cDNA Cloning and Partial Characterization of the DJ-1 Gene from Tribolium castaneum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Identification of the TcDJ-1 Sequence and Bioinformatics Analysis
2.3. Immunoblotting
2.4. Total RNA Purification from Whole-Body Samples and cDNA Synthesis
2.5. Quantitative Real-Time PCR (qRT-PCR)
2.6. Synthesis and Injection of dsRNA
2.7. Determination of the LC50 of Paraquat in TcDJ-1 Knockdown Adults
2.8. Examination of the Relationship of TcDJ-1 with the Antioxidant Proteins, Superoxide Dismutases (SODs)
3. Results
3.1. Identification and Characterization of the TcDJ-1 Nucleotide Sequence
3.2. Examination of Cross-Reactivity in TcDJ-1, and BmDJ-1
3.3. Examination of TcDJ-1 mRNA Expression during Development
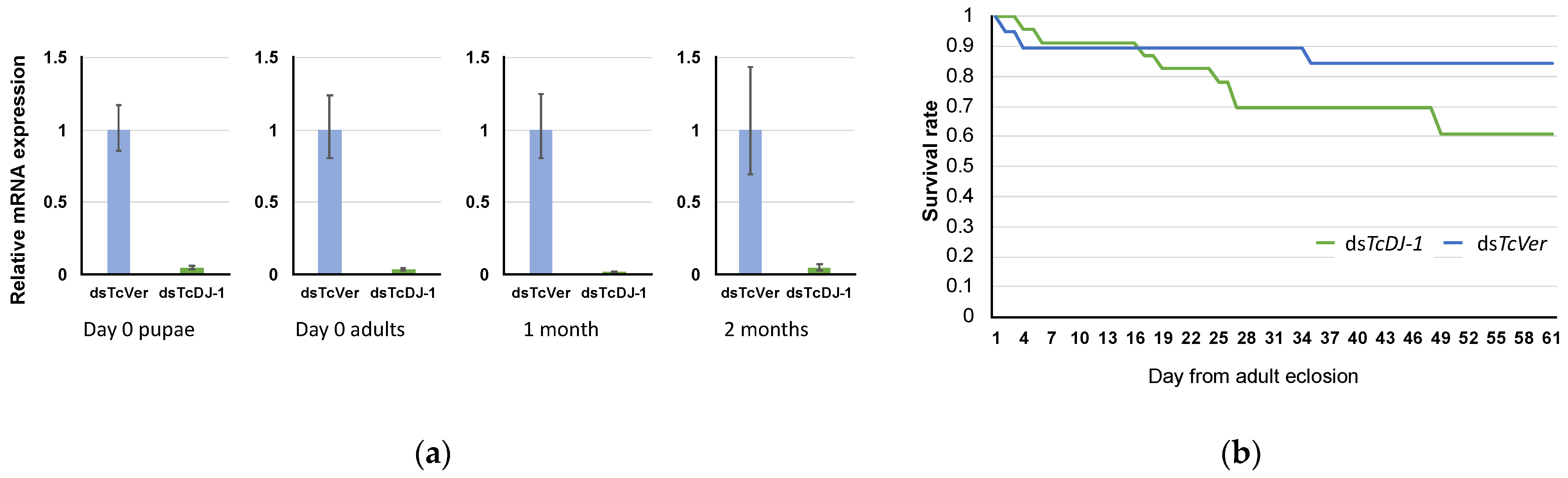
3.4. Evaluation of TcDJ-1 Knockdown Efficiency
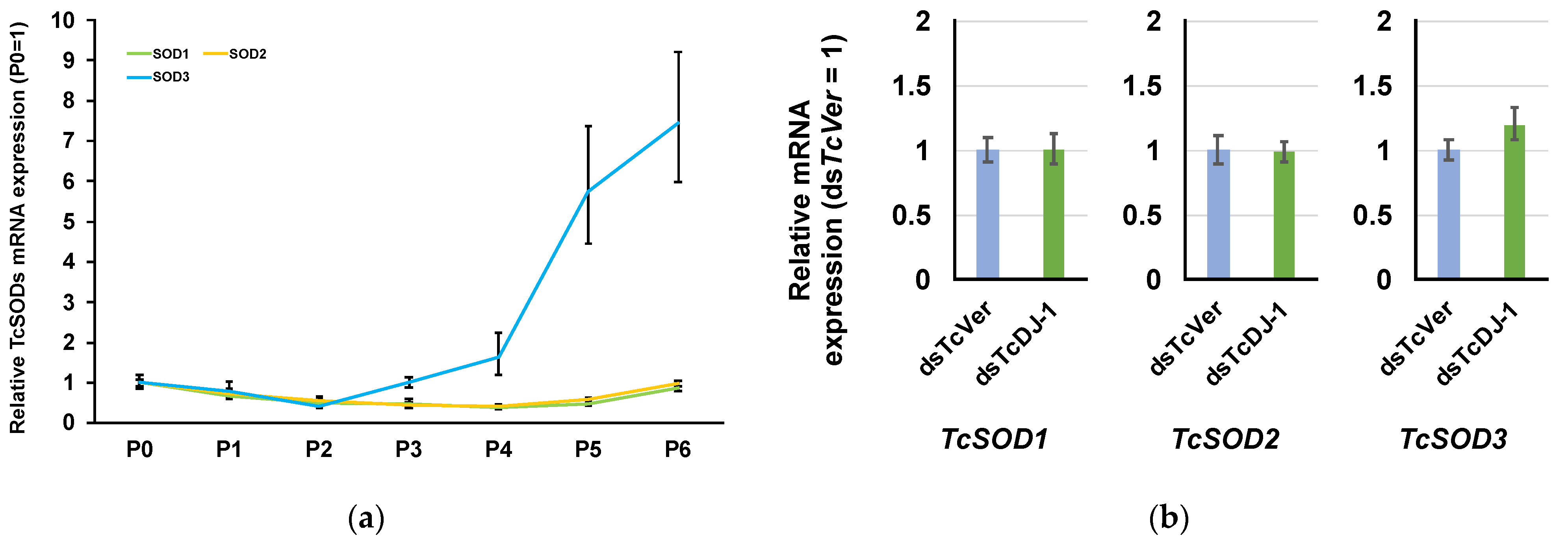
3.5. Examination of the Relationship between TcSOD Expression and TcDJ-1 Knockdown
3.6. Examination of Paraquat-Induced Oxidative Stress in TcDJ-1 Knockdown Beetles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nagakubo, D.; Taira, T.; Kitaura, H.; Ikeda, M.; Tamai, K.; Iguchi-Ariga, S.M.M.; Ariga, H. DJ-1, a Novel Oncogene Which Transforms Mouse NIH3T3 Cells in Cooperation with ras. Biochem. Biophys. Res. Commun. 1997, 231, 509–513. [Google Scholar] [CrossRef]
- Lang, A.E.; Lozano, A.M. Parkinson’s disease, First of two parts. N. Engl. J. Med. 1998, 339, 1044–1053. [Google Scholar] [CrossRef]
- Lang, A.E.; Lozano, A.M. Parkinson’s disease, Second of two parts. N. Engl. J. Med. 1998, 339, 1130–1143. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, S.; Cookson, M.R. Evolutionary and functional relationships within the DJ1 superfamily. BMC Evol. 2004, 4, 6. [Google Scholar]
- Clements, C.M.; McNally, R.S.; Conti, B.J.; Mak, T.W.; Ting, J.P.-Y. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. USA 2006, 103, 15091–15096. [Google Scholar] [CrossRef] [Green Version]
- Klinefelter, G.R.; Welch, J.E.; Perreault, S.D.; Moore, H.D.; Zucker, R.M.; Suarez, J.D.; Roberts, N.L.; Bobseine, K.; Jeffay, S. Localization of the Sperm Protein SP22 and Inhibition of Fertility In Vivo and In Vitro. J. Androl. 2002, 23, 48–63. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, L.; Chin, L.-S. Parkinson disease protein DJ-1 converts from a zymogen to a protease by carboxyl-terminal cleavage. Hum. Mol. Genet. 2010, 19, 2395–2408. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wang, J.; Wang, J.; Yang, B.; He, Q.; Weng, Q. Role of DJ-1 in Immune and Inflammatory Diseases. Front. Immunol. 2020, 11, 994. [Google Scholar] [CrossRef] [PubMed]
