Distinction between 2′- and 3′-Phosphate Isomers of a Fluorescent NADPH Analogue Led to Strong Inhibition of Cancer Cells Migration
Abstract
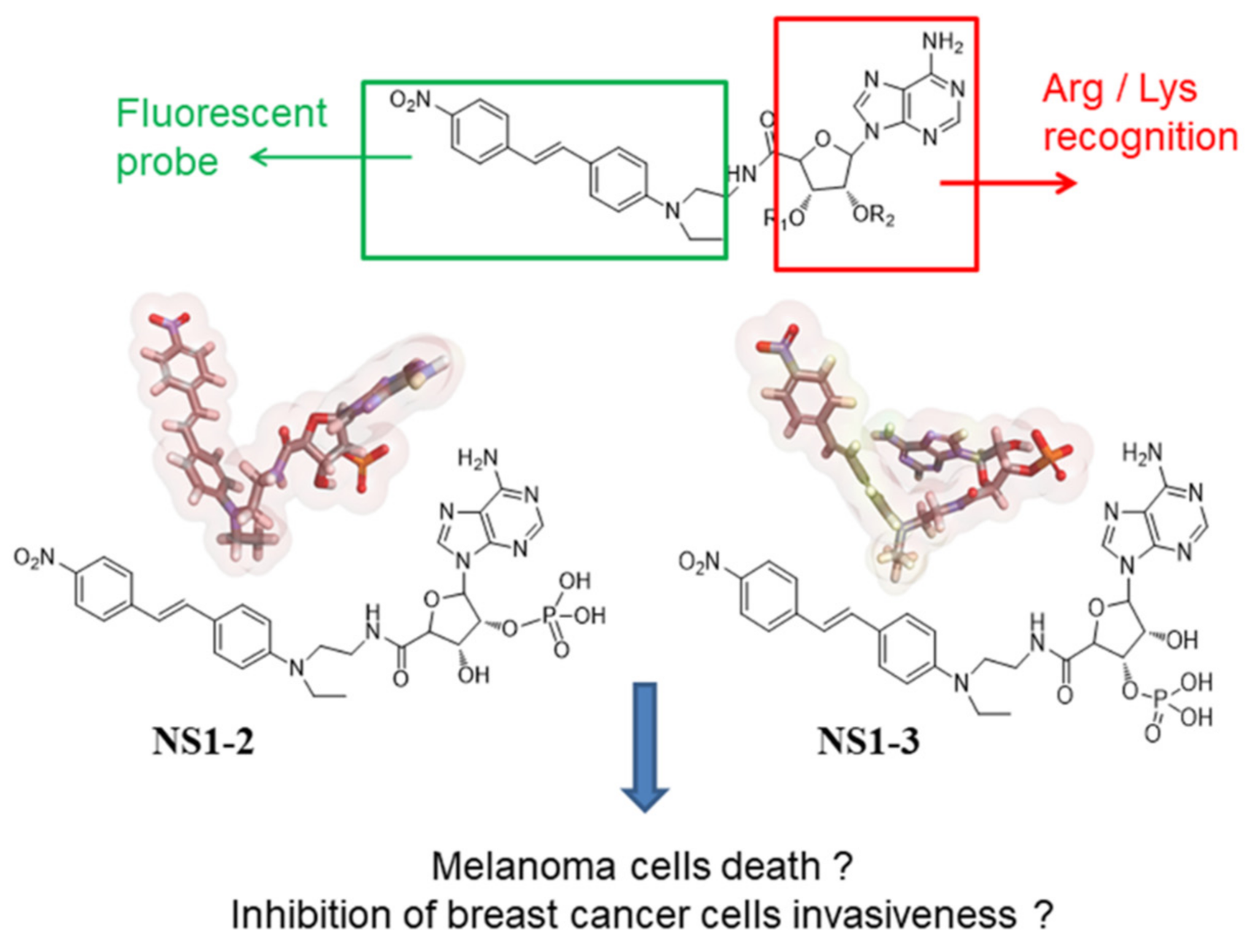
1. Introduction
2. Experimental Procedures
2.1. General Methods
2.2. Materials
2.3. Recombinant NOX2 C-Terminal Expression and Purification
2.4. Synthesis of NS1 and Separation of Isomers
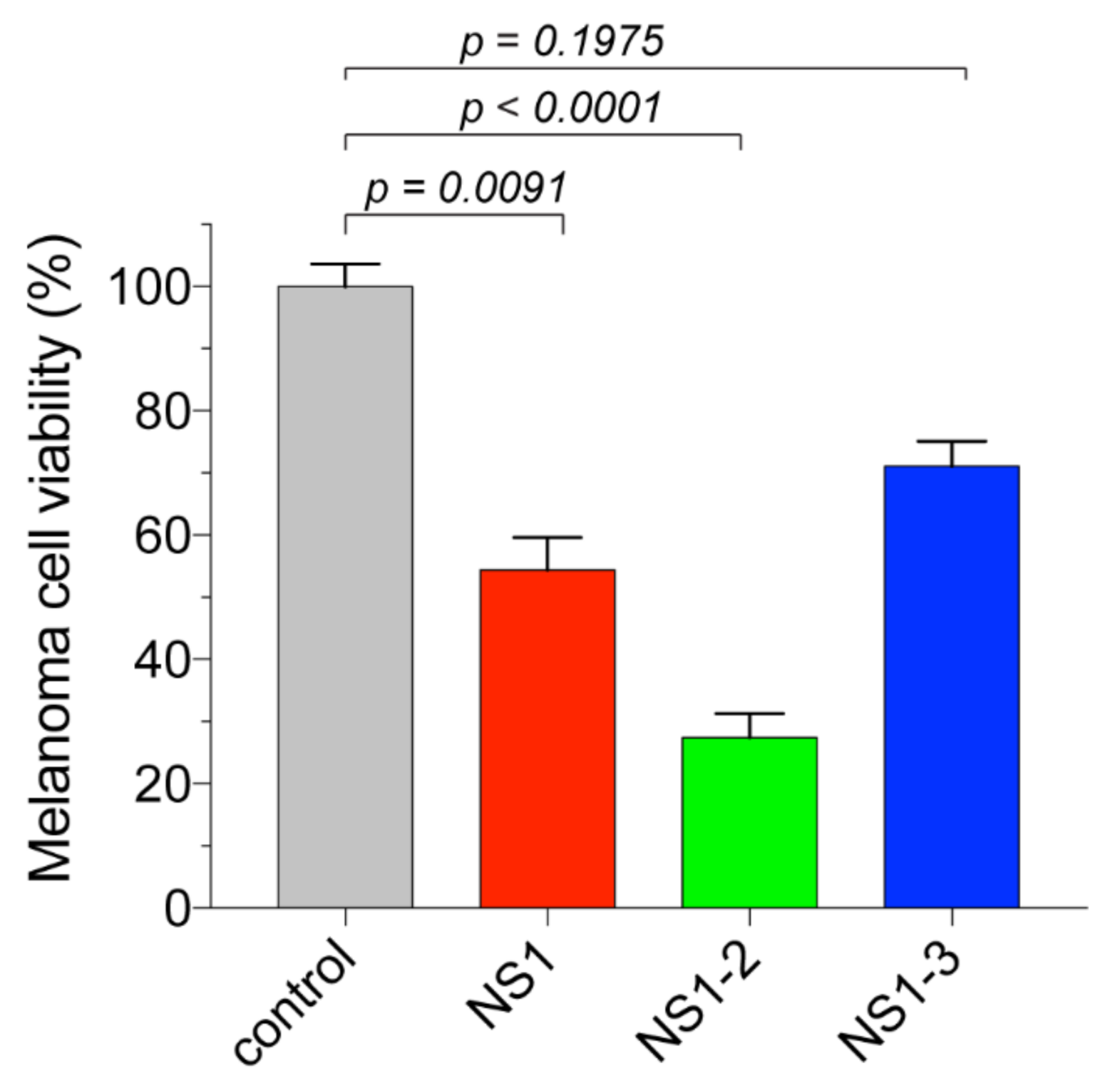
2.5. Viability of Melanoma Cells
2.6. Viability of Breast Cells, Multiple Myeloma and Colon Cancer Cells
2.7. Melanoma Cell Migration in a Boyden Chamber
2.8. Breast Cancer Cells Migration
2.9. Statistical Analysis
2.10. Effect of NS1-2 and NS1-3 on eNOS and nNOS Activity
2.11. Effect of NS1-2 or NS1-3 on Recombinant NOX2 C-Terminal Activity Determined by Fluorescence Measurements
2.12. Docking and MD Simulations
2.13. Molecular Docking of the Ligand (NS1) to NADPH Oxidase-2 NOX2 (gp91phox DH Domain) and Molecular Dynamics (MD) Simulations of the Resulting Docked Complex
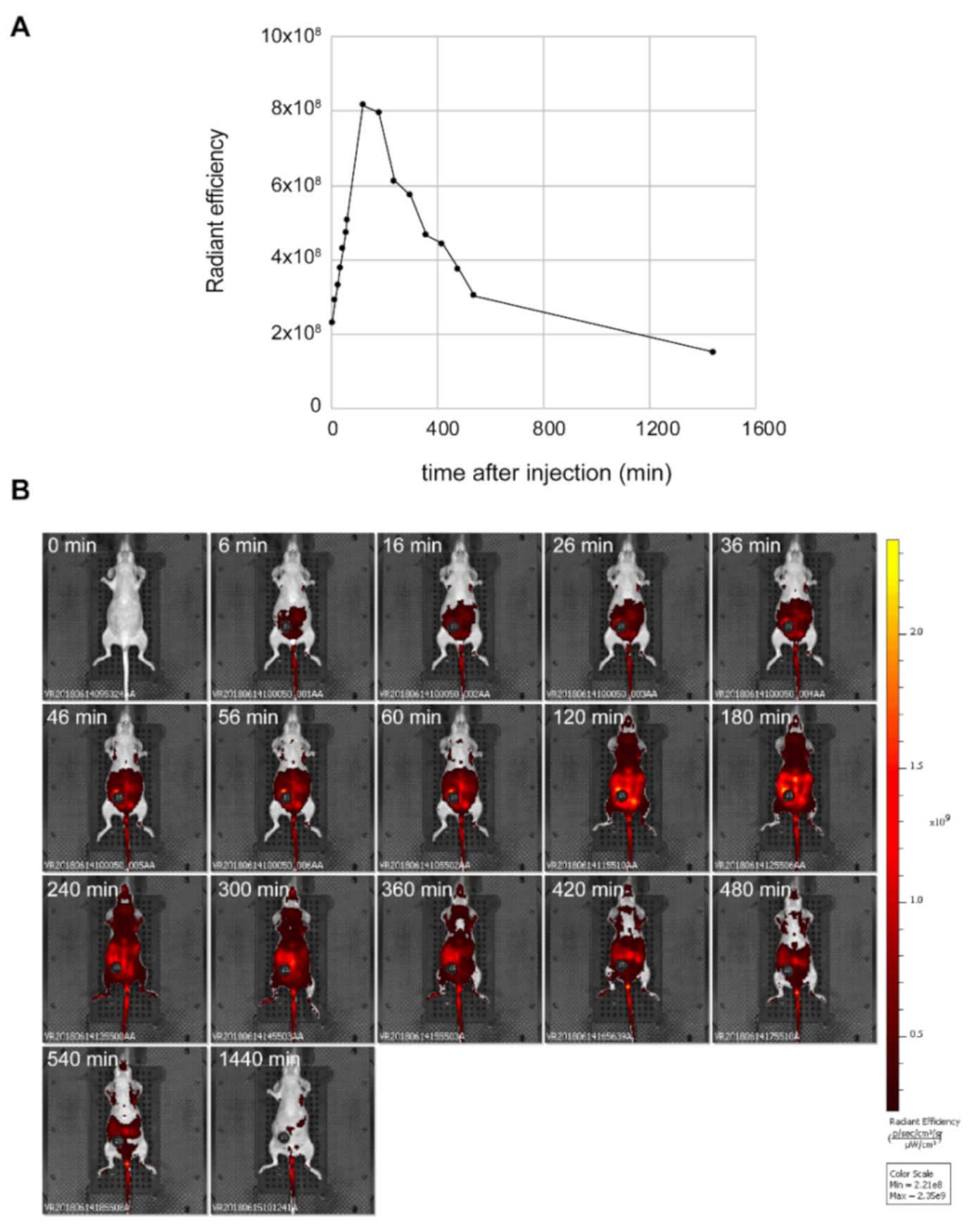
2.14. In Vivo Experiments
2.15. Western Blots
3. Results and Discussion
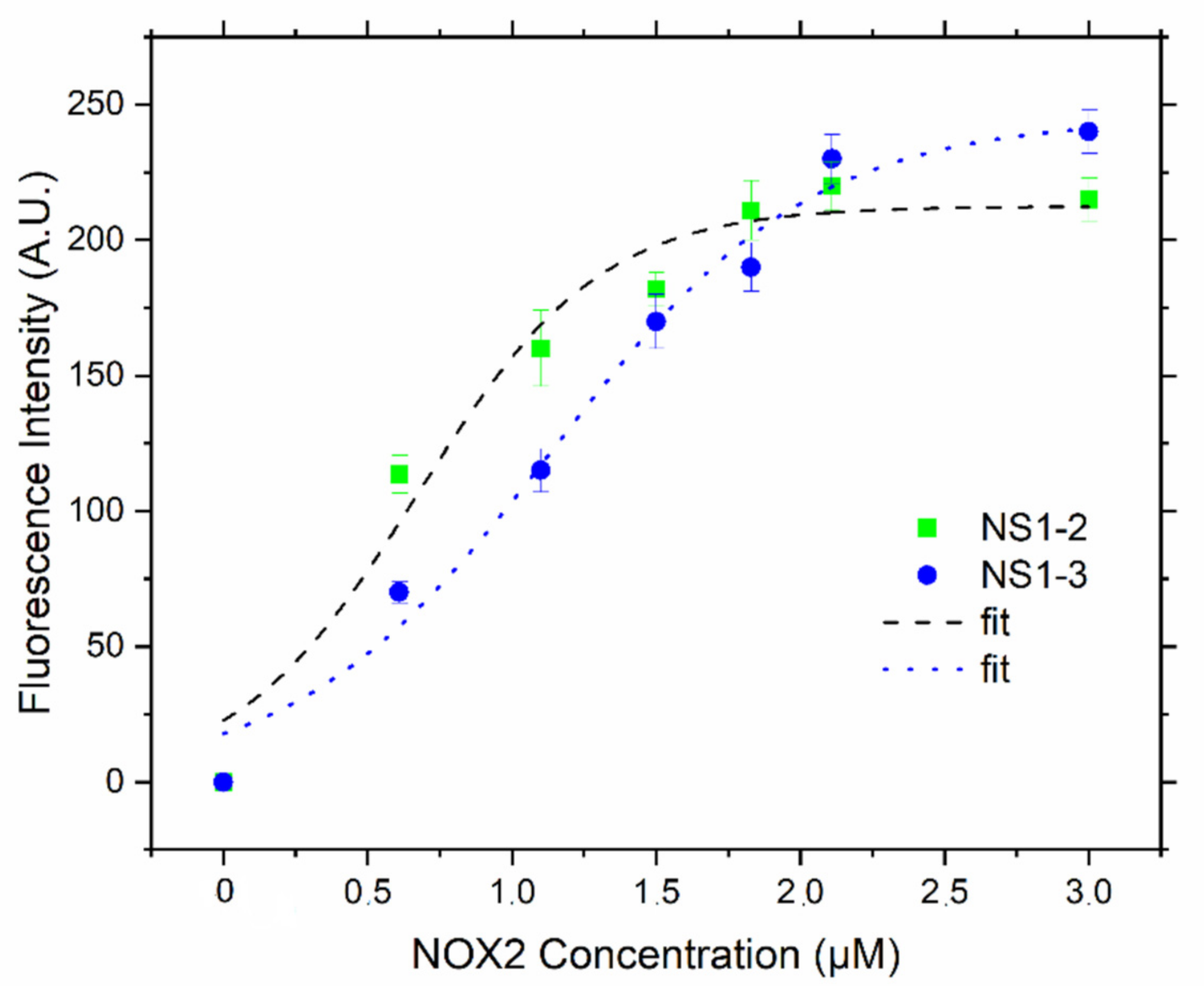
3.1. NS1-2 and NS1-3 as Fluorescence Probes In Vitro
3.2. Fluorescence Properties of NS1 In Vivo
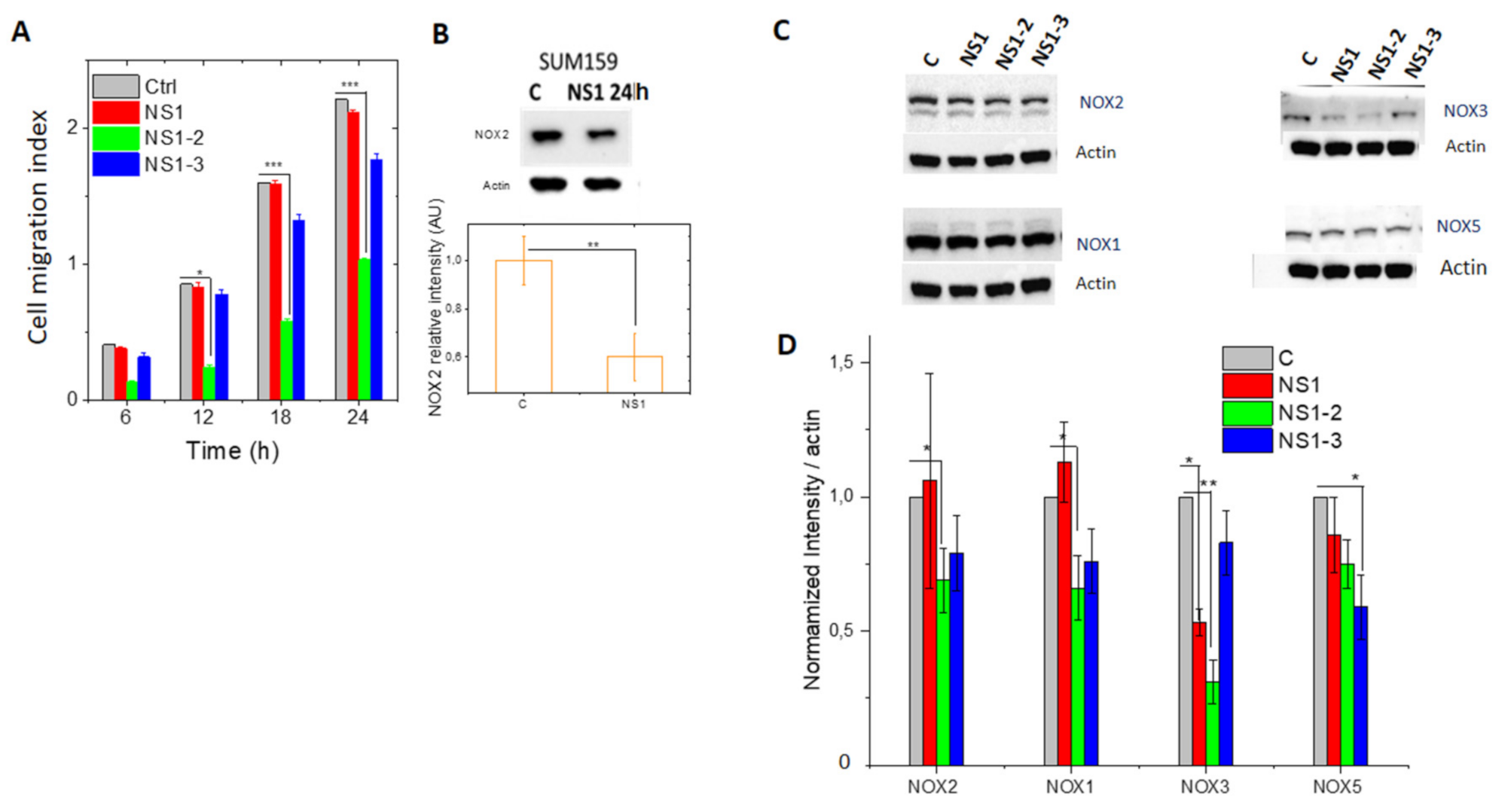
3.3. Cellular Studies
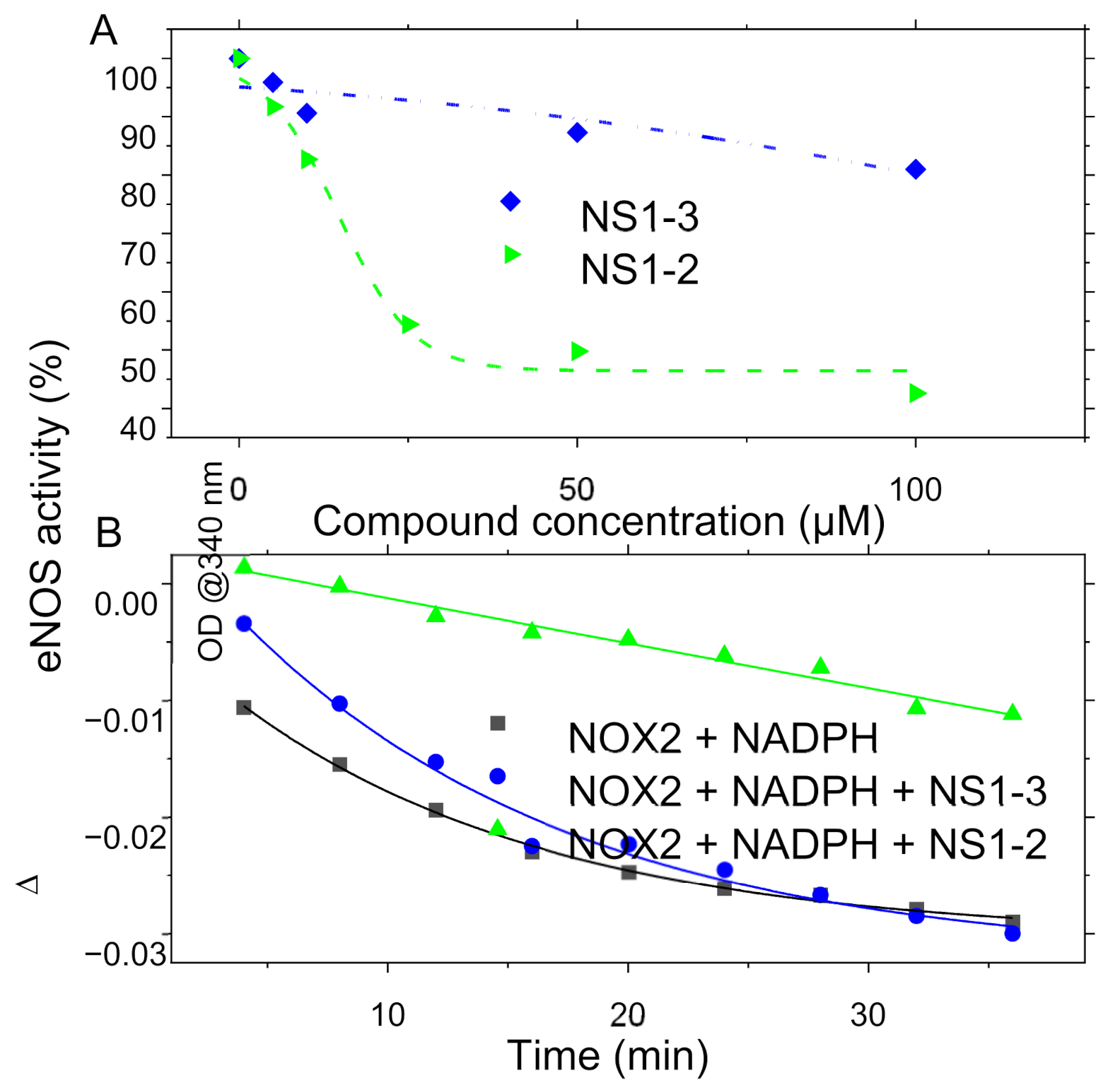
3.4. In Vitro Recombinant Enzymatic Activities Measurements
