Protein Persulfidation in Plants: Function and Mechanism
Abstract
:1. Introduction
2. H2S and Protein Persulfidation in Plants
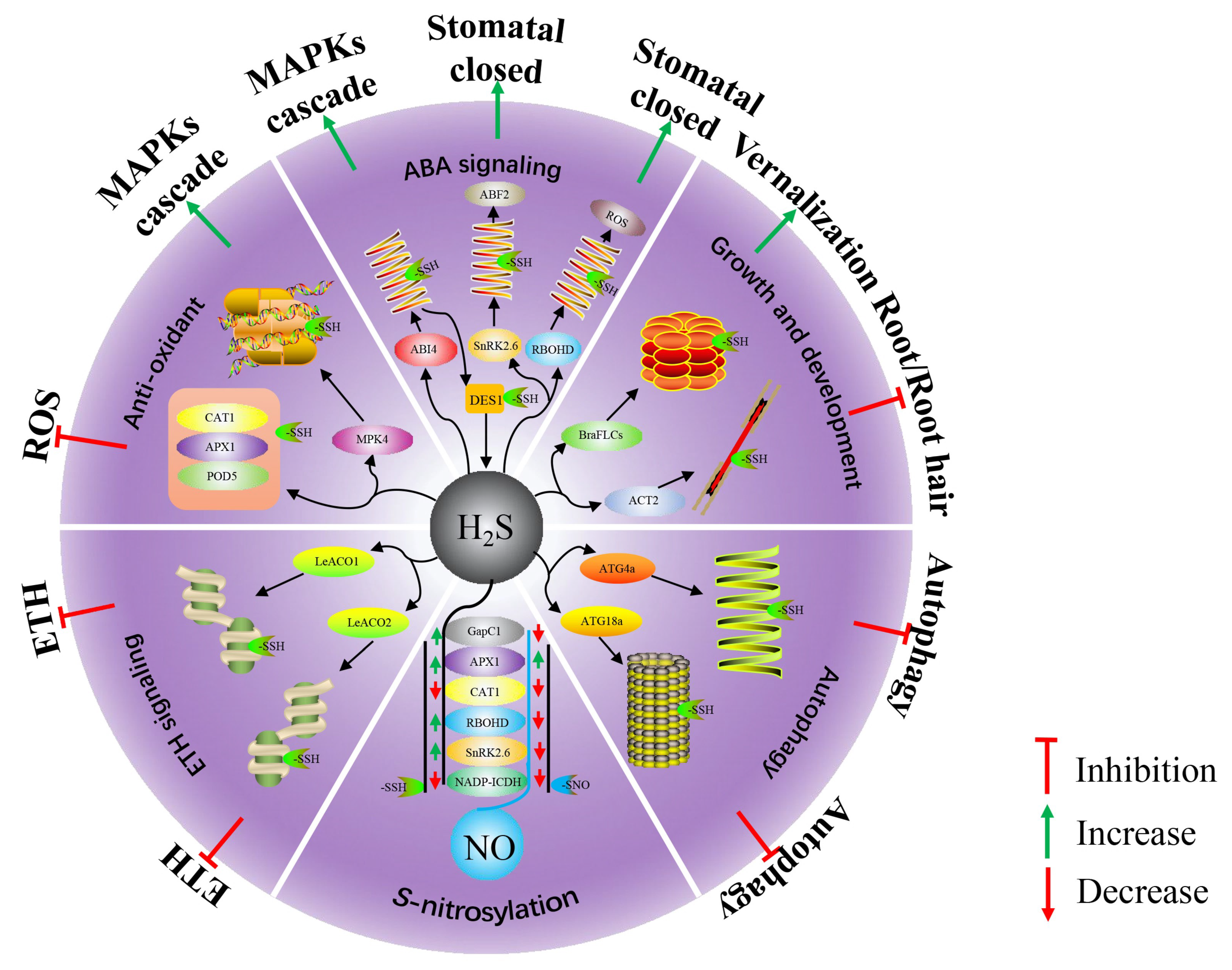
3. Roles of Protein Persulfidation in Plant Growth and Development
4. Roles of Protein Persulfidation in Plant Abiotic Stress
4.1. Antioxidant Protection Mechanism
4.2. Protein Persulfidation in Phytohormone Signal
4.3. Protein Persulfidation in Plant Autophagy
5. Protein Persulfidation and S-Nitrosylation
6. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010, 38, D142–D148. [Google Scholar] [CrossRef] [Green Version]
- Seet, B.T.; Dikic, I.; Zhou, M.M.; Pawson, T. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 2006, 7, 473–483. [Google Scholar] [CrossRef]
- Minguez, P.; Parca, L.; Diella, F.; Mende, D.R.; Kumar, R.; Helmer-Citterich, M.; Gavin, A.C.; van Noort, V.; Bork, P. Deciphering a global network of functionally associated post-translational modifications. Mol. Syst. Biol. 2012, 8, 599. [Google Scholar] [CrossRef]
- Aroca, A.; Serna, A.; Gotor, C.; Romero, L.C. S-sulfhydration: A cysteine posttranslational modification in plant systems. Plant Physiol. 2015, 168, 334–342. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Chen, S.; Wang, X.; Shi, C.; Liu, H.; Yang, J.; Shi, W.; Guo, J.; Jia, H. Hydrogen sulfide disturbs actin polymerization via S-sulfhydration resulting in stunted root hair growth. Plant Physiol. 2018, 178, 936–949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, X.; Zhang, L.; Pei, Z.; Zhang, L.; Liu, Z.; Liu, D.; Hao, X.; Jin, Z.; Pei, Y. Hydrogen sulfide promotes flowering in heading Chinese cabbage by S-sulfhydration of BraFLCs. Hortic. Res. 2021, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shi, C.; Wang, X.; Liu, C.; Ding, X.; Ma, P.; Wang, X.; Jia, H. Hydrogen sulfide regulates the activity of antioxidant enzymes through persulfidation and improves the resistance of tomato seedling to Copper Oxide nanoparticles (CuO NPs)-induced oxidative stress. Plant Physiol. Biochem. 2020, 156, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Jin, Z.; Liu, Z.; Liu, D.; Zhang, L.; Ma, X.; Yang, G.; Liu, S.; Guo, Y.; Pei, Y. H2S persulfidated and increased kinase activity of MPK4 to response cold stress in Arabidopsis. Front. Mol. Biosci. 2021, 8, 635470. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, J.; Shen, J.; Zhou, H.; Zhao, D.; Gotor, C.; Romero, L.C.; Fu, L.; Li, Z.; Yang, J.; et al. Hydrogen sulfide-linked persulfidation of ABI4 controls ABA responses through the transactivation of MAPKKK18 in Arabidopsis. Mol. Plant 2021, 14, 921–936. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Chen, S.; Liu, D.; Liesche, J.; Shi, C.; Wang, J.; Ren, M.; Wang, X.; Yang, J.; Shi, W.; et al. Ethylene-induced hydrogen sulfide negatively regulates ethylene biosynthesis by persulfidation of ACO in tomato under osmotic stress. Front. Plant Sci. 2018, 9, 1517. [Google Scholar] [CrossRef] [Green Version]
