Cyanidin-3-O-glucoside Regulates the Expression of Ucp1 in Brown Adipose Tissue by Activating Prdm16 Gene
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Cell Culture
2.3. Animals Studies
2.4. Isolation of Total RNA Extraction and Analysis by qPCR
2.5. Construction of Plasmid Vector
2.6. Cell Transfection and Luciferase Activity Detection
2.7. Yeast One-Hybrid Assay
2.8. shRNA-Mediated Gene Silencing of Prdm16
2.9. Microscale Thermophoresis (MST) Study
2.10. Analysis of Experimental Data
3. Results
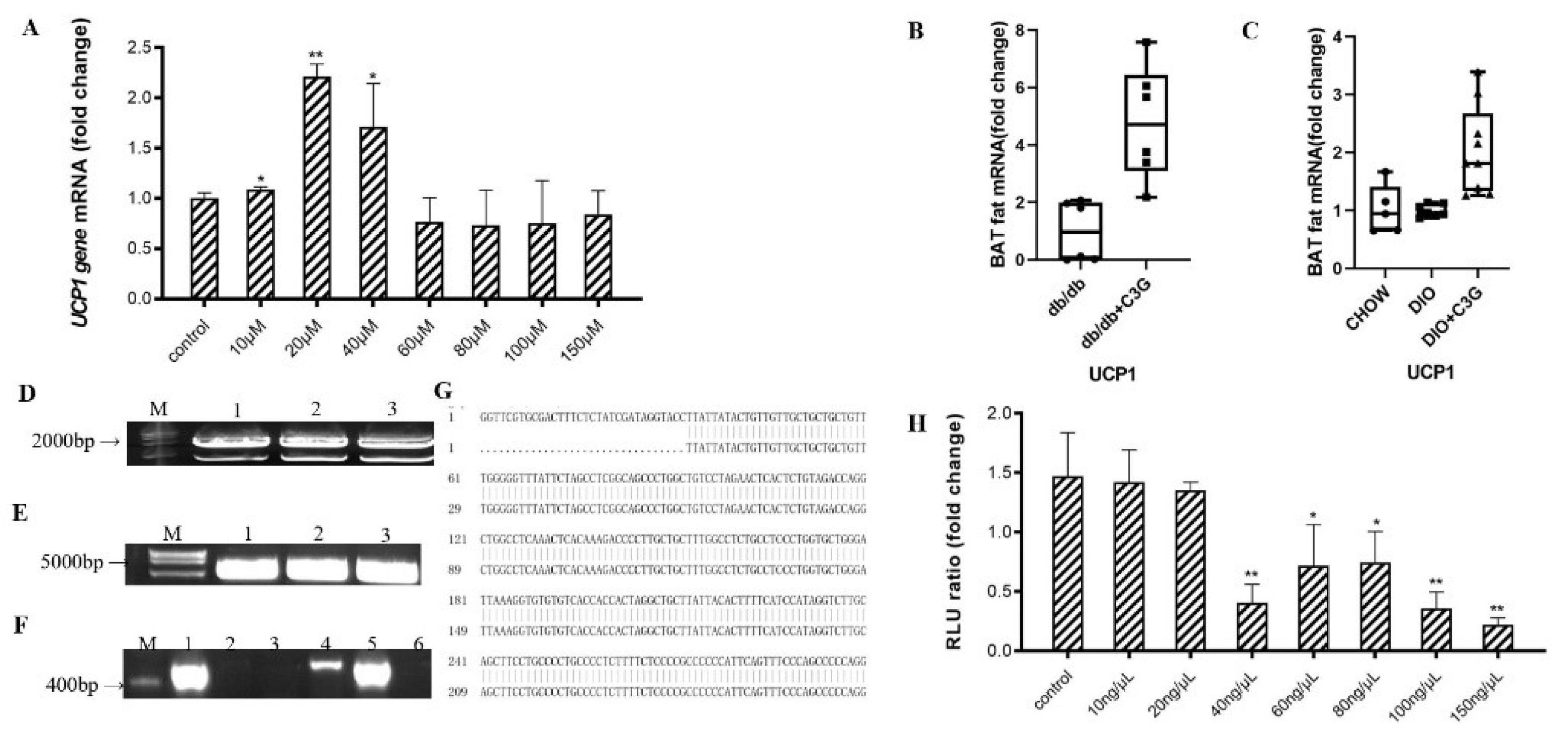
3.1. C3G Upregulates the Expression of Ucp1 Gene Both In Vitro and In Vivo
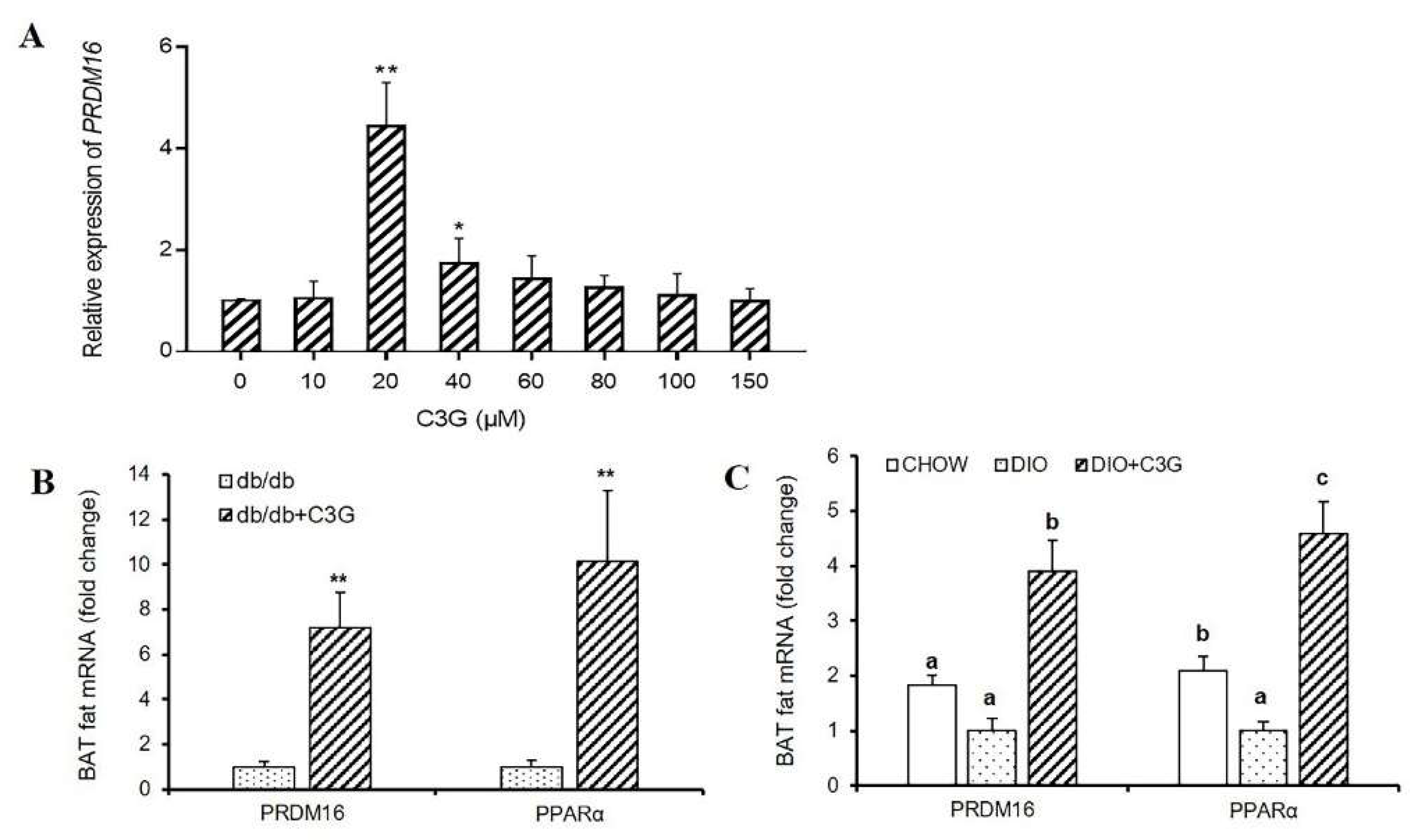
3.2. C3G Upregulates the Expression of Prdm16 Gene Both In Vitro and In Vivo
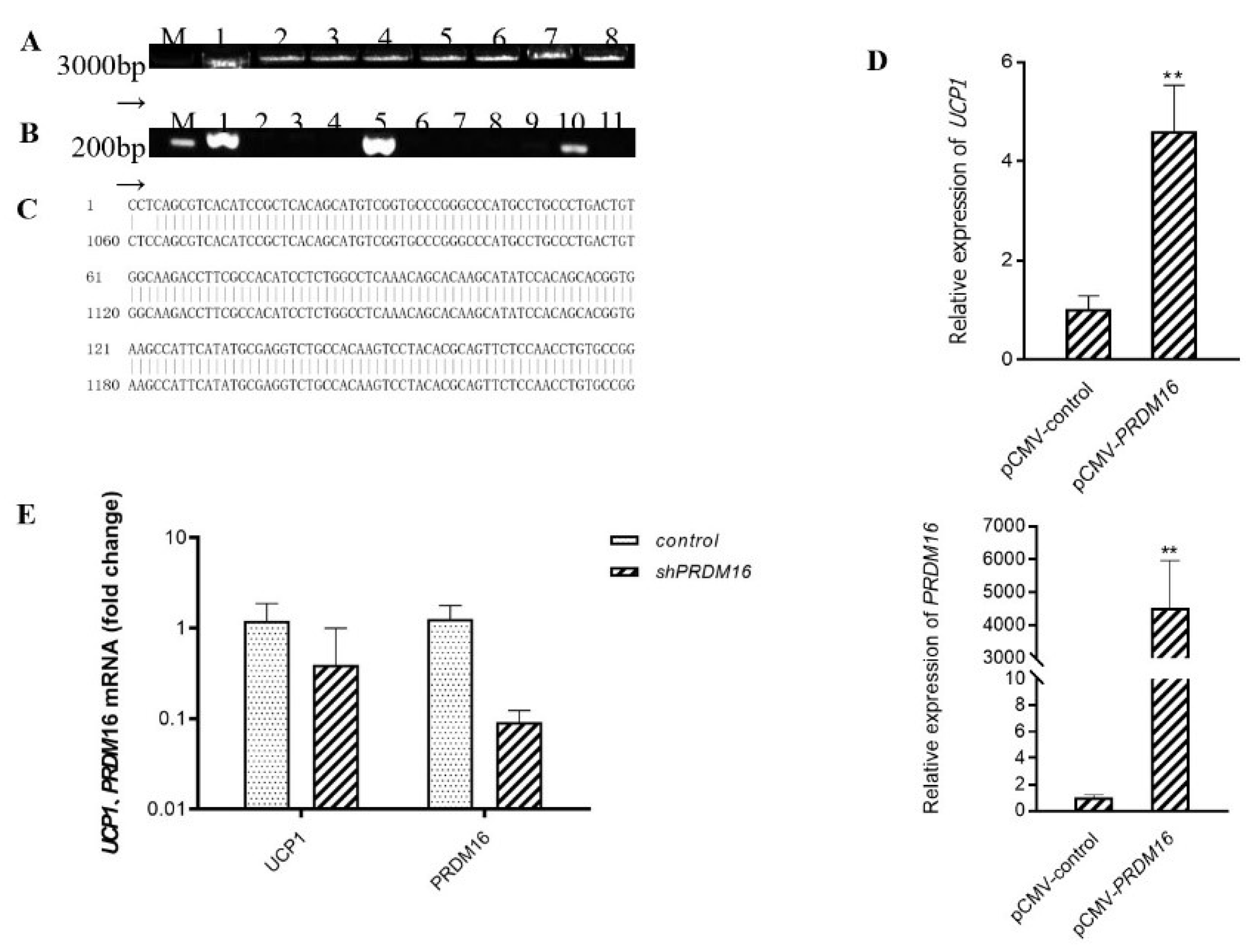
3.3. Prdm16 Regulates the Expression of Ucp1 Gene in Brown Adipocytes
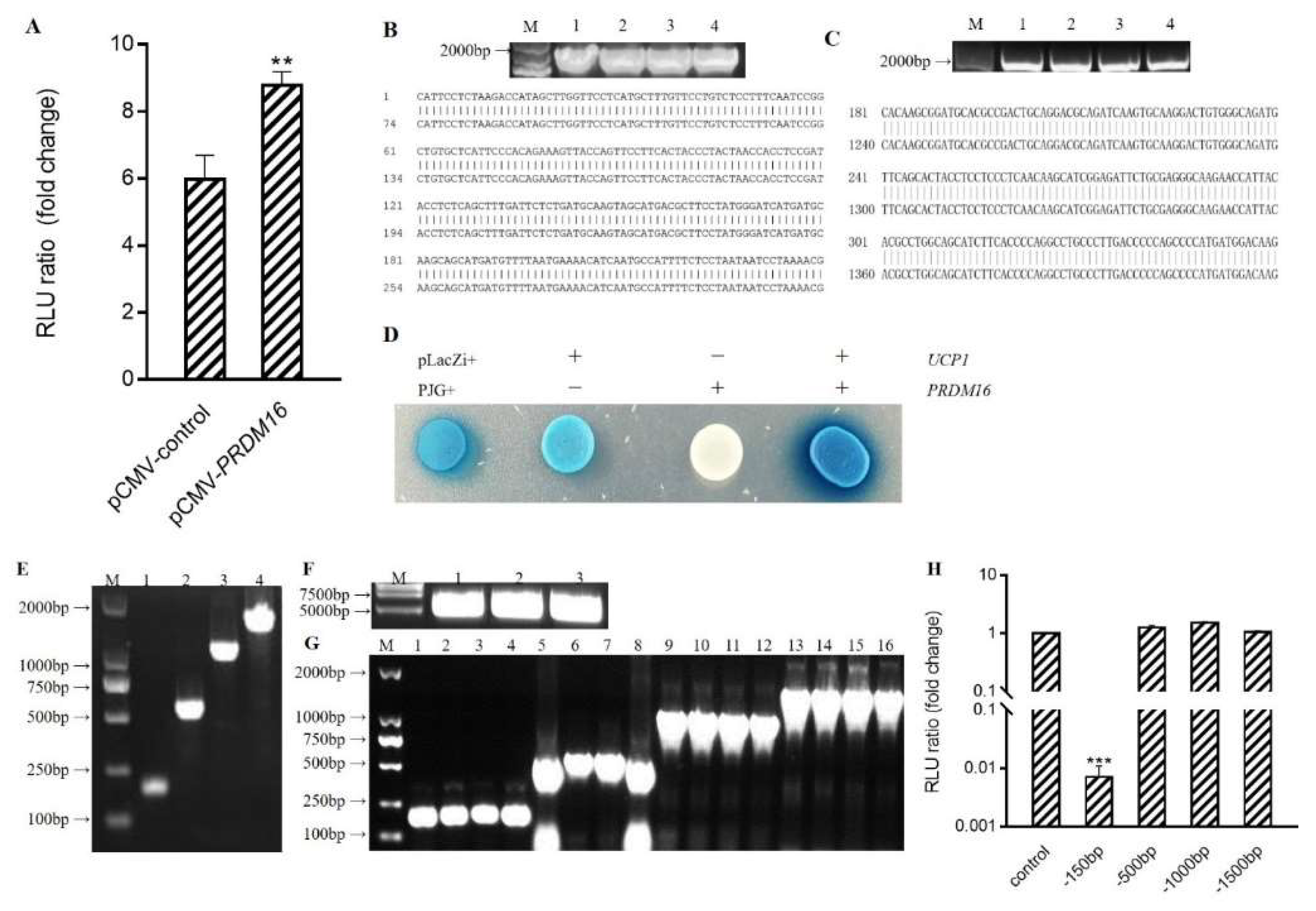
3.4. Prdm16 Directly Binds and Activates Ucp1 Promoter
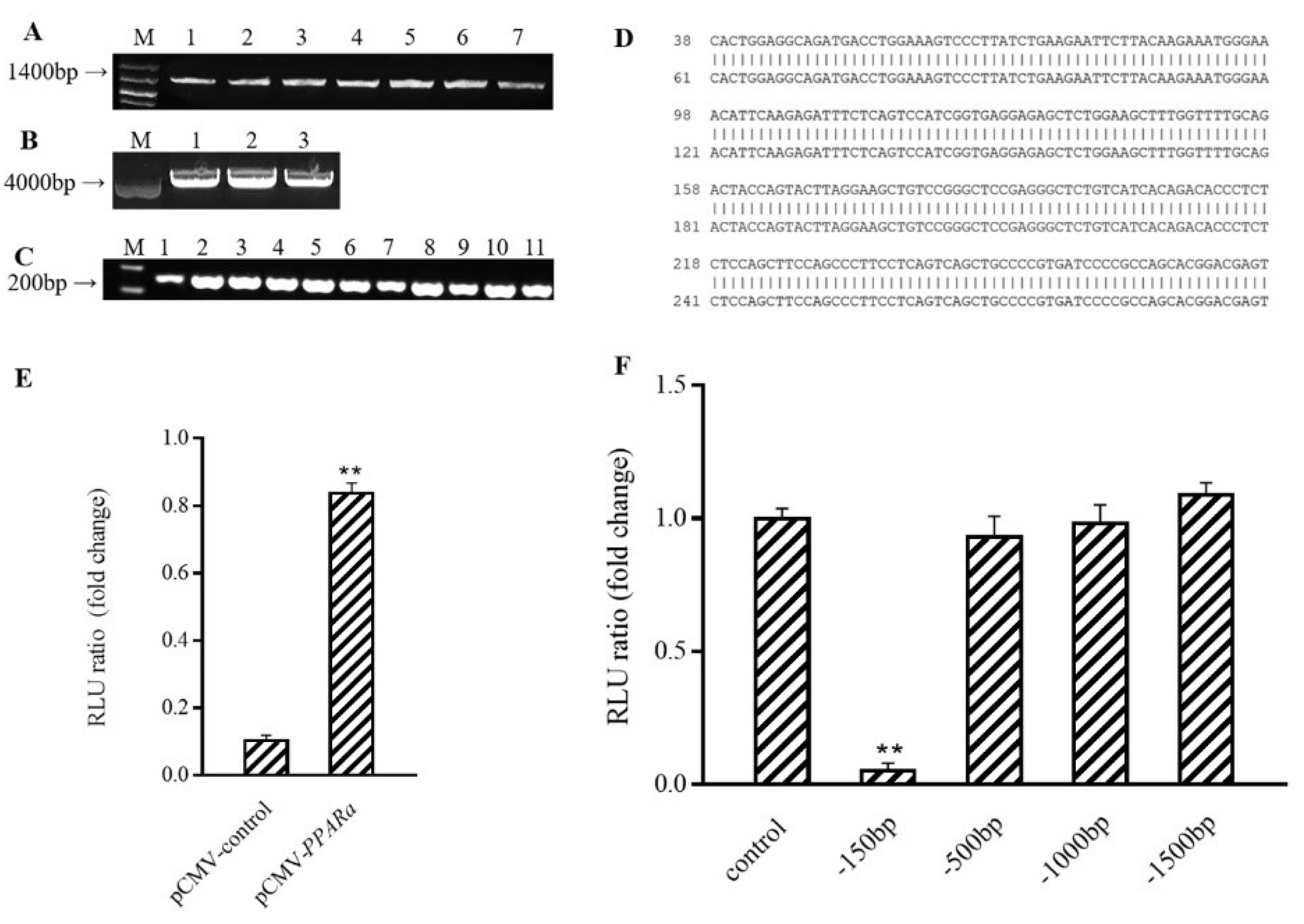
3.5. PPARα Binds to Ucp1 Promoter to Activate Transcription
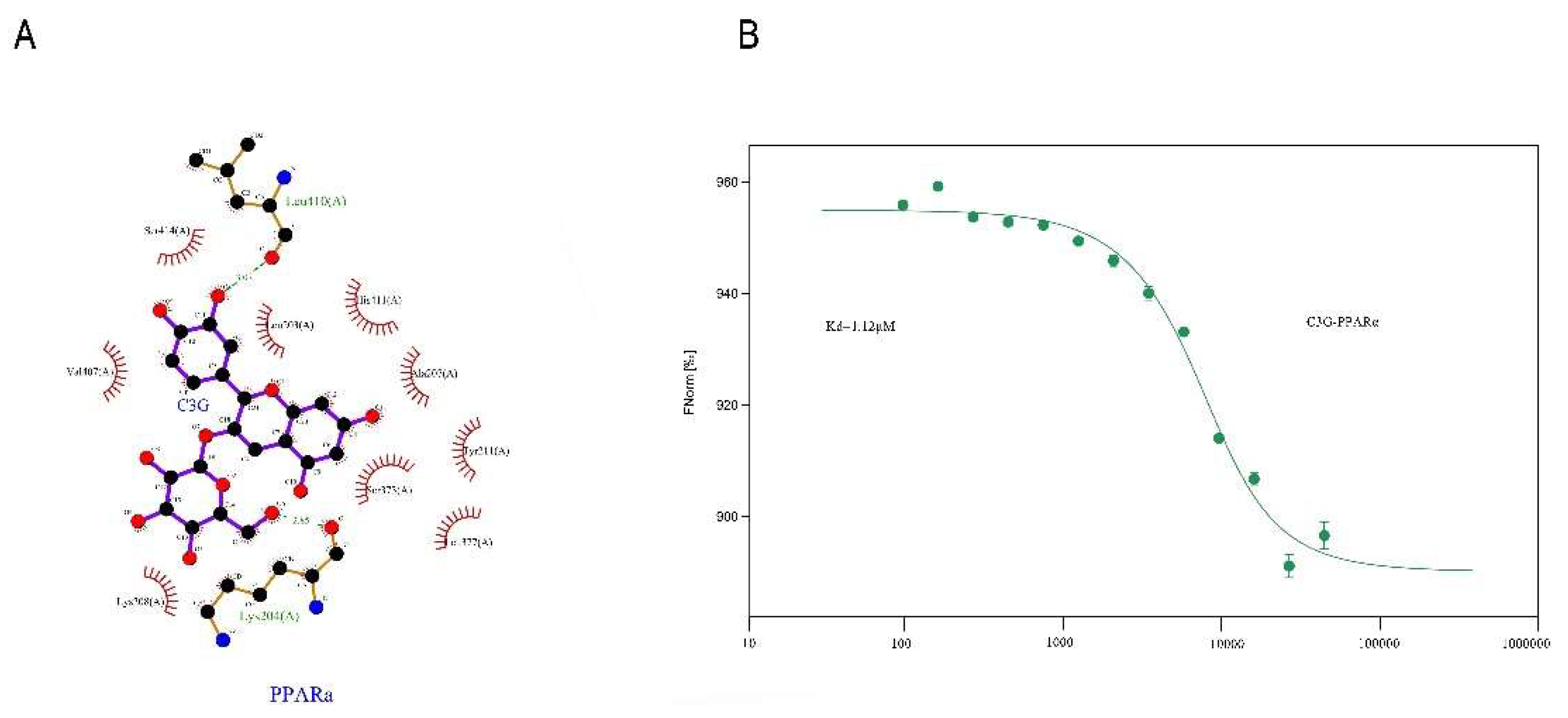
3.6. Conformational Investigation and Interaction Study of PPARα–C3G
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- You, Y.; Zhou, F.; Huang, D. Eating the Right Color: Dietary Anthocyanins and Obesity Control. Food Beverage Asia 2018, 12, 57–59. [Google Scholar]
- Duchowicz, P.R.; Szewczuk, N.A.; Pomilio, A.B. QSAR Studies of the Antioxidant Activity of Anthocyanins. J. Food Sci. Technol. -Mysore 2019, 56, 5518–5530. [Google Scholar] [CrossRef] [PubMed]
- Correia, P.; Araújo, P.; Ribeiro, C.; Oliveira, H.; Pereira, A.R.; Mateus, N.; de Freitas, V.; Brás, N.F.; Gameiro, P.; Coelho, P.; et al. Anthocyanin-Related Pigments: Natural Allies for Skin Health Maintenance and Protection. Antioxidants 2021, 10, 1038. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Yuan, X.; Liu, X.; Liang, C.; Zhan, J. Front Cover: Cyanidin-3-glucoside Increases Whole Body Energy Metabolism by Upregulating Brown Adipose Tissue Mitochondrial Function. Mol. Nutr. Food Res. 2017, 61, 1770111. [Google Scholar] [CrossRef]