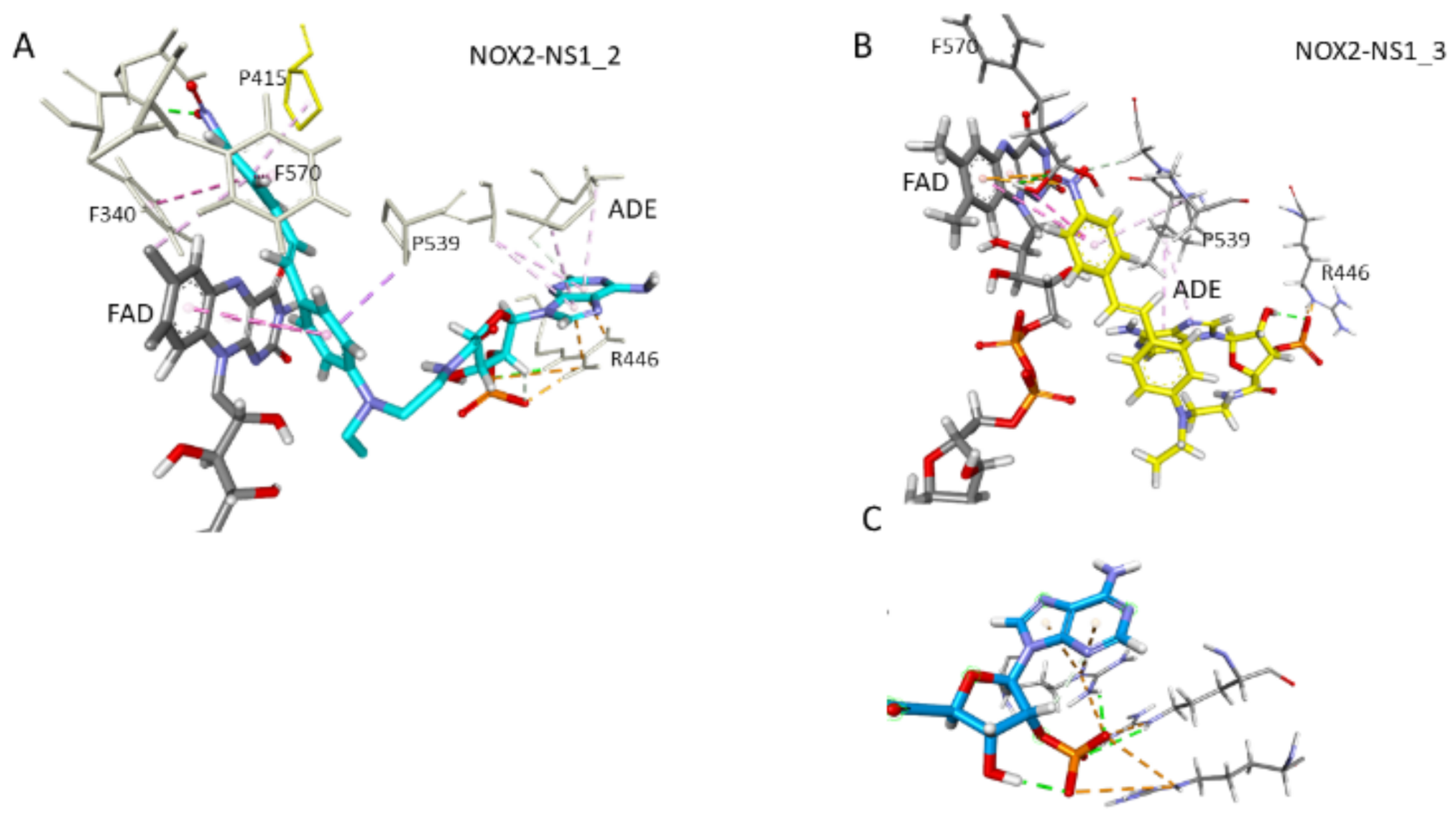
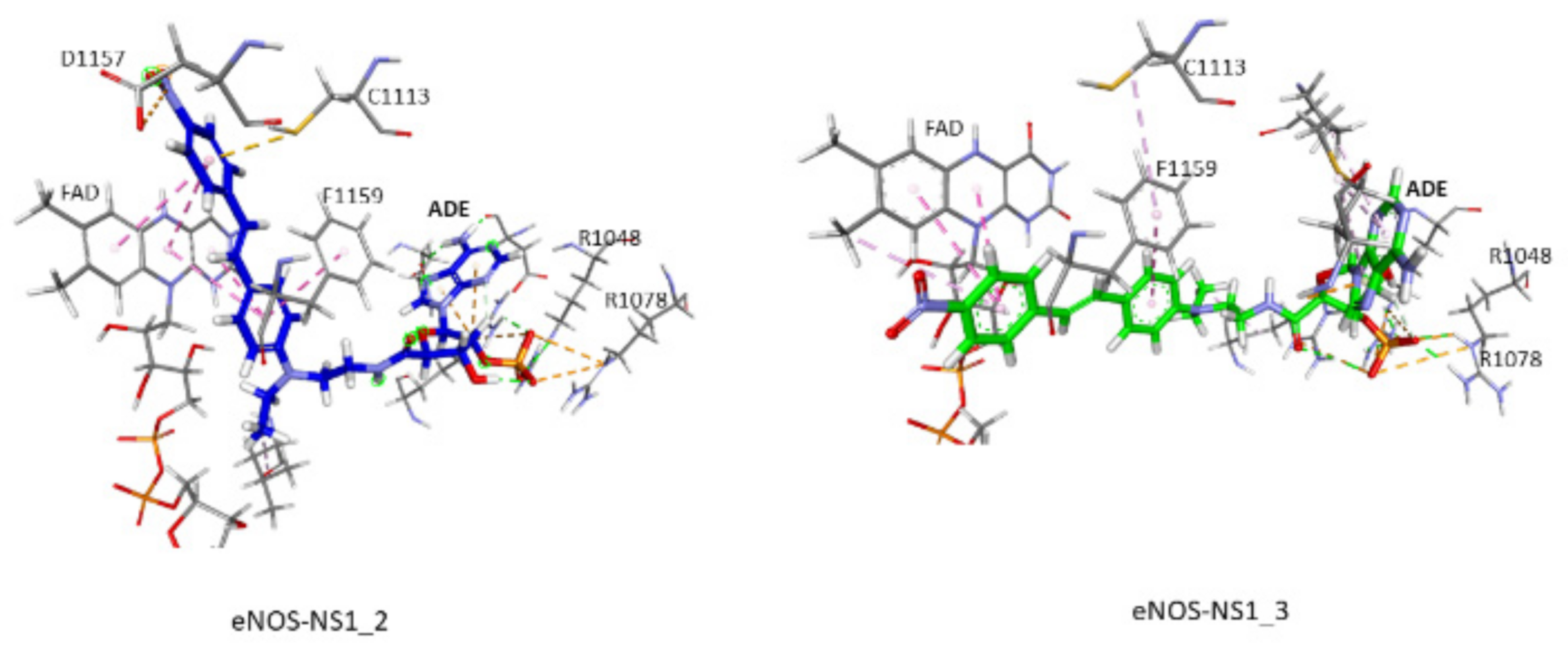
3.5. Docking and MD Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FAD | Flavin adenine dinucleotide |
| L-NAME | N(ω)-nitro-L-arginine methyl ester |
| MD | molecular dynamics |
| NOX | NADPH oxidase |
| NOS | NO-synthase |
| NS1 | nanoshutter 1 [11] |
References
- Cheng, H.; Wang, L.; Mollica, M.; Re, A.T.; Wu, S.; Zuo, L. Nitric oxide in cancer metastasis. Cancer Lett. 2014, 353, 1–7. [Google Scholar] [CrossRef]
- Lahdenranta, J.; Hagendoorn, J.; Padera, T.P.; Hoshida, T.; Nelson, G.; Kashiwagi, S.; Jain, R.K.; Fukumura, D. Endothelial nitric oxide synthase mediates lymphangiogenesis and lymphatic metastasis. Cancer Res. 2009, 69, 2801–2808. [Google Scholar] [CrossRef]
- Liu-Smith, F.; Dellinger, R.; Meyskens, F.L., Jr. Updates of reactive oxygen species in melanoma etiology and progression. Arch. Biochem. Biophys. 2014, 563, 51–55. [Google Scholar] [CrossRef] [PubMed]
- Parascandolo, A.; Laukkanen, M.O. Carcinogenesis and Reactive Oxygen Species Signaling: Interaction of the NADPH Oxidase NOX1-5 and Superoxide Dismutase 1–3 Signal Transduction Pathways. Antioxid. Redox Signal. 2019, 30, 443–486. [Google Scholar] [CrossRef] [PubMed]
- Scharping, N.E.; Rivadeneira, D.B.; Menk, A.V.; Vignali, P.D.A.; Ford, B.R.; Rittenhouse, N.L.; Peralta, R.; Wang, Y.; Wang, Y.; De Peaux, K.; et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat. Immunol. 2021, 22, 205–215. [Google Scholar] [CrossRef]
