The Modus Operandi of Hydrogen Sulfide(H2S)-Dependent Protein Persulfidation in Higher Plants
Abstract
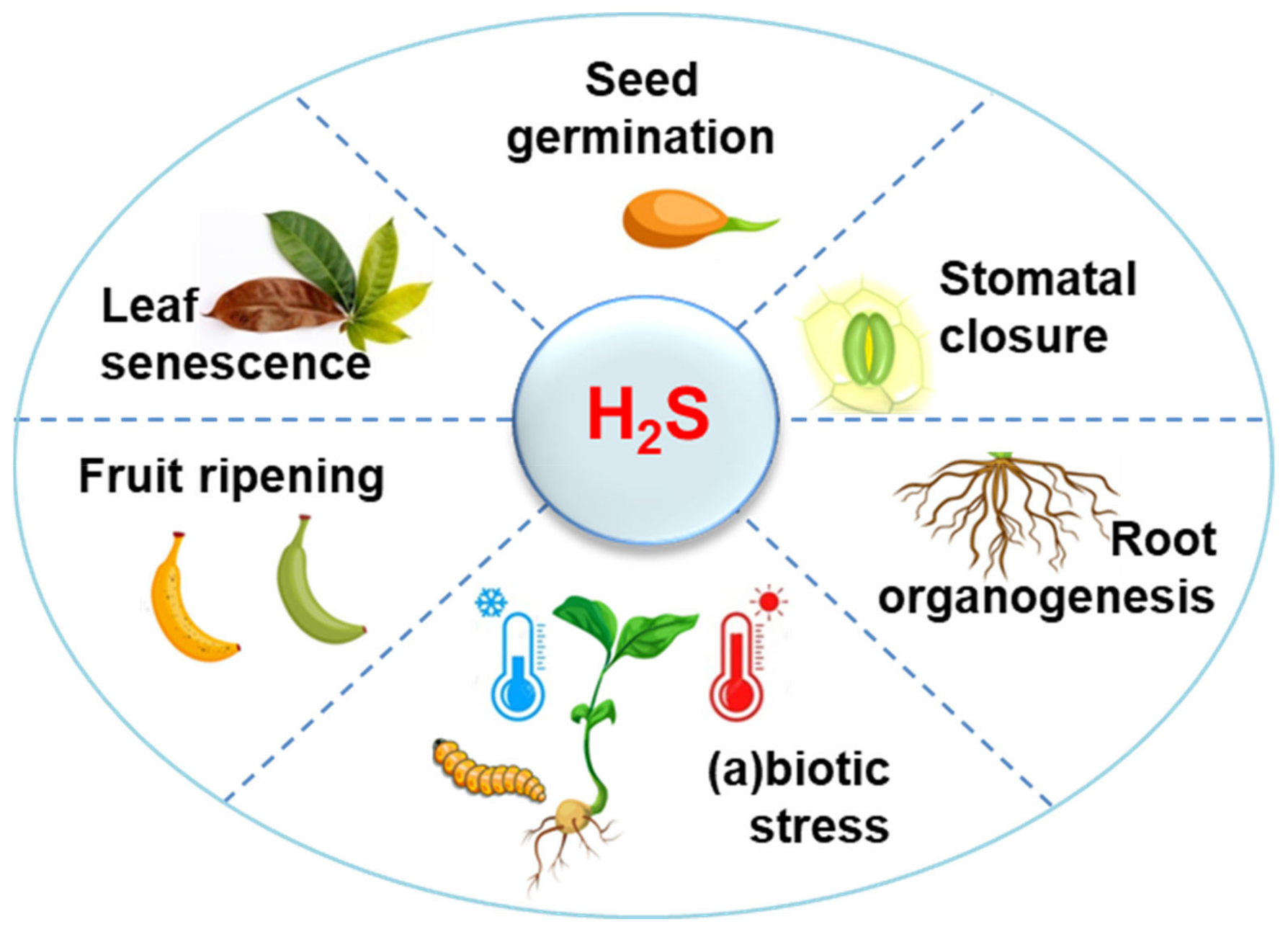
1. H2S Metabolism in Higher Plants: A Perspective
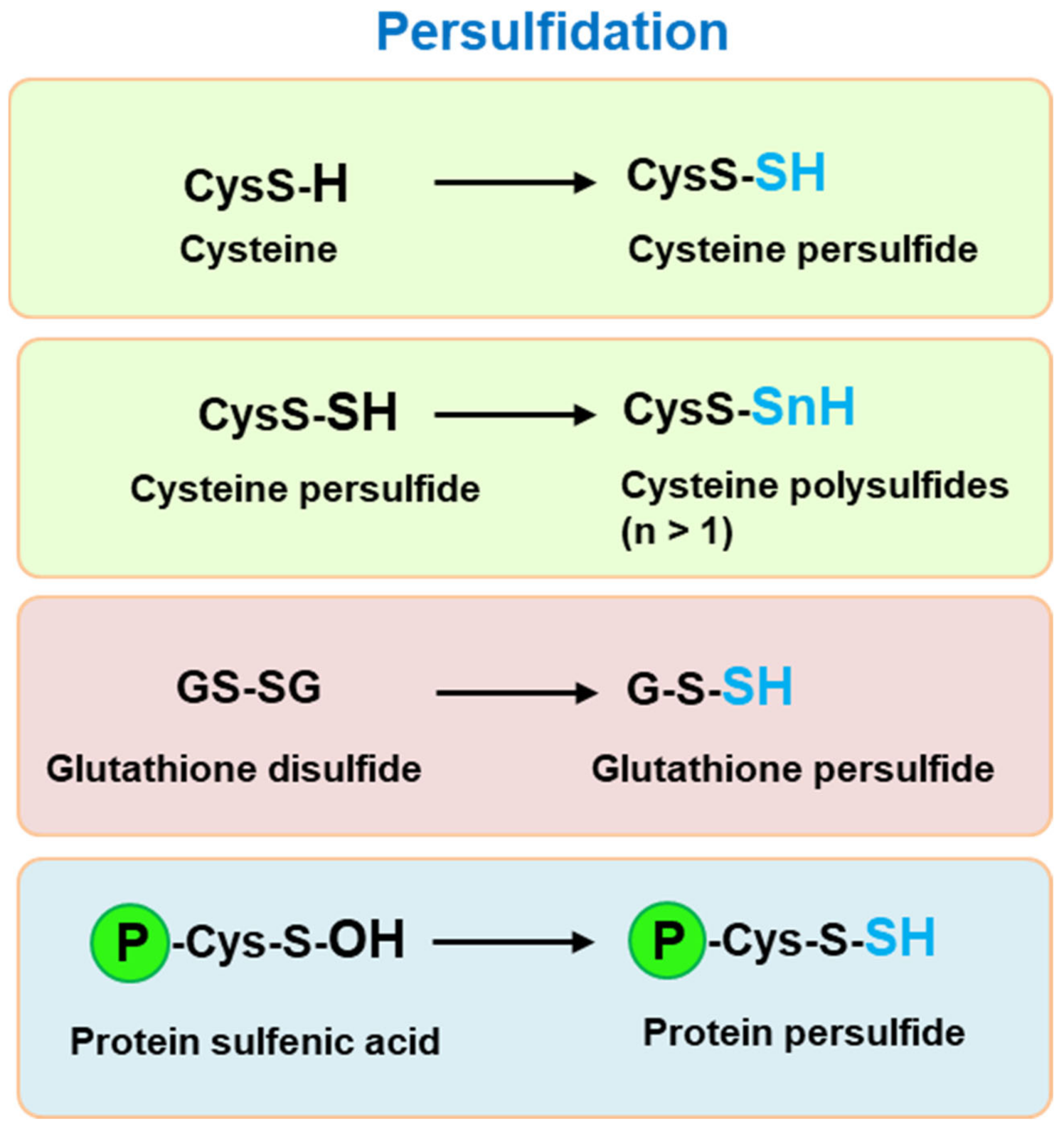
2. Persufidation: A Redox Regulation of Thiol Groups
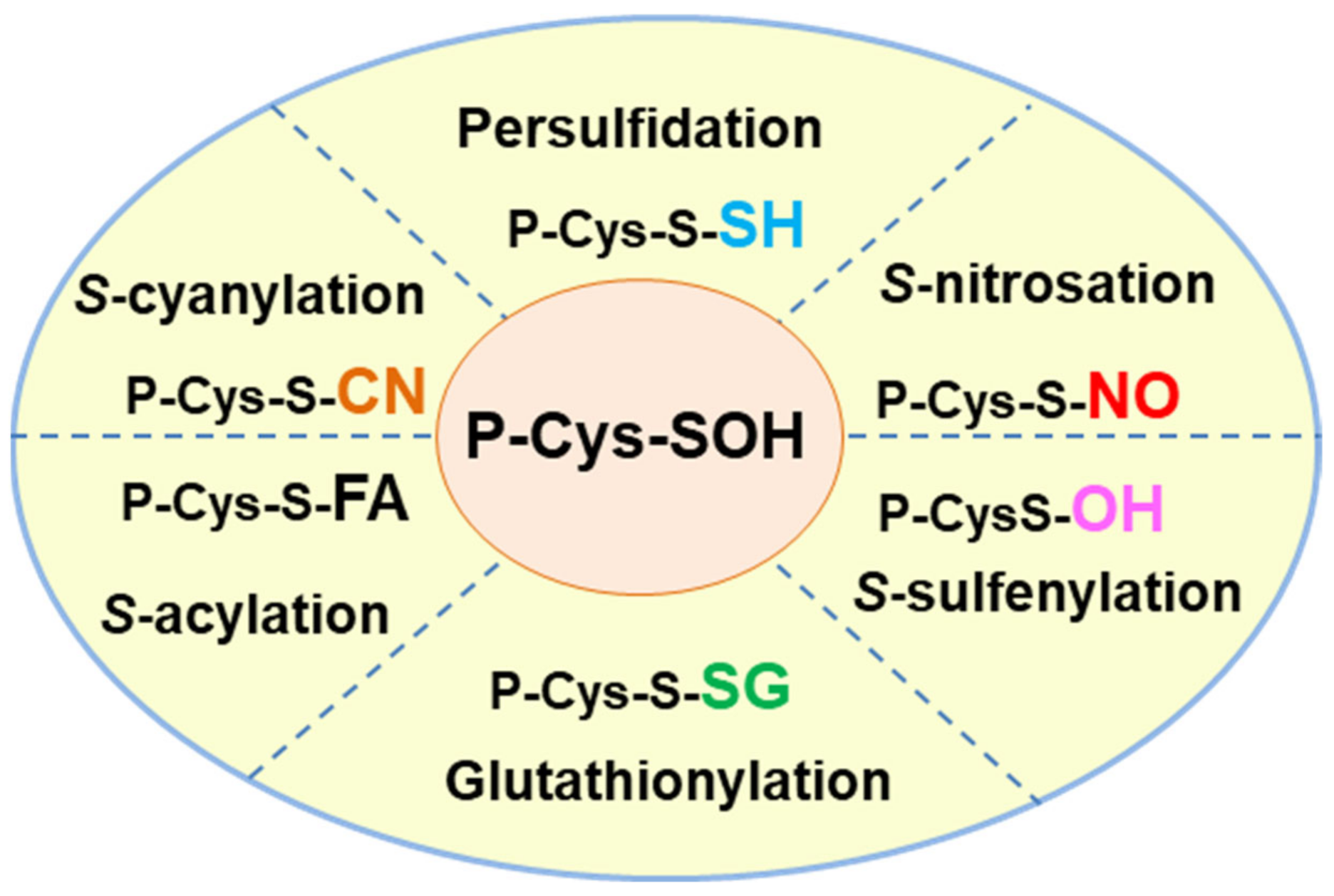
3. Protein Persulfidation and Other Cysteine Modifications
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Abe, K.; Kimura, H. The possible role of hydrogen sulfide as an endogenous neuromodulator. J. Neurosci. 1996, 16, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- Hosoki, R.; Matsuki, N.; Kimura, H. The possible role of hydrogen sulfide as an endogenous smooth muscle relaxant in synergy with nitric oxide. Biochem. Biophys. Res. Commun. 1997, 237, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Petersen, E. Preservation of ascorbic acid in vegetables by hydrogen sulphide during air-drying. Nature 1948, 157, 370. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.M.; Ibrahium, I.K.A.; Hollis, J.P. Hydrogen sulphide: Effects on the physiology of rice plants and relation to straight head disease. Phytophatology 1975, 65, 1165–1170. [Google Scholar] [CrossRef]
- Jin, Z.; Sun, L.; Yang, G.; Pei, Y. Hydrogen sulfide regulates energy production to delay leaf senescence induced by drought stress in Arabidopsis. Front. Plant Sci. 2018, 9, 1722. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B.; González-Gordo, S.; Muñoz-Vargas, M.A.; Palma, J.M. Hydrogen sulfide: A novel component in Arabidopsis peroxisomes which triggers catalase inhibition. J. Integral Plant Biol. 2019, 61, 871–883. [Google Scholar] [CrossRef]
- Pantaleno, R.; Scuffi, D.; García-Mata, C. Hydrogen sulfide as a guard cell network regulator. New Phytol. 2021, 230, 451–456. [Google Scholar] [CrossRef]
- Siodmak, A.; Hirt, H. New insights into stomatal regulation: Role of H2S induced persulfidation in ABA signaling. Mol. Plant. 2021, 14, 858–860. [Google Scholar] [CrossRef]
- Fu, L.; Liu, K.; He, J.; Tian, C.; Yu, X.; Yang, J. Direct proteomic mapping of cysteine persulfidation. Antioxid. Redox Signal. 2020, 33, 1061–1076. [Google Scholar] [CrossRef]
- Ma, D.; Ding, H.; Wang, C.; Qin, H.; Han, Q.; Hou, J.; Lu, H.; Xie, Y.; Guo, T. Alleviation of drought stress by hydrogen sulfide is partially related to the abscisic acid signaling pathway in wheat. PLoS ONE 2016, 11, e0163082. [Google Scholar] [CrossRef]
- Corpas, F.J. Hydrogen Sulfide: A New Warrior against Abiotic Stress. Trends Plant Sci. 2019, 24, 983–988. [Google Scholar] [CrossRef]