- Ng, T. Global, Regional, and National Prevalence of Overweight and Obesity in Children and Adults during 1980–2013: A Systematic Analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef]
- Al-Tahami, B.A.M.; Ismail, A.A.A.-S.; Bee, Y.T.G.; Awang, S.A.; Rani, W.R.S.W.A.; Sanip, Z.; Rasool, A.H.G. The effects of anti-obesity intervention with orlistat and sibutramine on microvascular endothelial function. Clin. Hemorheol. Microcirc. 2015, 59, 323–334. [Google Scholar] [CrossRef]
- Siebenhofer, A.; Jeitler, K.; Horvath, K.; Berghold, A.; Semlitsch, T. Long-Term Effects of Weight-Reducing Drugs in People with Hypertension. Cochrane Database Syst. Rev. 2016, 3, CD007654. [Google Scholar] [CrossRef]
- Cypess, A.M.; Chen, Y.C.; Sze, C.; Wang, K.; English, J.; Chan, O.; Holman, A.R.; Tal, M.; Palmer, M.R.; Kolodny, G.M. Cold but Not Sympathomimetics Activates Human Brown Adipose Tissue in Vivo. Proc. Natl. Acad. Sci. USA 2012, 109, 10001–10005. [Google Scholar] [CrossRef]
- van Marken Lichtenbelt, W.D.; Vanhommerig, J.W.; Smulders, N.M.; Drossaerts, J.M.; Kemerink, G.J.; Bouvy, N.D.; Teule, G.J. None Cold-Activated Brown Adipose Tissue in Healthy Men. N. Engl. J. Med. 2009, 360, 1917. [Google Scholar] [CrossRef]
- Villarroya, F.; Peyrou, M.; Giralt, M. Transcriptional Regulation of the Uncoupling Protein-1 Gene. Biochimie 2017, 134, 86–92. [Google Scholar] [CrossRef]
- Escher, P.; Braissant, O.; Basu-Modak, S.; Michalik, L.; Wahli, W.; Desvergne, B. Wahli Rat PPARs: Quantitative Analysis in Adult Rat Tissues and Regulation in Fasting and Refeeding. Endocrinology 2001, 142, 4195–4202. [Google Scholar] [CrossRef]