- Rouaud, F.; Boucher, J.L.; Slama-Schwok, A.; Rocchi, S. Mechanism of melanoma cells selective apoptosis induced by a photoactive NADPH analogue. Oncotarget 2016, 7, 82804–82819. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Zhang, Y.; Lin, H.; Kang, S.; Li, Y.; Sun, D.; Chen, M.; Wang, Z.; Jiao, Z.; Wang, Y.; et al. Ultrasound and Near-Infrared Light Dual-Triggered Upconversion Zeolite-Based Nanocomposite for Hyperthermia-Enhanced Multimodal Melanoma Therapy via a Precise Apoptotic Mechanism. ACS Appl. Mater. Interfaces 2020, 12, 32420–32431. [Google Scholar] [CrossRef] [PubMed]
- Panday, A.; Sahoo, M.K.; Osorio, D.; Batra, S. NADPH oxidases: An overview from structure to innate immunity-associated pathologies. Cell Mol. Immunol. 2015, 12, 5–23. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; Wu, Y.; Meitzler, J.L.; Juhasz, A.; Liu, H.; Jiang, G.; Lu, J.; Antony, S.; Doroshow, J.H. NADPH oxidases and cancer. Clin. Sci. (Lond.) 2015, 128, 863–875. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Tarus, B.; Perez, M.R.; Morellato, L.; Henry, E.; Berka, V.; Tsai, A.L.; Ramassamy, B.; Dhimane, H.; et al. Rational design of a fluorescent NADPH derivative imaging constitutive nitric-oxide synthases upon two-photon excitation. Proc. Natl. Acad. Sci. USA 2012, 109, 12526–12531. [Google Scholar] [CrossRef] [PubMed]