- Kharbech, O.; Houmani, H.; Chaoui, A.; Corpas, F.J. Alleviation of Cr(VI)-induced oxidative stress in maize (Zea mays L.) seedlings by NO and H2S donors through differential organ-dependent regulation of ROS and NADPH-recycling metabolisms. J. Plant Physiol. 2017, 219, 71–80. [Google Scholar] [CrossRef]
- Kharbech, O.; Sakouhi, L.; Ben Massoud, M.; Jose Mur, L.A.; Corpas, F.J.; Djebali, W.; Chaoui, A. Nitric oxide and hydrogen sulfide protect plasma membrane integrity and mitigate chromium-induced methylglyoxal toxicity in maize seedlings. Plant Physiol. Biochem. 2020, 157, 244–255. [Google Scholar] [CrossRef]
- Li, J.; Shi, C.; Wang, X.; Liu, C.; Ding, X.; Ma, P.; Wang, X.; Jia, H. Hydrogen sulfide regulates the activity of antioxidant enzymes through persulfidation and improves the resistance of tomato seedling to copper oxide nanoparticles (CuO NPs)-induced oxidative stress. Plant Physiol. Biochem. 2020, 156, 257–266. [Google Scholar] [CrossRef]
- Corpas, F.J.; Palma, J.M. H2S signaling in plants and applications in agriculture. J. Adv. Res. 2020, 24, 131–137. [Google Scholar] [CrossRef]
- Goyal, V.; Jhanghel, D.; Mehrotra, S. Emerging warriors against salinity in plants: Nitric oxide and hydrogen sulphide. Physiol. Plant. 2021, 171, 896–908. [Google Scholar] [CrossRef]
- Liu, H.; Xue, S. Interplay between hydrogen sulfide and other signaling molecules in the regulation of guard cell signaling and abiotic/biotic stress response. Plant Commun. 2021, 2, 100179. [Google Scholar] [CrossRef]
- Zhou, L.; Gao, X.; Weits, D.A.; Zeng, P.; Wang, X.; Cai, J. H2S regulates low oxygen signaling via integration with the unfolded protein response in Arabidopsis thaliana. Plant Soil 2021, 467, 531–547. [Google Scholar] [CrossRef]
- Iqbal, N.; Umar, S.; Khan, N.A.; Corpas, F.J. Nitric oxide and hydrogen sulfide coordinately reduce glucose sensitivity and decrease oxidative stress via ascorbate-glutathione cycle in heat-stressed wheat (Triticum aestivum L.) plants. Antioxidants 2021, 10, 108. [Google Scholar] [CrossRef]
- Gotor, C.; Laureano-Marín, A.M.; Moreno, I.; Aroca, Á.; García, I.; Romero, L.C. Signaling in the plant cytosol: Cysteine or sulfide? Amino Acids 2015, 47, 2155–2164. [Google Scholar] [CrossRef]
- Selles, B.; Moseler, A.; Rouhier, N.; Couturier, J. Rhodanese domain-containing sulfurtransferases: Multifaceted proteins involved in sulfur trafficking in plants. J. Exp. Bot. 2109, 70, 4139–4154. [Google Scholar] [CrossRef]
