Enzyme-Assisted Release of Antioxidant Peptides from Porphyra dioica Conchocelis
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Algal Biomass
2.3. Determination of Nitrogenous Composition
2.4. Hydrolysate Generation
2.5. Characterization of the Hydrolysates
2.6. Total and Free Amino Acid Analysis
2.7. In Vitro Antioxidant Bioassays
2.8. Statistical Analysis
3. Results and Discussion
3.1. Protein Composition, Amino Acid Profile and Extent of Hydrolysis
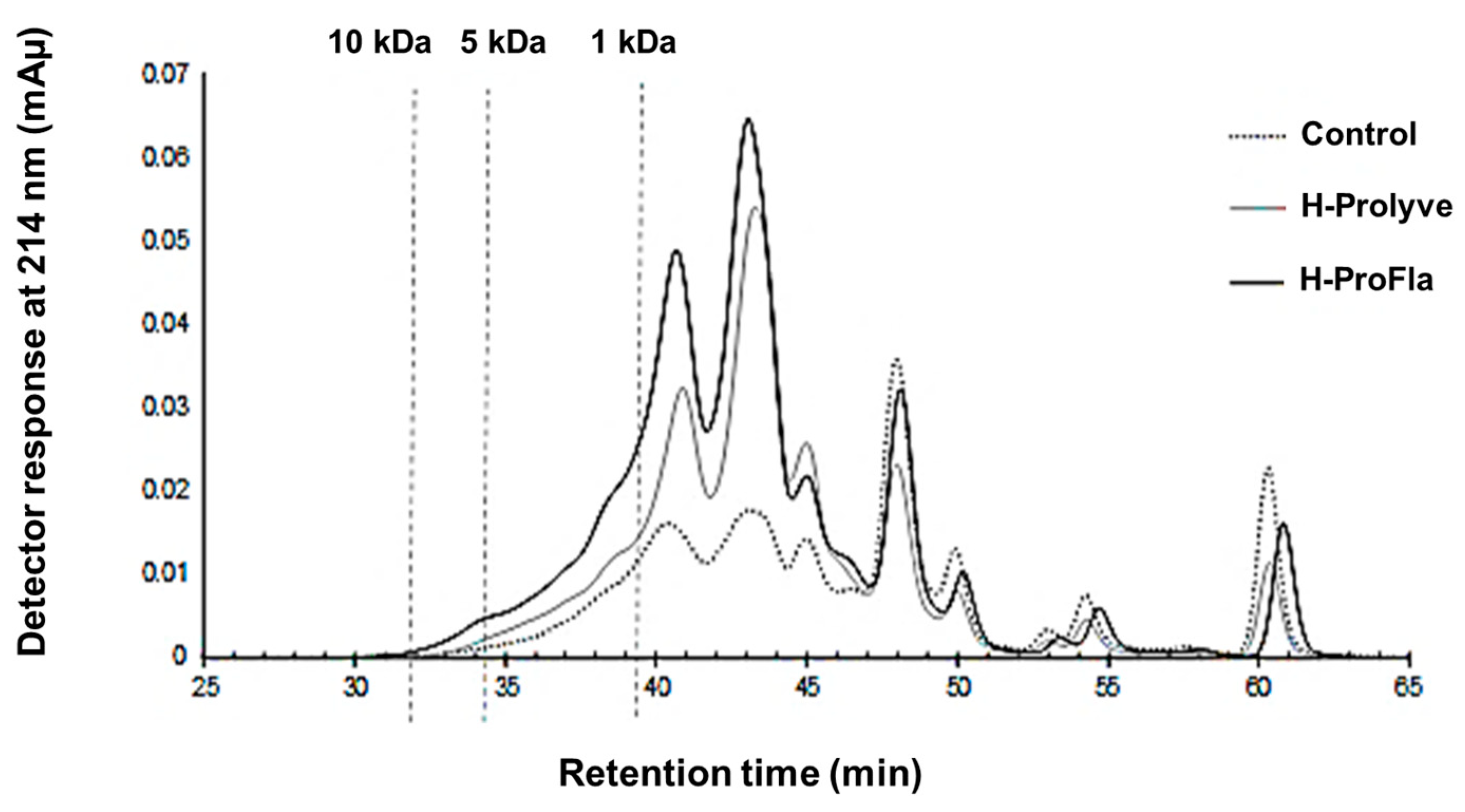
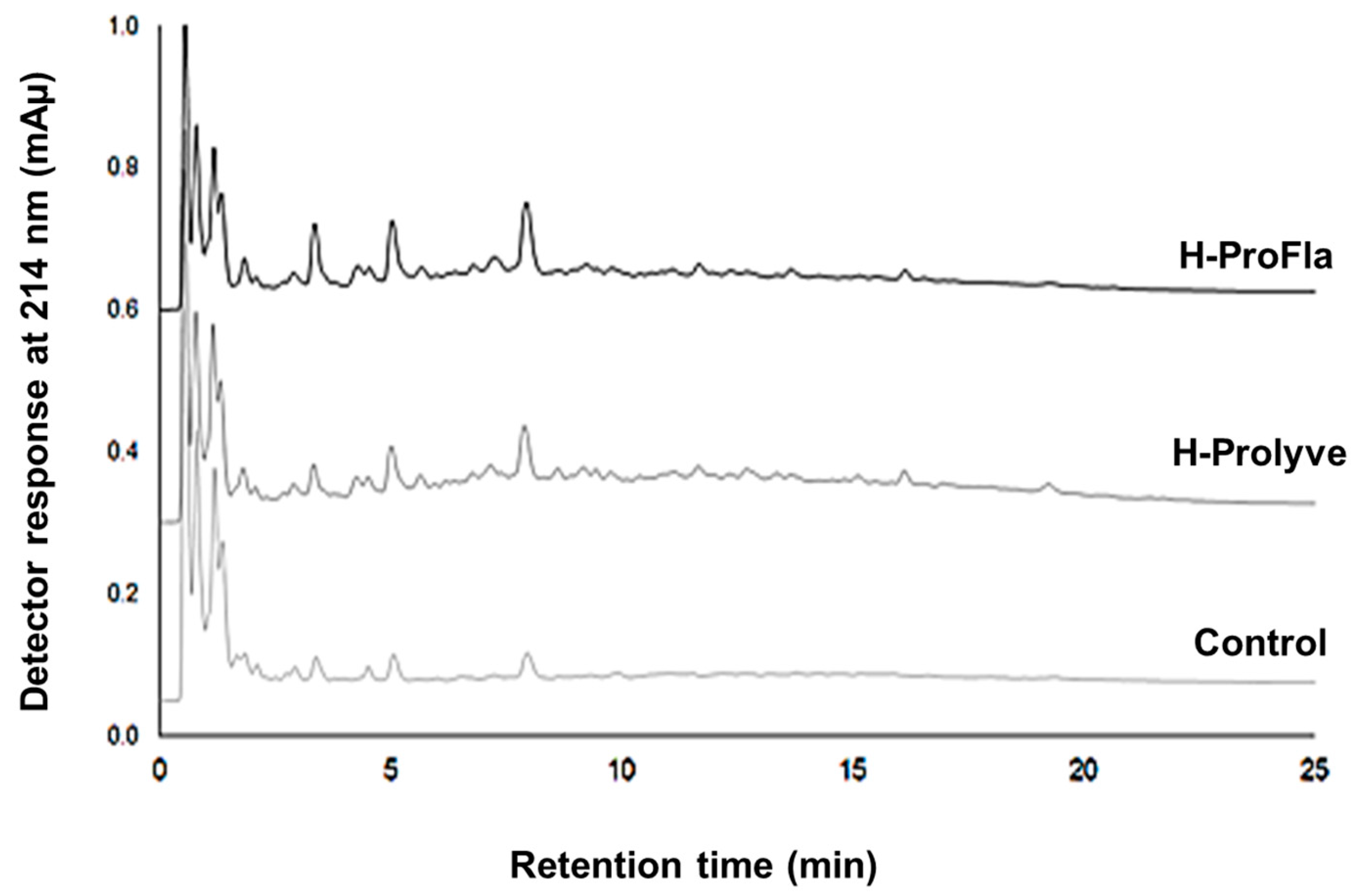
3.2. Peptide Profile and Molecular Mass Distribution
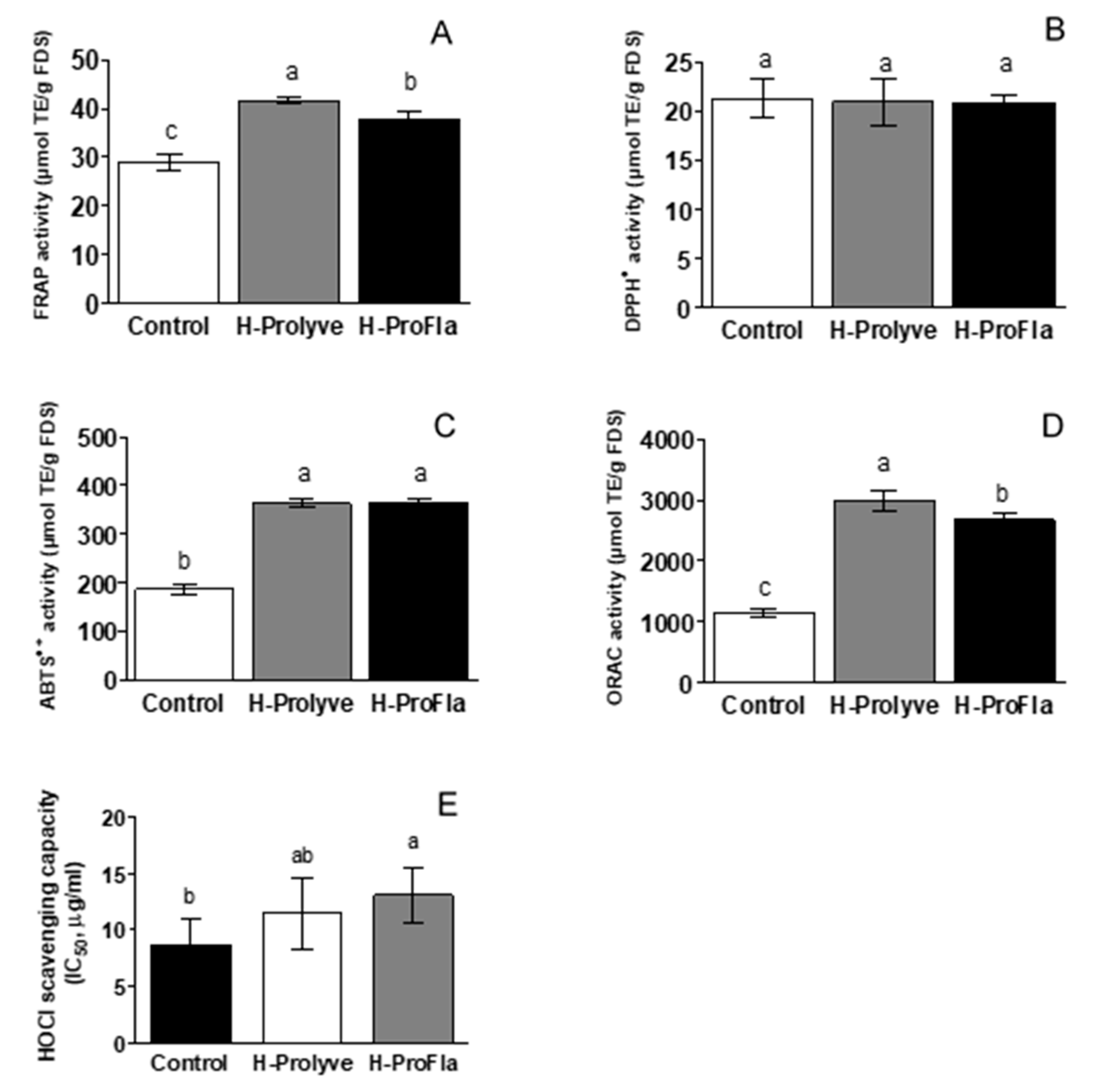
3.3. In Vitro Assessment of Antioxidant Activity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Corsetto, P.A.; Montorfano, G.; Zava, S.; Colombo, I.; Ingadottir, B.; Jonsdottir, R.; Sveinsdottir, K.; Rizzo, A.M. Characterization of antioxidant potential of seaweed extracts for enrichment of convenience food. Antioxidants 2020, 9, 249. [Google Scholar] [CrossRef] [PubMed]
- Khairy, H.M.; El-Sheikh, M.A. Antioxidant activity and mineral composition of three Mediterranean common seaweeds from Abu-Qir Bay, Egypt. Saudi J. Biol. Sci. 2015, 22, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, S.A.; Emire, S.A. Production and processing of antioxidant bioactive peptides: A driving force for the functional food market. Heliyon 2020, 6, e04765. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.B.; Wu, C.L.; Liu, D.; Yang, X.H.; Huang, J.F.; Zhang, J.; Liao, B.; He, H.L.; Li, H. Overview of antioxidant peptides derived from marine resources: The sources, characteristic, purification, and evaluation methods. Appl. Biochem. Biotechnol. 2015, 176, 1815–1833. [Google Scholar] [CrossRef] [PubMed]