- Rouaud, F.; Romero-Perez, M.; Wang, H.; Lobysheva, I.; Ramassamy, B.; Henry, E.; Tauc, P.; Giacchero, D.; Boucher, J.L.; Deprez, E.; et al. Regulation of NADPH-dependent Nitric Oxide and reactive oxygen species signalling in endothelial and melanoma cells by a photoactive NADPH analogue. Oncotarget 2014, 5, 10650–10664. [Google Scholar] [CrossRef]
- Lejal, N.; Truchet, S.; Bechor, E.; Bouguyon, E.; Khedkar, V.; Bertho, N.; Vidic, J.; Adenot, P.; Solier, S.; Pick, E.; et al. Turning off NADPH oxidase-2 by impeding p67(phox) activation in infected mouse macrophages reduced viral entry and inflammation. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Moali, C.; Boucher, J.L.; Sari, M.A.; Stuehr, D.J.; Mansuy, D. Substrate specificity of NO synthases: Detailed comparison of L-arginine, homo-L-arginine, their N omega-hydroxy derivatives, and N omega-hydroxynor-L-arginine. Biochemistry 1998, 37, 10453–10460. [Google Scholar] [CrossRef] [PubMed]
- Hevel, J.M.; Marletta, M.A. Nitric-oxide synthase assays. Methods Enzymol. 1994, 233, 250–258. [Google Scholar] [CrossRef]
- Lambry, J.C.; Beaumont, E.; Tarus, B.; Blanchard-Desce, M.; Slama-Schwok, A. Selective probing of a NADPH site controlled light-induced enzymatic catalysis. J. Mol. Recognit. 2010, 23, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Garcin, E.D.; Bruns, C.M.; Lloyd, S.J.; Hosfield, D.J.; Tiso, M.; Gachhui, R.; Stuehr, D.J.; Tainer, J.A.; Getzoff, E.D. Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase. J. Biol. Chem. 2004, 279, 37918–37927. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Open Source Drug Discovery, C.; Lynn, A. g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Gorin, Y.; Block, K. Nox4 and diabetic nephropathy: With a friend like this, who needs enemies? Free Radic. Biol. Med. 2013, 61, 130–142. [Google Scholar] [CrossRef]
- Plecita-Hlavata, L.; Jaburek, M.; Holendova, B.; Tauber, J.; Pavluch, V.; Berkova, Z.; Cahova, M.; Schroder, K.; Brandes, R.P.; Siemen, D.; et al. Glucose-Stimulated Insulin Secretion Fundamentally Requires H2O2 Signaling by NADPH Oxidase 4. Diabetes 2020, 69, 1341–1354. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Li, B.; Brun, T.; Deffert-Delbouille, C.; Mahiout, Z.; Daali, Y.; Ma, X.J.; Krause, K.H.; Maechler, P. NADPH oxidase NOX2 defines a new antagonistic role for reactive oxygen species and cAMP/PKA in the regulation of insulin secretion. Diabetes 2012, 61, 2842–2850. [Google Scholar] [CrossRef]
- Smeda, M.; Kieronska, A.; Adamski, M.G.; Proniewski, B.; Sternak, M.; Mohaissen, T.; Przyborowski, K.; Derszniak, K.; Kaczor, D.; Stojak, M.; et al. Nitric oxide deficiency and endothelial-mesenchymal transition of pulmonary endothelium in the progression of 4T1 metastatic breast cancer in mice. Breast Cancer Res. 2018, 20, 86. [Google Scholar] [CrossRef] [PubMed]