- González-Gordo, S.; Palma, J.M.; Corpas, F.J. Appraisal of H2S metabolism in Arabidopsis thaliana: In silico analysis at the subcellular level. Plant Physiol. Biochem. 2020, 155, 579–588. [Google Scholar] [CrossRef]
- Kumar, R.; Banerjee, R. Regulation of the redox metabolome and thiol proteome by hydrogen sulfide. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 221–235. [Google Scholar] [CrossRef]
- Mishra, V.; Singh, P.; Tripathi, D.K.; Corpas, F.J.; Singh, V.P. Nitric oxide and hydrogen sulfide: An indispensable combination for plant functioning. Trends Plant Sci. 2021, in press. [Google Scholar] [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Rodriguez-Ruiz, M.; Muñoz-Vargas, M.A.; Palma, J.M. Nitric oxide and hydrogen sulfide share regulatory functions in higher plant events. BioCell 2022, 46, 1–5. [Google Scholar] [CrossRef]
- Rudyk, O.; Eaton, P. Biochemical methods for monitoring protein thiol redox states in biological systems. Redox Biol. 2014, 2, 803–813. [Google Scholar] [CrossRef]
- Bak, D.W.; Bechtel, T.J.; Falco, J.A.; Weerapana, E. Cysteine reactivity across the subcellular universe. Curr. Opin. Chem. Biol. 2019, 48, 96–105. [Google Scholar] [CrossRef]
- Filipovic, M.R.; Zivanovic, J.; Alvarez, B.; Banerjee, R. Chemical biology of H2S signaling through persulfidation. Chem. Rev. 2018, 118, 1253–1337. [Google Scholar] [CrossRef]
- Benchoam, D.; Semelak, J.A.; Cuevasanta, E.; Mastrogiovanni, M.; Grassano, J.S.; Ferrer-Sueta, G.; Zeida, A.; Trujillo, M.; Möller, M.N.; Estrin, D.A.; et al. Acidity and nucleophilic reactivity of glutathione persulfide. J. Biol. Chem. 2020, 295, 15466–15481. [Google Scholar] [CrossRef]
- Fu, L.H.; Hu, K.D.; Hu, L.Y.; Li, Y.H.; Hu, L.B.; Yan, H.; Liu, Y.S.; Zhang, H. An antifungal role of hydrogen sulfide on the postharvest pathogens Aspergillus niger and Penicillium italicum. PLoS ONE 2014, 9, e104206. [Google Scholar] [CrossRef]
- Echizen, H.; Hanaoka, K. Recent advances in probe design to detect reactive sulfur species and in the chemical reactions employed for fluorescence switching. J Clin. Biochem. Nutr. 2021, 68, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Abiko, Y.; Katayama, Y.; Akiyama, M.; Kumagai, Y. Lipophilic compounds in garlic decrease the toxicity of methylmercury by forming sulfur adducts. Food Chem. Toxicol. 2021, 150, 112061. [Google Scholar] [CrossRef] [PubMed]
- Akaike, T.; Ida, T.; Wei, F.Y.; Nishida, M.; Kumagai, Y.; Alam, M.M.; Ihara, H.; Sawa, T.; Matsunaga, T.; Kasamatsu, S.; et al. Cysteinyl-tRNA synthetase governs cysteine polysulfidation and mitochondrial bioenergetics. Nat. Commun. 2017, 8, 1177. [Google Scholar] [CrossRef] [PubMed]