- Collins, S.; Yehuda-Shnaidman, E.; Wang, H. Positive and Negative Control of Ucp1 Gene Transcription and the Role of β-Adrenergic Signaling Networks. Int. J. Obes. 2010, 34, S28–S33. [Google Scholar] [CrossRef]
- Harms, M.J.; Ishibashi, J.; Wang, W.; Lim, H.W.; Seale, P. Prdm16 Is Required for the Maintenance of Brown Adipocyte Identity and Function in Adult Mice. Cell Metab. 2014, 19, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Kajimura, S.; Seale, P.; Kubota, K.; Lunsford, E.; Frangioni, J.V.; Gygi, S.P.; Spiegelman, B.M. Initiation of Myoblast to Brown Fat Switch by a Prdm16–C/EBP-β Transcriptional Complex. Nature 2009, 460, 1154–1158. [Google Scholar] [CrossRef]
- Matsukawa, T.; Inaguma, T.; Han, J.; Villareal, M.O.; Isoda, H. Cyanidin-3-Glucoside Derived from Black Soybeans Ameliorate Type 2 Diabetes through the Induction of Differentiation of Preadipocytes into Smaller and Insulin-Sensitive Adipocytes. J. Nutr. Biochem. 2015, 26, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Yu, Z.; Tang, Q.; Song, H.; Gao, Z.; Chen, W.; Zheng, X. Honeysuckle Anthocyanin Supplementation Prevents Diet-Induced Obesity in C57BL/6 Mice. Food Funct. 2013, 4, 1654–1661. [Google Scholar] [CrossRef]
- You, Y.; Liang, C.; Han, X.; Guo, J.; Ren, C.; Liu, G.; Huang, W.; Zhan, J. Mulberry Anthocyanins, Cyanidin 3-Glucoside and Cyanidin 3-Rutinoside, Increase the Quantity of Mitochondria during Brown Adipogenesis. J. Funct. Foods 2017, 36, 348–356. [Google Scholar] [CrossRef]
- You, Y.; Han, X.; Guo, J.; Guo, Y.; Yin, M.; Liu, G.; Zhan, J. Cyanidin-3-Glucoside Attenuates High-Fat and High-Fructose Diet-Induced Obesity by Promoting the Thermogenic Capacity of Brown Adipose Tissue. J. Funct. Foods 2018, 41, 62–71. [Google Scholar] [CrossRef]
- Becerril, S.; Gómez-Ambrosi, J.; Martín, M.; Moncada, R.; Frühbeck, G. Role of Prdm16 in the Activation of Brown Fat Programming. Relevance to the Development of Obesity. Histol. Histopathol. 2013, 28, 28. Available online: http://hdl.handle.net/10201/61619 (accessed on 18 May 2021).