- Zaoui, M.; Morel, M.; Ferrand, N.; Fellahi, S.; Bastard, J.P.; Lamaziere, A.; Larsen, A.K.; Bereziat, V.; Atlan, M.; Sabbah, M. Breast-Associated Adipocytes Secretome Induce Fatty Acid Uptake and Invasiveness in Breast Cancer Cells via CD36 Independently of Body Mass Index, Menopausal Status and Mammary Density. Cancers (Basel) 2019, 11, 2012. [Google Scholar] [CrossRef] [PubMed]
- Magnani, F.; Nenci, S.; Millana Fananas, E.; Ceccon, M.; Romero, E.; Fraaije, M.W.; Mattevi, A. Crystal structures and atomic model of NADPH oxidase. Proc. Natl. Acad. Sci. USA 2017, 114, 6764–6769. [Google Scholar] [CrossRef]
- Beaumont, E.; Lambry, J.C.; Gautier, C.; Robin, A.C.; Gmouh, S.; Berka, V.; Tsai, A.L.; Blanchard-Desce, M.; Slama-Schwok, A. Synchronous photoinitiation of endothelial NO synthase activity by a nanotrigger targeted at its NADPH site. J. Am. Chem. Soc. 2007, 129, 2178–2186. [Google Scholar] [CrossRef]
- Dilly, S.; Roman, L.J.; Bogliotti, N.; Xie, J.; Deprez, E.; Slama-Schwok, A. Design of Light-Sensitive Triggers for Endothelial NO-Synthase Activation. Antioxidants (Basel) 2020, 9, 89. [Google Scholar] [CrossRef] [PubMed]
- Laporte, F.; Doussiere, J.; Vignais, P.V. Characterization of multiple active forms of the NADPH dehydrogenase component of the oxidase complex from rabbit peritoneal neutrophils by photolabeling with an arylazido derivative of NADP+. Biochem. Biophys. Res. Commun. 1990, 168, 78–84. [Google Scholar] [CrossRef]