- Pimentel, F.B.; Cermeño, M.; Kleekayai, T.; Harnedy-Rothwell, P.A.; Fernandes, E.; Alves, R.C.; Beatriz, P.P.O.M.; FitzGerald, R.J. Enzymatic modification of Porphyra dioica-derived proteins to improve their antioxidant potential. Molecules 2020, 25, 2838. [Google Scholar] [CrossRef]
- Admassu, H.; Gasmalla, M.A.A.; Yang, R.; Zhao, W. Bioactive peptides derived from seaweed protein and their health benefits: Antihypertensive, antioxidant, and antidiabetic properties. J. Food Sci. 2018, 83, 6–16. [Google Scholar] [CrossRef]
- Bhat, Z.; Kumar, S.; Bhat, H.F. Bioactive peptides of animal origin: A review. J. Food Sci. Technol. 2015, 52, 5377–5392. [Google Scholar] [CrossRef]
- Pimentel, F.B.; Alves, R.C.; Harnedy, P.A.; FitzGerald, R.J.; Oliveira, M.B.P. Macroalgal-derived protein hydrolysates and bioactive peptides: Enzymatic release and potential health enhancing properties. Trends Food Sci. Technol. 2019, 93, 106–124. [Google Scholar] [CrossRef]
- Pavlicevic, M.; Maestri, E. Marine bioactive peptides—an overview of generation, structure and application with a focus on food sources. Mar. Drugs 2020, 18, 424. [Google Scholar] [CrossRef]
- Montesano, D.; Gallo, M.; Blasi, F.; Cossignani, L. Biopeptides from vegetable proteins: New scientific evidences. Curr. Opin. Food Sci. 2020, 31, 31–37. [Google Scholar] [CrossRef]
- Stefanucci, A.; Mollica, A.; Macedonio, G.; Zengin, G.; Ahmed, A.A.; Novellino, E. Exogenous opioid peptides derived from food proteins and their possible uses as dietary supplements: A critical review. Food Rev. Int. 2018, 34, 70–86. [Google Scholar] [CrossRef]
- Lafarga, T.; Acién-Fernández, F.G.; Garcia-Vaquero, M. Bioactive peptides and carbohydrates from seaweed for food applications: Natural occurrence, isolation, purification, and identification. Algal Res. 2020, 48, 101909. [Google Scholar] [CrossRef]