- Sawa, T.; Motohashi, H.; Ihara, H.; Akaike, T. Enzymatic regulation and biological functions of reactive cysteine persulfides and polysulfides. Biomolecules 2020, 10, 1245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Tsutsuki, H.; Ono, K.; Akaike, T.; Sawa, T. Antioxidative and anti-inflammatory actions of reactive cysteine persulfides. J. Clin. Biochem. Nutr. 2021, 68, 5–8. [Google Scholar] [CrossRef]
- Chen, J.; Liu, T.W.; Hu, W.J.; Simon, M.; Wang, W.H.; Chen, J.; Liu, X.; Zheng, H.L. Comparative proteomic analysis of differentially expressed proteins induced by hydrogen sulfide in Spinacia oleracea leaves. PLoS ONE 2014, 9, e105400. [Google Scholar] [CrossRef][Green Version]
- Aroca, Á.; Serna, A.; Gotor, C.; Romero, L.C. S-sulfhydration: A cysteine posttranslational modification in plant systems. Plant Physiol. 2015, 168, 334–342. [Google Scholar] [CrossRef]
- Aroca, A.; Benito, J.M.; Gotor, C.; Romero, L.C. Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J. Exp. Bot. 2017, 68, 4915–4927. [Google Scholar] [CrossRef]
- Jurado-Flores, A.; Romero, L.C.; Gotor, C. Label-Free quantitative proteomic analysis of nitrogen starvation in arabidopsis root reveals new aspects of H2S signaling by protein persulfidation. Antioxidants 2021, 10, 508. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, M.; Zhou, H.; Zhao, D.; Gotor, C.; Romero, L.C.; Shen, J.; Ge, Z.; Zhang, Z.; Shen, W.; et al. Hydrogen sulfide (H2S), a signaling molecule in plant stress responses. J. Integral Plant Biol. 2020, 63, 146–160. [Google Scholar] [CrossRef]
- Chen, J.; Wu, F.H.; Wang, W.H.; Zheng, C.J.; Lin, G.H.; Dong, X.J.; He, J.X.; Pei, Z.M.; Zheng, H.L. Hydrogen sulphide enhances photosynthesis through promoting chloroplast biogenesis, photosynthetic enzyme expression, and thiol redox modification in Spinacia oleracea seedlings. J. Exp. Bot. 2011, 62, 4481–4493. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, J.; Zhou, M.; Zhou, H.; Cui, B.; Gotor, C.; Romero, L.C.; Fu, L.; Yang, J.; Foyer, C.H.; et al. Persulfidation-based modification of cysteine desulfhydrase and the NADPH oxidase RBOHD controls guard cell abscisic acid signaling. Plant Cell. 2020, 32, 1000–1017. [Google Scholar] [CrossRef]
- Li, J.; Chen, S.; Wang, X.; Shi, C.; Liu, H.; Yang, J.; Shi, W.; Guo, J.; Jia, H. Hydrogen sulfide disturbs actin polymerization via S-Sulfhydration resulting in stunted root hair growth. Plant Physiol. 2018, 178, 936–949. [Google Scholar] [CrossRef]