- Chen, K.; Craige, S.E.; Keaney, J.F., Jr. Downstream targets and intracellular compartmentalization in Nox signaling. Antioxid. Redox Signal. 2009, 11, 2467–2480. [Google Scholar] [CrossRef] [PubMed]
- Laothamatas, I.; Gao, P.; Wickramaratne, A.; Quintanilla, C.G.; Dino, A.; Khan, C.A.; Liou, J.; Green, C.B. Spatiotemporal regulation of NADP(H) phosphatase Nocturnin and its role in oxidative stress response. Proc. Natl. Acad. Sci. USA 2020, 117, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, C.; Kawai, S.; Murata, K. NADP(H) phosphatase activities of archaeal inositol monophosphatase and eubacterial 3’-phosphoadenosine 5’-phosphate phosphatase. Appl. Environ. Microbiol. 2007, 73, 5447–5452. [Google Scholar] [CrossRef]
- Cahn, J.K.; Werlang, C.A.; Baumschlager, A.; Brinkmann-Chen, S.; Mayo, S.L.; Arnold, F.H. A General Tool for Engineering the NAD/NADP Cofactor Preference of Oxidoreductases. ACS Synth. Biol. 2017, 6, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Buchwalow, I.; Schnekenburger, J.; Samoilova, V.; Boecker, W.; Neumann, J.; Tiemann, K. New Insight into the Role of Nitric Oxide Pathways in Pancreas. Acta Histochem. Cytochem. 2018, 51, 167–172. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manuel, R.; Lima, M.d.S.; Dilly, S.; Daunay, S.; Abbe, P.; Pramil, E.; Solier, S.; Guillaumond, F.; Tubiana, S.-S.; Escargueil, A.; et al. Distinction between 2′- and 3′-Phosphate Isomers of a Fluorescent NADPH Analogue Led to Strong Inhibition of Cancer Cells Migration. Antioxidants 2021, 10, 723. https://doi.org/10.3390/antiox10050723