- Kinumi, T.; Kimata, J.; Taira, T.; Ariga, H.; Nikia, E. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2004, 317, 722–728. [Google Scholar] [CrossRef]
- Bové, J.; Prou, D.; Perier, C.; Przedborski, S. Toxin-induced models of Parkinson’s disease. NeuroRx 2005, 2, 484–494. [Google Scholar] [CrossRef] [Green Version]
- Terzioglu, M.; Galter, D. Parkinson’s disease: Genetic versus toxin-induced rodent models. FEBS J. 2008, 275, 1384–1391. [Google Scholar] [CrossRef]
- Meulener, M.; Whitworth, A.J.; Armstrong-Gold, C.E.; Rizzu, P.; Heutink, P.; Wes, P.; Pallanck, L.J.; Bonini, N. Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson’s disease. Curr. Biol. 2007, 15, 1572–1577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavara-Culebras, E.; Paricio, N. Drosophila DJ-1 mutants are sensitive to oxidative stress and show reduced lifespan and motor deficits. Gene 2007, 400, 158–165. [Google Scholar] [CrossRef]
- Tabunoki, H.; Ode, H.; Banno, Y.; Katsuma, S.; Shimada, T.; Mita, K.; Yamamoto, K.; Sato, R.; Ishii-Nozawa, R.; Satoh, J. BmDJ-1 is a key regulator of oxidative modification in the development of the silkworm, Bombyx mori. PLoS ONE 2011, 6, e17683. [Google Scholar] [CrossRef]
- Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature 2008, 452, 06784. [Google Scholar]
- Schmitt-Engel, C.; Schultheis, D.; Schwirz, J.; Ströhlein, N.; Troelenberg, N.; Majumdar, U.; Dao, V.A.; Grossmann, D.; Richter, T.; Tech, M.; et al. The iBeetle large-scale RNAi screen reveals gene functions for insect development and physiology. Nat. Commun. 2015, 6, 7822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bucher, G.; Scholten, J.; Klingler, M. Parental RNAi in Tribolium (Coleoptera). Curr. Biol. 2002, 12, R85–R86. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, J.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, T.; Boutros, M. E-RNA i: A web application for the multi-species design of RNAi reagents—2010 update. Nucleic Acids Res. 2010, 38, W332–W339. [Google Scholar] [CrossRef]