- Lin, R.; Stekoll, M.S. Responses of chlorophyll a content for conchocelis phase of Alaskan Porphyra (Bangiales, Rhodophyta) species to environmental factors. Adv. Biosci. Bioeng. 2013, 1, 28–39. [Google Scholar] [CrossRef][Green Version]
- Machado, M.; Machado, S.; Pimentel, F.B.; Freitas, V.; Alves, R.C.; Oliveira, M.B.P.P. Amino acid profile and protein quality assessment of macroalgae produced in an integrated multi-trophic aquaculture system. Foods 2020, 9, 1382. [Google Scholar] [CrossRef]
- Pimentel, F.B.; Cermeño, M.; Kleekayai, T.; Machado, S.; Rego, A.; Fernandes, E.; Alves, R.C.; Oliveira, M.B.P.P.; FitzGerald, R.J. Contribution of in vitro simulated gastrointestinal digestion to the antioxidant activity of Porphyra dioica conchocelis. Algal Res. 2020, 51, 102085. [Google Scholar] [CrossRef]
- Connolly, A.; Piggott, C.O.; FitzGerald, R.J. Characterisation of protein-rich isolates and antioxidative phenolic extracts from pale and black brewers’ spent grain. Int. J. Food Sci. Technol. 2013, 48, 1670–1681. [Google Scholar] [CrossRef]
- Angell, A.R.; Mata, L.; de Nys, R.; Paul, N.A. The protein content of seaweeds: A universal nitrogen-to-protein conversion factor of five. J. Appl. Phycol. 2016, 28, 511–524. [Google Scholar] [CrossRef]
- Le Maux, S.; Nongonierma, A.B.; Barre, C.; FitzGerald, R.J. Enzymatic generation of whey protein hydrolysates under pH-controlled and non pH-controlled conditions: Impact on physicochemical and bioactive properties. Food Chem. 2016, 199, 246–251. [Google Scholar] [CrossRef]
- Spellman, D.; O’Cuinn, G.; FitzGerald, R.J. Bitterness in Bacillus proteinase hydrolysates of whey proteins. Food Chem. 2009, 114, 440–446. [Google Scholar] [CrossRef]
- Cermeño, M.; Stack, J.; Tobin, P.R.; O’Keeffe, M.B.; Harnedy, P.A.; Stengel, D.B.; FitzGerald, R.J. Peptide identification from a Porphyra dioica protein hydrolysate with antioxidant, angiotensin converting enzyme and dipeptidyl peptidase IV inhibitory activities. Food Funct. 2019, 10, 3421–3429. [Google Scholar] [CrossRef]