- Hondares, E.; Rosell, M.; Diaz-Delfin, J.; Olmos, Y.; Monsalve, M.; Iglesias, R.; Villarroya, F.; Giralt, M. Peroxisome Proliferator-Activated Receptor α (PPARα) Induces PPARγ Coactivator 1α (PGC-1α) Gene Expression and Contributes to Thermogenic Activation of Brown Fat: Involvement of Prdm16. J. Biol. Chem. 2011, 286, 43112. [Google Scholar] [CrossRef]
- Ohno, H.; Shinoda, K.; Ohyama, K.; Sharp, L.Z.; Kajimura, S. EHMT1 Controls Brown Adipose Cell Fate and Thermogenesis through the Prdm16 Complex. Nature 2013, 504, 163–167. [Google Scholar] [CrossRef]
- Seale, P.; Kajimura, S.; Yang, W.; Chin, S.; Rohas, L.; Uldry, M.; Tavernier, G.; Langin, D.; Spiegelman, B. Transcriptional Control of Brown Fat Determination by Prdm16. Cell Metab. 2007, 6, 38–54. [Google Scholar] [CrossRef]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.H.; Doria, A. Identification and Importance of Brown Adipose Tissue in Adult Humans. Obstet. Gynecol. Surv. 2009, 64, 519–520. [Google Scholar] [CrossRef]
- Himms-Hagen, J. Brown Adipose Tissue Thermogenesis: Interdisciplinary Studies. Faseb. J. 1990, 4, 2890–2898. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, S.; Kakuma, T.; Yoshimatsu, H.; Yasunaga, S.; Kurokawa, M.; Sakata, T. Molecular Cloning of Rat Uncoupling Protein 2 CDNA and Its Expression in Genetically Obese Zucker Fatty (Fa/Fa) Rats. Biochim. Biophys. Acta 1998, 1389, 178–186. [Google Scholar] [CrossRef]
- Mao, W.; Yu, X.X.; Zhong, A.; Li, W.; Brush, J.; Sherwood, S.W.; Adams, S.H.; Pan, G. UCP4, a Novel Brain-Specific Mitochondrial Protein That Reduces Membrane Potential in Mammalian Cells. FEBS Lett. 1999, 443, 326–330. [Google Scholar] [CrossRef]
- Solanes, G.; Vidal-Puig, A.; Grujic, D.; Flier, J.S.; Lowell, B.B. The Human Uncoupling Protein-3 Gene: Genomic structure, chromosomal localization, and genetic basis for short and long form transcripts *. J. Biol. Chem. 1997, 272, 25433–25436. [Google Scholar] [CrossRef]