- Tabunoki, H.; Gorman, M.J.; Dittmer, N.T.; Kanost, M.R. Superoxide dismutase 2 knockdown leads to defects in locomotor activity, sensitivity to paraquat, and increased cuticle pigmentation in Tribolium castaneum. Sci. Rep. 2016, 8, 29583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finny, D.J. Probit Analysis: A Statistical Treatment of the Sigmoid Response Curve; Cambridge University Press: Cambridge, NY, USA, 1947; p. 20. [Google Scholar]
- Halio, S.B.; Bauer, M.W.; Mukund, S.; Adams, M.; Kelly, R.M. Purification and characterization of two functional forms of intracellular protease PfpI from the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 1997, 63, 289–295. [Google Scholar] [CrossRef] [Green Version]
- Halio, S.B.; Blumentals, I.I.; Short, S.A.; Merrill, B.M.; Kelly, R.M. Sequence, expression in Escherichia coli, and analysis of the gene encoding a novel intracellular protease (PfpI) from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 1996, 178, 2605–2612. [Google Scholar] [CrossRef] [Green Version]
- Fridovich, I. Superoxide dismutases. Annu. Rev. Biochem. 1975, 44, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Kiyotake, H.; Matsumoto, H.; Nakayama, S.; Sakai, M.; Miyatake, T.; Ryuda, M.; Hayakawa, Y. Gain of long tonic immobility behavioral trait causes the red flour beetle to reduce anti-stress capacity. J. Insect Physiol. 2014, 60, 92–97. [Google Scholar] [CrossRef]
- Zhang, J.; Lu, A.; Kong, L.; Zhang, Q.; Ling, E. Functional analysis of insect molting fluid proteins on the protection and regulation of ecdysis. J. Biol. Chem. 2014, 289, 35891–35906. [Google Scholar] [CrossRef] [Green Version]
- Nishinaga, H.; Takahashi-Niki, K.; Taira, T.; Andreadis, A.; Iguchi-Ariga, S.M.M.; Ariga, H. Expression profiles of genes in DJ-1-knockdown and L166P DJ-1 mutant cells. Neurosci. Lett. 2005, 390, 54–59. [Google Scholar] [CrossRef]
- Huai, Q.; Sun, Y.; Wang, H.; Chin, L.S.; Li, L.; Robinson, H.; Ke, H. Crystal structure of DJ-1/RS and implication on familial Parkinson’s disease. FEBS Lett. 2003, 549, 171–175. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, S.; Taira, T.; Niki, T.; Takahashi-Niki, K.; Maita, C.; Maita, H.; Ariga, H.; Iguchi-Ariga, S.M. Oxidative status of DJ-1-dependent activation of dopamine synthesis through interaction of tyrosine hydroxylase and 4-dihydroxy-L-phenylalanine (L-DOPA) decarboxylase with DJ-1. J. Biol. Chem. 2009, 284, 28832–28844. [Google Scholar] [CrossRef] [Green Version]