- Machado, S.; Costa, A.S.G.; Pimentel, F.; Beatriz, M.P.P.; Alves, R.C. A study on the protein fraction of coffee silverskin: Protein/non-protein nitrogen and free and total amino acid profiles. Food Chem. 2020, 326, 126940. [Google Scholar] [CrossRef]
- Harnedy, P.A.; FitzGerald, R.J. In vitro assessment of the cardioprotective, anti-diabetic and antioxidant potential of Palmaria palmata protein hydrolysates. J. Appl. Phycol. 2013, 25, 1793–1803. [Google Scholar] [CrossRef]
- Barizão, É.O.; Visentainer, J.V.; de Cinque Almeida, V.; Ribeiro, D.; Chisté, R.C.; Fernandes, E. Citharexylum solanaceum fruit extracts: Profiles of phenolic compounds and carotenoids and their relation with ROS and RNS scavenging capacities. Food Res. Int. 2016, 86, 24–33. [Google Scholar] [CrossRef]
- Mao, S.; Wang, K.; Lei, Y.; Yao, S.; Lu, B.; Huang, W. Antioxidant synergistic effects of Osmanthus fragrans flowers with green tea and their major contributed antioxidant compounds. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kleekayai, T.; Harnedy, P.A.; O’Keeffe, M.B.; Poyarkov, A.A.; Cunhaneves, A.; Suntornsuk, W.; FitzGerald, R.J. Extraction of antioxidant and ACE inhibitory peptides from Thai traditional fermented shrimp pastes. Food Chem. 2015, 176, 441–447. [Google Scholar] [CrossRef]
- Biancarosa, I.; Espe, M.; Bruckner, C.G.; Heesch, S.; Liland, N.; Waagbø, R.; Torstensen, B.; Lock, E.J. Amino acid composition, protein content, and nitrogen-to-protein conversion factors of 21 seaweed species from Norwegian waters. J. Appl. Phycol. 2017, 29, 1001–1009. [Google Scholar] [CrossRef]
- Cian, R.E.; Alaiz, M.; Vioque, J.; Drago, S.R. Enzyme proteolysis enhanced extraction of ACE inhibitory and antioxidant compounds (peptides and polyphenols) from Porphyra columbina residual cake. J. Appl. Phycol. 2013, 25, 1197–1206. [Google Scholar] [CrossRef]