- Jia, H.; Chen, S.; Liu, D.; Liesche, J.; Shi, C.; Wang, J.; Ren, M.; Wang, X.; Yang, J.; Shi, W.; et al. Ethylene-induced hydrogen sulfide negatively regulates ethylene biosynthesis by persulfidation of ACO in tomato under osmotic stress. Front. Plant Sci. 2018, 9, 1517. [Google Scholar] [CrossRef]
- Muñoz-Vargas, M.A.; González-Gordo, S.; Cañas, A.; López-Jaramillo, J.; Palma, J.M.; Corpas, F.J. Endogenous hydrogen sulfide (H2S) is up-regulated during sweet pepper (Capsicum annuum L.) fruit ripening. In vitro analysis shows that NADP-dependent isocitrate dehydrogenase (ICDH) activity is inhibited by H2S and NO. Nitric Oxide 2018, 81, 36–45. [Google Scholar] [CrossRef]
- Muñoz-Vargas, M.A.; González-Gordo, S.; Palma, J.M.; Corpas, F.J. Inhibition of NADP-malic enzyme activity by H2S and NO in sweet pepper (Capsicum annuum L.) fruits. Physiol. Plant. 2020, 168, 278–288. [Google Scholar]
- Chen, S.; Jia, H.; Wang, X.; Shi, C.; Wang, X.; Ma, P.; Wang, J.; Ren, M.; Li, J. Hydrogen sulfide positively regulates abscisic acid signaling through persulfidation of SnRK2.6 in guard cells. Mol. Plant 2020, 13, 732–744. [Google Scholar] [CrossRef]
- Chen, S.; Wang, X.; Jia, H.; Li, F.; Ma, Y.; Liesche, J.; Liao, M.; Ding, X.; Liu, C.; Chen, Y.; et al. Persulfidation-induced structural change in SnRK2.6 establishes intramolecular interaction between phosphorylation and persulfidation. Mol. Plant 2021, in press. [Google Scholar] [CrossRef]
- Laureano-Marín, A.M.; Aroca, A.; Perez-Perez, M.E.; Yruela, I.; Jurado-Flores, A.; Moreno, I.; Crespo, J.L.; Romero, L.C.; Gotor, C. Abscisic acid-triggered persulfidation of cysteine protease ATG4 mediates regulation of autophagy by sulfide. Plant Cell 2020, 32, 3902–3920. [Google Scholar] [CrossRef]
- Aroca, A.; Yruela, I.; Gotor, C.; Bassham, D.C. Persulfidation of ATG18a regulates autophagy under ER stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2023604118. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, L.; Pei, Z.; Zhang, L.; Liu, Z.; Liu, D.; Hao, X.; Jin, Z.; Pei, Y. Hydrogen sulfide promotes flowering in heading Chinese cabbage by S-sulfhydration of BraFLCs. Hortic. Res. 2021, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Jin, Z.; Liu, Z.; Liu, D.; Zhang, L.; Ma, X.; Yang, G.; Liu, S.; Guo, Y.; Pei, Y. H2S persulfidated and increased kinase activity of MPK4 to response cold stress in Arabidopsis. Front. Mol. Biosci. 2021, 8, 635470. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhang, J.; Shen, J.; Zhou, H.; Zhao, D.; Gotor, C.; Romero, L.C.; Fu, L.; Li, Z.; Yang, J.; et al. Hydrogen sulfide-linked persulfidation of ABSCISIC INSENSITIVE 4 Controls Arabidopsis ABA responses through the transactivation of mitogen-activated protein kinase kinase kinase 18. Mol. Plant 2021, 14, 921–936. [Google Scholar] [CrossRef] [PubMed]