- Miyazaki, S.; Yanagida, T.; Nunome, K.; Ishikawa, S.; Inden, M.; Kitamura, Y.; Nakagawa, S.; Taira, T.; Hirita, K.; Niwa, M.; et al. DJ-1-binding compounds prevent oxidative stress-induced cell death and movement defect in Parkinson’s disease model rats. J. Neurochem. 2008, 105, 2418–2434. [Google Scholar] [CrossRef]
- Anderson, P.C.; Daggett, V. Molecular basis for the structural instability of human DJ-1 induced by the L166P mutation associated with Parkinson’s disease. Biochemistry 2008, 47, 9380–9393. [Google Scholar] [CrossRef] [Green Version]
- Canet-Avilés, R.M.; Wilson, M.A.; Miller, D.W.; Ahmad, R.; McLendon, C.; Bandyopadhyay, S.; Cookson, M.R. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA 2004, 101, 9103–9108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdallah, J.; Kern, R.; Malki, A.; Eckey, V.; Richarme, G. Cloning, expression, and purification of the general stress protein YhbO from Escherichia coli. Protein Expr. Purif. 2006, 47, 455–460. [Google Scholar] [CrossRef] [Green Version]
- Lucas, J.I.; Marín, I. A new evolutionary paradigm for the Parkinson disease gene DJ-1. Mol. Biol. Evol. 2007, 24, 551–561. [Google Scholar] [CrossRef]
- Honbou, K.; Suzuki, N.N.; Horiuchi, M.; Niki, T.; Taira, T.; Ariga, H.; Inagaki, F. The crystal structure of DJ-1, a protein related to male fertility and Parkinson’s disease. J. Biol. Chem. 2003, 278, 31380–31384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, X.; Tong, L. Crystal structure of human DJ-1, a protein associated with early onset Parkinson’s disease. J. Biol. Chem. 2003, 278, 31372–31379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stennicke, H.R.; Salvesen, G.S. Catalytic properties of the caspases. Cell Death Differ. 1999, 6, 1054–1059. [Google Scholar] [CrossRef]
- Walker, R.K.; Krishnaswamy, S. The activation of prothrombin by the prothrombinase complex. The contribution of the substrate-membrane interaction to catalysis. J. Biol. Chem. 1994, 269, 27441–27450. [Google Scholar] [CrossRef]
- Wilson, K.P.; Black, J.A.F.; Thomson, J.A.; Kim, E.E.; Griffith, J.P.; Navia, M.A.; Murcko, M.A.; Chambers, S.P.; Aldape, R.A.; Raybuck, S.A.; et al. Structure and mechanism of interleukin-lβ converting enzyme. Nature 1994, 370, 270–275. [Google Scholar] [CrossRef]
- Lin, J.; Prahlad, J.; Wilson, M.A. Conservation of oxidative protein stabilization in an insect homologue of parkinsonism-associated protein DJ-1. Biochemistry 2012, 51, 3799–3807. [Google Scholar] [CrossRef] [Green Version]