- Tapal, A.; Tiku, P.K. Nutritional and nutraceutical improvement by enzymatic modification of food proteins. In Enzymes in Food Biotechnology; Kuddus, M., Ed.; Academic Press: Cambridge, MA, USA, 2019; pp. 471–481. [Google Scholar]
- Saidi, S.; Saoudi, M.; Amar, R.B. Valorisation of tuna processing waste biomass: Isolation, purification and characterisation of four novel antioxidant peptides from tuna by-product hydrolysate. Environ. Sci. Pollut. Res. 2018, 25, 17383–17392. [Google Scholar] [CrossRef]
- Peixoto, J.; Álvarez-Rivera, G.; Alves, R.C.; Costa, A.S.G.; Andrade, N.; Moreira, A.; Cifuentes, A.; Martel, F.; Oliveira, M.B.P.P.; Ibáñez, E. Cherry stem infusions: Antioxidante potential and phenolic profile by UHPLC-ESI-QTOF-MS. Food Funct. 2020, 11, 3471–3482. [Google Scholar] [CrossRef]
- Costa, A.S.G.; Alves, R.C.; Vinha, A.F.; Costa, E.; Costa, C.S.G.; Nunes, M.A.; Almeida, A.; Santos-Silva, A.; Oliveira, M.B.P.P. Nutritional, chemical and antioxidante/pro-oxidant profiles of silverskin, a coffee roasting by-product. Food Chem. 2018, 267, 28–35. [Google Scholar] [CrossRef]
- Prior, R.L.; Wu, X.; Schaich, K. Standardized methods for the determination of antioxidant capacity and phenolics in foods and dietary supplements. J. Agric. Food Chem. 2005, 53, 4290–4302. [Google Scholar] [CrossRef]
- Harnedy, P.A.; O’Keeffe, M.B.; FitzGerald, R.J. Fractionation and identification of antioxidant peptides from an enzymatically hydrolysed Palmaria palmata protein isolate. Food Res. Int. 2017, 100, 416–422. [Google Scholar] [CrossRef]
- Carvalho, L.C.; Estevão, M.S.; Ferreira, L.M.; Fernandes, E.; Marques, M.M.B. A new insight on the hypochlorous acid scavenging mechanism of tryptamine and tryptophan derivatives. Bioorganic Med. Chem. Lett. 2010, 20, 6475–6478. [Google Scholar] [CrossRef]