- Yu, X.X.; Mao, W.; Zhong, A.; Schow, P.; Brush, J.; Sherwood, S.W.; Adams, S.H.; Pan, G. Characterization of Novel UCP5/BMCP1 Isoforms and Differential Regulation of UCP4 and UCP5 Expression through Dietary or Temperature Manipulation. Faseb. J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2000, 14, 1611. [Google Scholar] [CrossRef]
- Rosen, E.; Spiegelman, B. What We Talk About When We Talk About Fat. Cell 2014, 156, 20–44. [Google Scholar] [CrossRef]
- Rehnmark, S.; Bianco, A.C.; Kieffer, J.D.; Silva, J.E. Transcriptional and Posttranscriptional Mechanisms in Uncoupling Protein MRNA Response to Cold. Am. J. Physiol. 1992, 262, 58–67. [Google Scholar] [CrossRef] [PubMed]
- You, Y.; Yuan, X.; Lee, H.J.; Huang, W.; Jin, W.; Zhan, J. Zhan Mulberry and Mulberry Wine Extract Increase the Number of Mitochondria during Brown Adipogenesis. Food Funct. 2015, 6, 401–408. [Google Scholar] [CrossRef]
- Han, X.; Guo, J.; You, Y.; Yin, M.; Liang, J.; Ren, C.; Huang, W. Juan Vanillic Acid Activates Thermogenesis in Brown and White Adipose Tissue. Food Funct. 2018, 9, 4366–4375. [Google Scholar] [CrossRef]
- Han, X.; Zhang, Y.; Guo, J.; You, Y.; Huang, W. Chlorogenic Acid Stimulates the Thermogenesis of Brown Adipocytes by Promoting the Uptake of Glucose and the Function of Mitochondria. J. Food Sci. 2019, 84, 3815–3824. [Google Scholar] [CrossRef]
- Echtay, K. Mitochondrial Uncoupling Proteins—What Is Their Physiological Role? Free Radic. Biol. Med. 2007, 43, 1351–1371. [Google Scholar] [CrossRef]
- Jastroch, M.; Withers, K.; Klingenspor, M. Klingenspor Uncoupling Protein 2 and 3 in Marsupials: Identification, Phylogeny, and Gene Expression in Response to Cold and Fasting in Antechinus Flavipes. Physiol. Genom. 2004, 17, 130–139. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Collins, S.; Daniel, K.W.; Petro, A.E.; Surwit, R.S. Strain-Specific Response to Beta 3-Adrenergic Receptor Agonist Treatment of Diet-Induced Obesity in Mice. Endocrinology 1997, 138, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M. Differential Effects of Retinoic Acid on Uncoupling Protein-1 and Leptin Gene Expression. J. Endocrinol. 1998, 157, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Rabelo, R.; Camirand, A.; Silva, J.E. Silva 3′,5′-Cyclic Adenosine Monophosphate-Response Sequences of the Uncoupling Protein Gene Are Sequentially Recruited during Darglitazone-Induced Brown Adipocyte Differentiation. Endocrinology 1997, 138, 5325–5332. [Google Scholar] [CrossRef][Green Version]
- Sasaki, N.; Uchida, E.; Niiyama, M.; Yoshida, T.; Saito, M. Anti-Obesity Effects of Selective Agonists to the Beta 3-Adrenergic Receptor in Dogs. II. Recruitment of Thermogenic Brown Adipocytes and Reduction of Adiposity after Chronic Treatment with a Beta 3-Adrenergic Agonist. J. Vet. Med. Sci. 1998, 60, 465. [Google Scholar] [CrossRef][Green Version]
- Savontaus, E.; Rouru, J.; Boss, O.; Huupponen, R.; Koulu, M. Differential Regulation of Uncoupling Proteins by Chronic Treatments with Beta 3-Adrenergic Agonist BRL 35135 and Metformin in Obese Fa/Fa Zucker Rats. Biochem. Biophys. Res. Commun. 1998, 246, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Bajnok, L.; Longo, K.A.; Petersen, R.K.; Hansen, J.B.; Kristiansen, K.; Macdougald, O.A. Effects of Wnt Signaling on Brown Adipocyte Differentiation and Metabolism Mediated by PGC-1α. Mol. Cell. Biol. 2005, 25, 1272–1282. [Google Scholar] [CrossRef]
- Seale, P.; Conroe, H.M.; Estall, J.; Kajimura, S.; Frontini, A.; Ishibashi, J.; Cohen, P.; Cinti, S.; Spiegelman, B.M. Prdm16 Determines the Thermogenic Program of Subcutaneous White Adipose Tissue in Mice. J. Clin. Investig. 2011, 121, 96–105. [Google Scholar] [CrossRef]