- Tissot, M.; Stocker, R.F. Metamorphosis in Drosophila and other insects: The fate of neurons throughout the stages. Prog. Neurobiol. 2000, 62, 89–111. [Google Scholar] [CrossRef]
- Truman, J.W.; Riddiford, L.M. The evolution of insect metamorphosis: A developmental and endocrine view. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20190070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahle, P.J.; Waak, J.; Gasser, T. DJ-1 and prevention of oxidative stress in Parkinson’s disease and other age-related disorders. Free Radic. Biol. Med. 2009, 47, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Nojima, Y.; Bono, H.; Yokoyama, T.; Iwabuchi, K.; Sato, R.; Arai, K.; Tabunoki, H. Superoxide dismutase down-regulation and the oxidative stress is required to initiate pupation in Bombyx mori. Sci. Rep. 2019, 9, 14693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cochemé, H.M.; Murphy, M.P. Complex I is the major site of mitochondrial superoxide production by paraquat. J. Biol. Chem. 2008, 283, 1786–1798. [Google Scholar] [CrossRef] [Green Version]
- Ramachandiran, S.; Hansen, J.M.; Jones, D.P.; Richardson, J.R.; Miller, G.W. Divergent mechanisms of paraquat, MPP+, and rotenone toxicity: Oxidation of thioredoxin and caspase-3 activation. Toxicol. Sci. 2007, 95, 163–171. [Google Scholar] [CrossRef] [Green Version]
- Cassar, M.; Issa, A.R.; Riemensperger, T.; Petitgas, C.; Rival, T.; Coulom, H.; Birman, S. A dopamine receptor contributes to paraquat-induced neurotoxicity in Drosophila. Hum. Mol. Genet. 2015, 24, 197–212. [Google Scholar] [CrossRef] [Green Version]
- Colle, D.; Farina, M.; Ceccatelli, S.; Raciti, M. Paraquat and maneb exposure alters rat neural stem cell proliferation by inducing oxidative stress: New insights on pesticide-induced neurodevelopmental toxicity. Neurotox. Res. 2018, 34, 820–833. [Google Scholar] [CrossRef]
- McCormack, A.L.; Atienza, J.G.; Johnston, L.C.; Andersen, J.K.; Vu, S.; Di Monte, D.A. Role of oxidative stress in paraquat-induced dopaminergic cell degeneration. J. Neurochem. 2005, 93, 1030–1037. [Google Scholar] [CrossRef]
- Yang, W.; Chen, L.; Ding, Y.; Zhuang, X.; Kang, U.J. Paraquat induces dopaminergic dysfunction and proteasome impairment in DJ-1-deficient mice. Hum. Mol. Genet. 2007, 16, 2900–2910. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Perez, D.A.; Jimenez-Del-Rio, M.; Velez-Pardo, C. Epigallocatechin-3-gallate protects and prevents paraquat-induced oxidative stress and neurodegeneration in knockdown dj-1-β Drosophila melanogaster. Neurotox. Res. 2018, 34, 401–416. [Google Scholar] [CrossRef] [PubMed]