- Kimatu, B.M.; Zhao, L.; Biao, Y.; Ma, G.; Yang, W.; Pei, F.; Hu, Q. Antioxidant potential of edible mushroom (Agaricus bisporus) protein hydrolysates and their ultrafiltration fractions. Food Chem. 2017, 230, 58–67. [Google Scholar] [CrossRef]
- Zou, T.-B.; He, T.P.; Li, H.B.; Tang, H.W.; Xia, E.Q. The structure-activity relationship of the antioxidant peptides from natural proteins. Molecules 2016, 21, 72. [Google Scholar] [CrossRef] [PubMed]
- Nwachukwu, I.D.; Aluko, R.E. Structural and functional properties of food protein-derived antioxidant peptides. J. Food Biochem. 2019, 43, e12761. [Google Scholar] [CrossRef]
- Sheih, I.C.; Wu, T.K.; Fang, T.J. Antioxidant properties of a new antioxidative peptide from algae protein waste hydrolysate in different oxidation systems. Bioresour. Technol. 2009, 100, 3419–3425. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, E.; Costa, D.; Toste, S.A.; Lima, J.L.; Reis, S. In vitro scavenging activity for reactive oxygen and nitrogen species by nonsteroidal anti-inflammatory indole, pyrrole, and oxazole derivative drugs. Free Radic. Biol. Med. 2004, 37, 1895–1905. [Google Scholar] [CrossRef]
- Hawkins, C.; Pattison, D.; Davies, M. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids 2003, 25, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Esfandi, R.; Walters, M.E.; Tsopmo, A. Antioxidant properties and potential mechanisms of hydrolyzed proteins and peptides from cereals. Heliyon 2019, 5, e01538. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhao, M.; Lin, L.; Wang, J.; Sun-Waterhouse, D.; Dong, Y.; Zhuang, M.; Su, G. Identification of antioxidative peptides from defatted walnut meal hydrolysate with potential for improving learning and memory. Food Res. Int. 2015, 78, 216–223. [Google Scholar] [CrossRef]