- Moseler, A.; Dhalleine, T.; Rouhier, N.; Couturier, J. Arabidopsis thaliana 3-mercaptopyruvate sulfurtransferases interact with and are protected by reducing systems. J. Biol. Chem. 2021, 296, 100429. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, L.; Tan, D.; Sun, W.; Ke, Q.; Yue, X.; Bai, B. S-desulfurization: A different covalent modification mechanism from persulfidation by GSH. Free Radic. Biol. Med. 2021, 167, 54–65. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Bedhomme, M.; Lemaire, S.D.; Trost, P. The emerging roles of protein glutathionylation in chloroplasts. Plant Sci. 2012, 185–186, 86–96. [Google Scholar] [CrossRef]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef]
- Corpas, F.J.; Alché, J.D.; Barroso, J.B. Current overview of S-nitrosoglutathione (GSNO) in higher plants. Front. Plant Sci. 2013, 4, 126. [Google Scholar] [CrossRef]
- Huang, J.; Willems, P.; Wei, B.; Tian, C.; Ferreira, R.B.; Bodra, N.; Martínez Gache, S.A.; Wahni, K.; Liu, K.; Vertommen, D.; et al. Mining for protein S-sulfenylation in Arabidopsis uncovers redox-sensitive sites. Proc. Natl. Acad. Sci. USA 2019, 116, 21256–21261. [Google Scholar] [CrossRef]
- Wei, B.; Willems, P.; Huang, J.; Tian, C.; Yang, J.; Messens, J.; Van Breusegem, F. Identification of sulfenylated cysteines in Arabidopsis thaliana proteins using a disulfide-linked peptide reporter. Front. Plant Sci. 2020, 11, 777. [Google Scholar] [CrossRef]
- Hurst, C.H.; Wright, K.M.; Turnbull, D.; Leslie, K.; Jones, S.; Hemsley, P.A. Juxta-membrane S-acylation of plant receptor-like kinases is likely fortuitous and does not necessarily impact upon function. Sci. Rep. 2019, 9, 12818. [Google Scholar] [CrossRef]
- Hemsley, P.A. S-acylation in plants: An expanding field. Biochem. Soc. Trans. 2020, 48, 529–536. [Google Scholar] [CrossRef]
- García, I.; Arenas-Alfonseca, L.; Moreno, I.; Gotor, C.; Romero, L.C. HCN regulates cellular processes through post-translational modification of proteins by S-cyanylation. Plant Physiol. 2019, 179, 107–123. [Google Scholar] [CrossRef]
- Li, S.; Yu, K.; Wu, G.; Zhang, Q.; Wang, P.; Zheng, J.; Liu, Z.X.; Wang, J.; Gao, X.; Cheng, H. pCysMod: Prediction of Multiple Cysteine Modifications Based on Deep Learning Framework. Front. Cell Dev. Biol. 2021, 9, 617366. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, Q.; Li, S.; Cheng, B.; Xue, H.; Wei, Z.; Shao, T.; Liu, Z.X.; Cheng, H.; Wang, Z. iCysMod: An integrative database for protein cysteine modifications in eukaryotes. Brief. Bioinform. 2021, 7, bbaa400. [Google Scholar] [CrossRef]