- Menzies, F.M.; Yenisetti, S.C.; Min, K.T. Roles of Drosophila DJ-1 in survival of dopaminergic neurons and oxidative stress. Curr. Biol. 2005, 15, 1578–1582. [Google Scholar] [CrossRef] [PubMed]

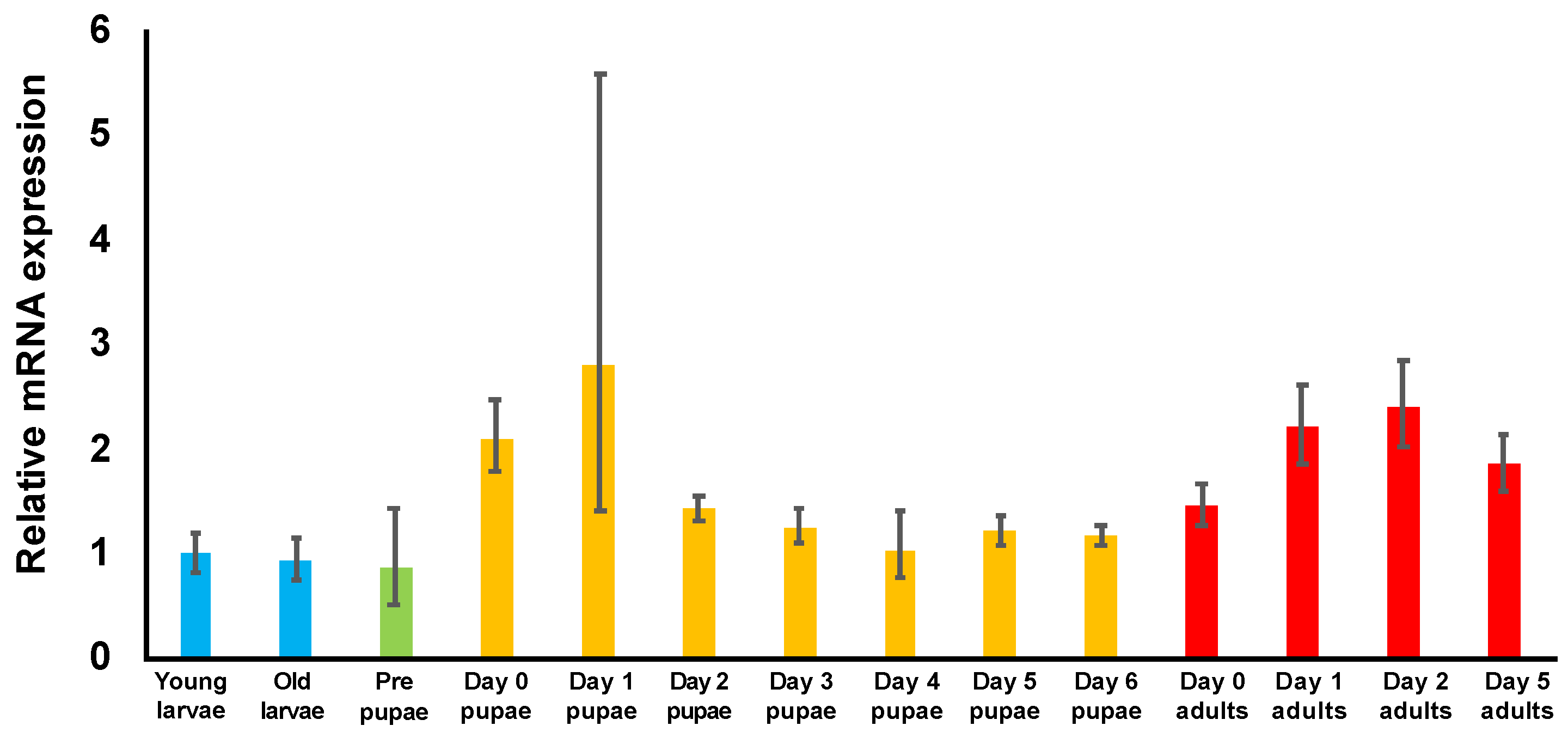
| Scientific Name | 46 | 53 | 106 | 126 |
|---|---|---|---|---|
| Tribolium castaneum | C | K | C | Y |
| Agrilus planipennis | C | K | C | Y |
| Anoplophora glabripennisX1 | C | K | C | Y |
| Anoplophora glabripennisX2 | C | N | C | Y |
| Onthophagus taurus | C | K | C | Y |
| Dufourea novaeangliae | C | C | C | Y |
| Camponotus floridanus | C | C | C | Y |
| Melipona quadrifasciata | C | C | C | Y |
| Harpegnathos saltator | C | C | C | Y |
| Ooceraea biroi | C | C | C | Y |
| Trachymyrmex cornetzi | C | C | C | Y |
| Drosophila melanogaster alpha | C | V | C | H |
| Drosophila melanogaster beta | C | L | C | Y |
| Bactrocera latifrons | C | V | C | Y |
| Bactrocera dorsalis | C | V | C | Y |
| Papilio xuthus | C | V | C | Y |
| Papilio machaon | C | V | C | Y |
| Bombyx mori | C | V | C | Y |
| Homo sapiens | C | C | C | H |
| Mus musculus | C | C | C | H |
| Moesziomyces antarcticus | C | V | C | H |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sasaki, S.; Nishiko, M.; Sakamoto, T.; Kanost, M.R.; Tabunoki, H. cDNA Cloning and Partial Characterization of the DJ-1 Gene from Tribolium castaneum. Antioxidants 2021, 10, 1970. https://doi.org/10.3390/antiox10121970
Sasaki S, Nishiko M, Sakamoto T, Kanost MR, Tabunoki H. cDNA Cloning and Partial Characterization of the DJ-1 Gene from Tribolium castaneum. Antioxidants. 2021; 10(12):1970. https://doi.org/10.3390/antiox10121970
Chicago/Turabian StyleSasaki, Shunya, Maaya Nishiko, Takuma Sakamoto, Michael R. Kanost, and Hiroko Tabunoki. 2021. "cDNA Cloning and Partial Characterization of the DJ-1 Gene from Tribolium castaneum" Antioxidants 10, no. 12: 1970. https://doi.org/10.3390/antiox10121970
APA StyleSasaki, S., Nishiko, M., Sakamoto, T., Kanost, M. R., & Tabunoki, H. (2021). cDNA Cloning and Partial Characterization of the DJ-1 Gene from Tribolium castaneum. Antioxidants, 10(12), 1970. https://doi.org/10.3390/antiox10121970