- Karkouch, I.; Tabbene, O.; Gharbi, D.; Ben Mlouka, M.A.; Elkahoui, S.; Rihouey, C.; Coquet, L.; Cosette, P.; Jouenne, T.; Limam, F. Antioxidant, antityrosinase and antibiofilm activities of synthesized peptides derived from Vicia faba protein hydrolysate: A powerful agents in cosmetic application. Ind. Crops Prod. 2017, 109, 310–319. [Google Scholar] [CrossRef]
- Sonklin, C.; Laohakunjit, N.; Kerdchoechuen, O. Assessment of antioxidant properties of membrane ultrafiltration peptides from mungbean meal protein hydrolysates. PeerJ 2018, 6, e5337. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.H.; Luo, Q.B.; Pan, X.; Chi, C.F.; Sun, K.L.; Wang, B. Preparation, identification, and activity evaluation of ten antioxidant peptides from protein hydrolysate of swim bladders of miiuy croaker (Miichthys miiuy). J. Funct. Foods 2018, 47, 503–511. [Google Scholar] [CrossRef]
- Chan, C.X.; Blouin, N.A.; Zhuang, Y.; Zäuner, S.; Prochnik, S.E.; Lindquist, E.; Lin, S.; Benning, C.; Lohr, M.; Yarish, C.; et al. Porphyra (Bangiophyceae) transcriptomes provide insights into red algal development and metabolism. J. Phycol. 2012, 48, 1328–1342. [Google Scholar] [CrossRef] [PubMed]

| Amino Acids | Total Amino Acids | Free Amino Acids | |
|---|---|---|---|
| P. dioica-Conchocelis | H-Prolyve | H-ProFla | |
| Asp | 28.74 ± 0.98 | 2.26 ± 0.06 | 2.12 ± 0.09 |
| Glu | 27.55 ± 1.08 | 8.48 ± 0.21 | 7.45 ± 0.29 |
| Asn | n.d. | 0.63 ± 0.01 | 1.19 ± 0.05 |
| Gln | n.d. | 0.53 ± 0.01 | 1.04 ± 0.04 |
| Ala | 24.98 ± 0.96 | 9.40 ± 0.22 | 9.40 ± 0.40 |
| Arg | 18.84 ± 0.63 | 1.86 ± 0.04 | 3.95 ± 0.17 |
| Gly | 18.04 ± 0.58 | 0.71 ± 0.01 | 1.12 ± 0.05 |
| Ser | 14.08 ± 0.50 | 0.91 ± 0.02 | 1.79 ± 0.07 |
| Tyr | 9.17 ± 0.40 | 0.88 ± 0.02 | 1.81 ± 0.07 |
| Pro | 7.98 ± 0.48 | 0.26 ± 0.02 | 0.23 ± 0.02 |
