirano, A.; Takatsu, H.; Urano, S. Long-term vitamin E-deficient mice exhibit cognitive dysfunction via elevation of brain oxidation. J. Nutr. Sci. Vitaminol. (Tokyo) 2015, 61, 362–368. [Google Scholar] [CrossRef]
- McDougall, M.; Choi, J.; Truong, L.; Tanguay, R.; Traber, M.G. Vitamin E deficiency during embryogenesis in zebrafish causes lasting metabolic and cognitive impairments despite refeeding adequate diets. Free Radic. Biol. Med. 2017, 110, 250–260. [Google Scholar] [CrossRef]
- Hosomi, A.; Goto, K.; Kondo, H.; Iwatsubo, T.; Yokota, T.; Ogawa, M.; Arita, M.; Aoki, J.; Arai, H.; Inoue, K. Localization of alpha-tocopherol transfer protein in rat brain. Neurosci. Lett. 1998, 256, 159–162. [Google Scholar] [CrossRef]
- Yokota, T.; Igarashi, K.; Uchihara, T.; Jishage, K.; Tomita, H.; Inaba, A.; Li, Y.; Arita, M.; Suzuki, H.; Mizusawa, H.; et al. Delayed-onset ataxia in mice lacking alpha -tocopherol transfer protein: Model for neuronal degeneration caused by chronic oxidative stress. Proc. Natl. Acad. Sci. USA 2001, 98, 15185–15190. [Google Scholar] [CrossRef] [PubMed]
- Finno, C.J.; Bordbari, M.H.; Gianino, G.; Ming-Whitfield, B.; Burns, E.; Merkel, J.; Britton, M.; Durbin-Johnson, B.; Sloma, E.A.; McMackin, M.; et al. An innate immune response and altered nuclear receptor activation defines the spinal cord transcriptome during alpha-tocopherol deficiency in Ttpa-null mice. Free Radic. Biol. Med. 2018, 120, 289–302. [Google Scholar] [CrossRef] [PubMed]
- Finno, C.J.; Peterson, J.; Kang, M.; Park, S.; Bordbari, M.H.; Durbin-Johnson, B.; Settles, M.; Perez-Flores, M.C.; Lee, J.H.; Yamoah, E.N. Single-cell RNA-seq reveals profound alterations in mechanosensitive dorsal root ganglion neurons with vitamin e deficiency. iScience 2019, 21, 720–735. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, O.; Wall, J.B.J.; Zheng, M.; Zhou, Y.; Wang, L.; Vaseghi, H.R.; Qian, L.; Liu, J. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 2017, 7, 2193. [Google Scholar] [CrossRef]
- Cherian, G.; Sim, J.S. Egg yolk polyunsaturated fatty acids and vitamin E content alters the tocopherol status of hatched chicks. Poult. Sci. 1997, 76, 1753–1759. [Google Scholar] [CrossRef] [PubMed]
- McDougall, M.; Choi, J.; Magnusson, K.; Truong, L.; Tanguay, R.; Traber, M.G. Chronic vitamin E deficiency impairs cognitive function in adult zebrafish via dysregulation of brain lipids and energy metabolism. Free Radic. Biol. Med. 2017, 112, 308–317. [Google Scholar] [CrossRef]
- Miller, G.W.; Labut, E.M.; Lebold, K.M.; Floeter, A.; Tanguay, R.L.; Traber, M.G. Zebrafish (Danio rerio) fed vitamin E-deficient diets produce embryos with increased morphologic abnormalities and mortality. J. Nutr. Biochem. 2012, 23, 478–486. [Google Scholar] [CrossRef]


| Primer Purpose | Sequence (5′-3′) | Product Size (bp) |
|---|---|---|
| Left homology arm F | AGGCGTGTGTCTCTGTAAGG | 418 |
| Left homology arm R | GCCGTACATGAACTGAGGGG | |
| Right homology arm F | CGCTACCTGGCGGACTTC | 309 |
| Right homology arm R | AATCCGGGAGGAATCAACAGG | |
| mScarlet cassette F | CGCTCTGAGAACAACATGACAC | 121 no insertion 879 insertion |
| mScarlet cassette R | GACAAATATGGTGCAATCCGGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Head, B.; La Du, J.; Barton, C.; Zhang, J.; Wong, C.; Ho, E.; Tanguay, R.L.; Traber, M.G. RedEfish: Generation of the Polycistronic mScarlet: GSG-T2A: Ttpa Zebrafish Line. Antioxidants 2021, 10, 965. https://doi.org/10.3390/antiox10060965
Head B, La Du J, Barton C, Zhang J, Wong C, Ho E, Tanguay RL, Traber MG. RedEfish: Generation of the Polycistronic mScarlet: GSG-T2A: Ttpa Zebrafish Line. Antioxidants. 2021; 10(6):965. https://doi.org/10.3390/antiox10060965
Chicago/Turabian StyleHead, Brian, Jane La Du, Carrie Barton, Jie Zhang, Carmen Wong, Emily Ho, Robyn L. Tanguay, and Maret G. Traber. 2021. "RedEfish: Generation of the Polycistronic mScarlet: GSG-T2A: Ttpa Zebrafish Line" Antioxidants 10, no. 6: 965. https://doi.org/10.3390/antiox10060965
APA StyleHead, B., La Du, J., Barton, C., Zhang, J., Wong, C., Ho, E., Tanguay, R. L., & Traber, M. G. (2021). RedEfish: Generation of the Polycistronic mScarlet: GSG-T2A: Ttpa Zebrafish Line. Antioxidants, 10(6), 965. https://doi.org/10.3390/antiox10060965