- Asada, K. Ascorbate peroxidase: A hydrogen peroxide scavenging enzyme in plants. Physiol. Plant. 1992, 85, 235–241. [Google Scholar] [CrossRef]
- Corpas, F.J.; Trelease, R.N. Differential expression of ascorbate peroxidase and a putative molecular chaperone in the boundary membrane of differentiating cucumber seedling peroxisomes. J. Plant Physiol. 1998, 153, 332–338. [Google Scholar] [CrossRef]
- Shigeoka, S.; Ishikawa, T.; Tamoi, M.; Miyagawa, Y.; Takeda, T.; Yabuta, Y.; Yoshimura, K. Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 2002, 53, 1305–1319. [Google Scholar] [CrossRef]
- Kitajima, S.; Kurioka, M.; Yoshimoto, T.; Shindo, M.; Kanaori, K.; Tajima, K.; Oda, K. A cysteine residue near the propionate side chain of heme is the radical site in ascorbate peroxidase. FEBS J. 2008, 275, 470–480. [Google Scholar] [CrossRef]
- Qu, C.; Wang, L.; Zhao, Y.; Liu, C. Molecular evolution of maize ascorbate peroxidase genes and their functional divergence. Genes 2020, 11, 1204. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; Sánchez-Calvo, B.; Chaki, M.; Valderrama, R.; Mata-Pérez, C.; López-Jaramillo, J.; Padilla, M.N.; Carreras, A.; Corpas, F.J.; Barroso, J.B. Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J. Exp. Bot. 2014, 65, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Huang, X.; Chen, L.; Sun, X.; Lu, C.; Zhang, L.; Wang, Y.; Zuo, J. Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2015, 167, 1731–1746. [Google Scholar] [CrossRef] [PubMed]
- Palma, J.M.; Mateos, R.M.; López-Jaramillo, J.; Rodríguez-Ruiz, M.; González-Gordo, S.; Lechuga-Sancho, A.M.; Corpas, F.J. Plant catalases as NO and H2S targets. Redox Biol. 2020, 34, 101525. [Google Scholar] [CrossRef] [PubMed]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Nitric oxide (NO) and hydrogen sulfide (H2S) modulate the NADPH-generating enzymatic system in higher plants. J. Exp. Bot. 2021, 72, 830–847. [Google Scholar] [CrossRef]
| Enzyme/Protein | Function | Effect | Ref. |
|---|---|---|---|
| RuBISCO | Photosynthesis | Activity up-regulated | [41] |
| O-acetylserine(thiol)lyase (OAS-TL) | Sulfur metabolism | Activity up-regulated | [41] |
| L-cysteine desulphydrase (LCD) | Sulfur metabolism | Activity up-regulated | [42] |
| Ascorbate peroxidase (APX) | Antioxidant | Activity up-regulated | [14,37] |
| Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) | Production of energy in the glycolysis | Activity up-regulated | [37] |
| Glutamine synthetase (GS) | Metabolism of nitrogen | Activity down-regulated | [37] |
| Actin | Organelle movement, cell division and expansion | Inhibits actin polymerization | [43] |
| 1-aminocyclopropane-1-carboxylic acid oxidase (ACO) | Ethylene biosynthesis | Activity down-regulated | [44] |
| NADP-isocitrate dehydrogenase (NADP-ICDH) | Provides NADPH as a reducing agent | Activity down-regulated | [45] |
| NADP-malic enzyme (NADP-ME) | Provides NADPH as a reducing agent | Activity down-regulated | [46] |
| Catalase | Antioxidant | Activity down-regulated | [6,14] |
| SNF1-RELATED PROTEIN KINASE2.6 (SnRK2.6) | Promotes ABA signaling | Promotes stomatal closureActivity up-regulated | [47,48] |
| Respiratory burst oxidase homolog protein D (RBOHD) | Generation of superoxide radical | Activity up-regulated | [42] |
| Cysteine protease ATG4 | Autophagy | Inhibits autophagy | [49] |
| ATG18 | Autophagy | Negative regulation | [50] |
| Peroxidase | ROS metabolism | Activity up-regulated | [14] |
| Flowering Locus C protein (FLC1 and 3) | Flowering regulatory pathway | Reduces binding abilities of FLCs | [51] |
| Mitogen-activated protein kinase (MPK) 4 | Allivates cold stress | Activity up-regulated | [52] |
| Abscisic acid insensitive 4 (ABI4) | Regulates ABA signaling | Enhanced the transactivation activity of ABI4 towards MAPKKK18 | [53] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corpas, F.J.; González-Gordo, S.; Muñoz-Vargas, M.A.; Rodríguez-Ruiz, M.; Palma, J.M. The Modus Operandi of Hydrogen Sulfide(H2S)-Dependent Protein Persulfidation in Higher Plants. Antioxidants 2021, 10, 1686. https://doi.org/10.3390/antiox10111686
Corpas FJ, González-Gordo S, Muñoz-Vargas MA, Rodríguez-Ruiz M, Palma JM. The Modus Operandi of Hydrogen Sulfide(H2S)-Dependent Protein Persulfidation in Higher Plants. Antioxidants. 2021; 10(11):1686. https://doi.org/10.3390/antiox10111686
Chicago/Turabian StyleCorpas, Francisco J., Salvador González-Gordo, María A. Muñoz-Vargas, Marta Rodríguez-Ruiz, and José M. Palma. 2021. "The Modus Operandi of Hydrogen Sulfide(H2S)-Dependent Protein Persulfidation in Higher Plants" Antioxidants 10, no. 11: 1686. https://doi.org/10.3390/antiox10111686
APA StyleCorpas, F. J., González-Gordo, S., Muñoz-Vargas, M. A., Rodríguez-Ruiz, M., & Palma, J. M. (2021). The Modus Operandi of Hydrogen Sulfide(H2S)-Dependent Protein Persulfidation in Higher Plants. Antioxidants, 10(11), 1686. https://doi.org/10.3390/antiox10111686