| Hyp | 0.05 ± 0.00 | 0.01 ± 0.00 | 0.01 ± 0.00 |
| Phe | 9.64 ± 0.36 | 1.44 ± 0.03 | 2.30 ± 0.08 |
| His | 6.99 ± 0.22 | 0.10 ± 0.01 | 0.33 ± 0.01 |
| Ile | 8.98 ± 0.32 | 0.75 ± 0.02 | 1.85 ± 0.08 |
| Leu | 18.42 ± 0.69 | 3.48 ± 0.09 | 5.49 ± 0.22 |
| Lys | 17.91 ± 0.37 | 1.00 ± 0.02 | 2.36 ± 0.09 |
| Met | 5.12 ± 0.20 | 0.23 ± 0.01 | 0.66 ± 0.07 |
| Thr | 12.70 ± 0.51 | 1.10 ± 0.02 | 1.71 ± 0.07 |
| Trp | 1.31 ± 0.13 | 0.44 ± 0.02 | 0.67 ± 0.05 |
| Val | 14.52 ± 0.51 | 1.26 ± 0.03 | 2.88 ± 0.12 |
| ∑AA | 245.02 ± 8.47 | 35.73 ± 0.79 | 48.36 ± 1.94 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pimentel, F.B.; Machado, M.; Cermeño, M.; Kleekayai, T.; Machado, S.; Rego, A.M.; Abreu, M.H.; Alves, R.C.; Oliveira, M.B.P.P.; FitzGerald, R.J. Enzyme-Assisted Release of Antioxidant Peptides from Porphyra dioica Conchocelis. Antioxidants 2021, 10, 249. https://doi.org/10.3390/antiox10020249
Pimentel FB, Machado M, Cermeño M, Kleekayai T, Machado S, Rego AM, Abreu MH, Alves RC, Oliveira MBPP, FitzGerald RJ. Enzyme-Assisted Release of Antioxidant Peptides from Porphyra dioica Conchocelis. Antioxidants. 2021; 10(2):249. https://doi.org/10.3390/antiox10020249
Chicago/Turabian StylePimentel, Filipa B., Marlene Machado, Maria Cermeño, Thanyaporn Kleekayai, Susana Machado, Andreia M. Rego, Maria H. Abreu, Rita C. Alves, Maria Beatriz P. P. Oliveira, and Richard J. FitzGerald. 2021. "Enzyme-Assisted Release of Antioxidant Peptides from Porphyra dioica Conchocelis" Antioxidants 10, no. 2: 249. https://doi.org/10.3390/antiox10020249
APA StylePimentel, F. B., Machado, M., Cermeño, M., Kleekayai, T., Machado, S., Rego, A. M., Abreu, M. H., Alves, R. C., Oliveira, M. B. P. P., & FitzGerald, R. J. (2021). Enzyme-Assisted Release of Antioxidant Peptides from Porphyra dioica Conchocelis. Antioxidants, 10(2), 249. https://doi.org/10.3390/antiox10020249