- Ricquier, D.; Casteilla, L.; Bouillaud, F. Molecular Studies of the Uncoupling Protein. FASEB J. 1991, 5, 2237–2242. [Google Scholar] [CrossRef] [PubMed]
- Seale, P.; Bjork, B.; Yang, W.; Kajimura, S.; Chin, S.; Kuang, S.; Scimè, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. Prdm16 Controls a Brown Fat/Skeletal Muscle Switch. Nature 2008, 454, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhan, J.; Huang, W. Grape Seed Proanthocyanidins Induce Apoptosis and Cell Cycle Arrest of HepG2 Cells Accompanied by Induction of the MAPK Pathway and NAG-1. Antioxidants 2020, 9, 1200. [Google Scholar] [CrossRef] [PubMed]
- Kay, C.D.; Kroon, P.A.; Cassidy, A. The Bioactivity of Dietary Anthocyanins Is Likely to Be Mediated by Their Degradation Products. Mol. Nutr. Food Res. 2010, 53, 92–101. [Google Scholar] [CrossRef]
- Pace, E.; Jiang, Y.; Clemens, A.; Crossman, T.; Rupasinghe, H.V. Impact of Thermal Degradation of Cyanidin-3-O-glucoside of Haskap Berry on Cytotoxicity of Hepatocellular Carcinoma HepG2 and Breast Cancer MDA-MB-231 Cells. Antioxidants 2018, 7, 24. [Google Scholar] [CrossRef]
| Gene | Primer Sequence (5′→3′) |
|---|---|
| Ucp1 | F:GGCAAAAACAGAAGGATTGC R:TAAGCCGGCTGAGATCTTGT |
| Prdm16 | F:GAAGTCACAGGAGGACACGGR: CTCGCTCCTCAACACACCTC |
| Vector | Gene | Primer Sequence (5′→3′) |
|---|---|---|
| Double luciferase reporter gene vector of Ucp1 promoter | Ucp1-KPN I-F | GGGGTACCcattcctctaagaccatagctt |
| Ucp1-MluⅠ-R | CGACGCGTacttctgcgccctgacct | |
| Eukaryotic vector for overexpression of Prdm16 | Prdm16-EcoRI-F | tccaagcttctgcaggaattcA TGCGA TCCAAGGCGAGG |
| Prdm16-XbaI-R | accgggcccactagttctagaTCA TTGCA TA TGCCTCCGGG | |
| Double luciferase reporter gene vector of Ucp1 promoter with different fragment size | Ucp1(-150bp)-Kpn I-F | atttctctatcgataggtaccGAGTGACGCGCGGCTGGG |
| Ucp1(-150bp)-Mlu I-R | cgagcccgggctagcacgcgtCTGCGCCCTGACCTGGGA | |
| Ucp1(-500bp)-Kpn I-F | atttctctatcgataggtaccTCCAGTCACCCAAA TCTGAAGG | |