Manuel R, Lima MdS, Dilly S, Daunay S, Abbe P, Pramil E, Solier S, Guillaumond F, Tubiana S-S, Escargueil A, et al. Distinction between 2′- and 3′-Phosphate Isomers of a Fluorescent NADPH Analogue Led to Strong Inhibition of Cancer Cells Migration. Antioxidants. 2021; 10(5):723. https://doi.org/10.3390/antiox10050723
Chicago/Turabian StyleManuel, Raoul, Michelle de Souza Lima, Sébastien Dilly, Sylvain Daunay, Patricia Abbe, Elodie Pramil, Stéphanie Solier, Fabienne Guillaumond, Sarah-Simha Tubiana, Alexandre Escargueil, and et al. 2021. "Distinction between 2′- and 3′-Phosphate Isomers of a Fluorescent NADPH Analogue Led to Strong Inhibition of Cancer Cells Migration" Antioxidants 10, no. 5: 723. https://doi.org/10.3390/antiox10050723
APA StyleManuel, R., Lima, M. d. S., Dilly, S., Daunay, S., Abbe, P., Pramil, E., Solier, S., Guillaumond, F., Tubiana, S.-S., Escargueil, A., Pêgas Henriques, J. A., Ferrand, N., Erdelmeier, I., Boucher, J.-L., Bertho, G., Agranat, I., Rocchi, S., Sabbah, M., & Slama Schwok, A. (2021). Distinction between 2′- and 3′-Phosphate Isomers of a Fluorescent NADPH Analogue Led to Strong Inhibition of Cancer Cells Migration. Antioxidants, 10(5), 723. https://doi.org/10.3390/antiox10050723