| Ucp1(-500bp)-Mlu I-R | cgagcccgggctagcacgcgtCTGCGCCCTGACCTGGGA | |
| Ucp1(-1000bp)-Kpn I-F | atttctctatcgataggtaccAGCAGAACCTGGCCAACCA | |
| Ucp1(-1000bp)-Mlu I-R | cgagcccgggctagcacgcgtCTGCGCCCTGACCTGGGA | |
| Ucp1(-1500bp)-Kpn I-F | atttctctatcgataggtaccTTATTATACTGTTGTTGCTGCTGCT | |
| Ucp1(-1500bp)-Mlu I-R | cgagcccgggctagcacgcgtCTGCGCCCTGACCTGGGA | |
| pLacZi-Ucp1 | Ucp1-EcoR I-F | ctttgatattggatcgaattcCATTCCTCTAAGACCATAGCTTGGT |
| Ucp1-Xho I-R | atacagagcacatgcctcgagACTTCTGCGCCCTGACCTG | |
| Prdm16-EcoR I-F | gattatgcctctcccgaattcTACGCTAGGTTCCGCTCCC | |
| Prdm16-Xho I-R | agaagtccaaagcttctcgagTAGTAACGTA TACGGAGGCCCA T | |
| PJG-Prdm16 | Prdm16-EcoR I-F | gattatgcctctcccgaattcTACGCTAGGTTCCGCTCCC |
| Prdm16-Xho I-R | agaagtccaaagcttctcgagTAGTAACGTA TACGGAGGCCCA T |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, S.; Yang, Y.; Lu, Y.; Guo, J.; Han, X.; Gao, Y.; Huang, W.; You, Y.; Zhan, J. Cyanidin-3-O-glucoside Regulates the Expression of Ucp1 in Brown Adipose Tissue by Activating Prdm16 Gene. Antioxidants 2021, 10, 1986. https://doi.org/10.3390/antiox10121986
Han S, Yang Y, Lu Y, Guo J, Han X, Gao Y, Huang W, You Y, Zhan J. Cyanidin-3-O-glucoside Regulates the Expression of Ucp1 in Brown Adipose Tissue by Activating Prdm16 Gene. Antioxidants. 2021; 10(12):1986. https://doi.org/10.3390/antiox10121986
Chicago/Turabian StyleHan, Suping, Yafan Yang, Yanan Lu, Jielong Guo, Xue Han, Yunxiao Gao, Weidong Huang, Yilin You, and Jicheng Zhan. 2021. "Cyanidin-3-O-glucoside Regulates the Expression of Ucp1 in Brown Adipose Tissue by Activating Prdm16 Gene" Antioxidants 10, no. 12: 1986. https://doi.org/10.3390/antiox10121986
APA StyleHan, S., Yang, Y., Lu, Y., Guo, J., Han, X., Gao, Y., Huang, W., You, Y., & Zhan, J. (2021). Cyanidin-3-O-glucoside Regulates the Expression of Ucp1 in Brown Adipose Tissue by Activating Prdm16 Gene. Antioxidants, 10(12), 1986. https://doi.org/10.3390/antiox10121986