- Parker, E.T.; Cleaves, H.J.; Dworkin, J.P.; Glavin, D.P.; Callahan, M.; Aubrey, A.; Lazcano, A.; Bada, J.L. Primordial synthesis of amines and amino acids in a 1958 Miller H2S-rich spark discharge experiment. Proc. Natl. Acad. Sci. USA 2011, 108, 5526–5531. [Google Scholar] [CrossRef] [Green Version]
- Yamasaki, H.; Cohen, M.F. Biological consilience of hydrogen sulfide and nitric oxide in plants: Gases of primordial earth linking plant, microbial and animal physiologies. Nitric Oxide 2016, 55–56, 91–100. [Google Scholar] [CrossRef]
- Lisjak, M.; Teklic, T.; Wilson, I.D.; Whiteman, M.; Hancock, J.T. Hydrogen sulfide: Environmental factor or signalling molecule? Plant Cell Environ. 2013, 36, 1607–1616. [Google Scholar] [CrossRef]
- Nicholls, P.; Kim, J.K. Sulphide as an inhibitor and electron donor for the cytochrome c oxidase system. Can. J. Biochem. 1982, 60, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.A.; Schuler, M.M.; Prior, M.G.; Yong, S.; Coppock, R.W.; Florence, L.Z.; Lillie, L.E. Effects of hydrogen sulfide exposure on lung mitochondrial respiratory chain enzymes in rats. Toxicol. Appl. Pharmacol. 1990, 103, 482–490. [Google Scholar] [CrossRef]
- Dooley, F.D.; Nair, S.P.; Ward, P.D. Increased growth and germination success in plants following hydrogen sulfide administration. PLoS ONE 2013, 8, e62048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaichko, N.V.; Melnik, A.V.; Yoltukhivskyy, M.M.; Olhovskiy, A.S.; Palamarchuk, I.V. Hydrogen sulfide: Metabolism, biological and medical role. Biochem. J. 2014, 86, 5–25. [Google Scholar] [CrossRef]
- Mikami, Y.; Shibuya, N.; Kimura, Y.; Nagahara, N.; Ogasawara, Y.; Kimura, H. Thioredoxin and dihydrolipoic acid are required for 3-mercaptopyruvate sulfurtransferase to produce hydrogen sulfide. Biochem. J. 2011, 439, 479–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corpas, F.J.; Palma, J.M. H2S signaling in plants and applications in agriculture. J. Adv. Res. 2020, 24, 131–137. [Google Scholar] [CrossRef]
- Corpas, F.J.; Barroso, J.B.; Gonzalez-Gordo, S.; Munoz-Vargas, M.A.; Palma, J.M. Hydrogen sulfide: A novel component in Arabidopsis peroxisomes which triggers catalase inhibition. J. Integr. Plant Biol. 2019, 61, 871–883. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.G. Analysis of some enzyme activities of hydrogen sulfide metabolism in plants. Methods Enzymol. 2015, 555, 253–269. [Google Scholar] [PubMed]
- Filipovic, M.R.; Jovanovi, V.M. More than just an intermediate: Hydrogen sulfide signalling in plants. J. Exp. Bot. 2017, 68, 4733–4736. [Google Scholar] [CrossRef] [PubMed]
- Romero, L.C.; Aroca, M.A.; Laureano-Marín, A.M.; Moreno, I.; García, I.; Gotor, C. Cysteine and cysteine-related signaling pathways in Arabidopsis thaliana. Mol. Plant. 2014, 7, 264–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bilska-Wilkosz, A.; Górny, M.; Iciek, M. Inactivation of aldehyde dehydrogenase by disulfiram in the presence and absence of lipoic acid or dihydrolipoic acid: An in vitro study. Biomolecules 2019, 9, 375. [Google Scholar] [CrossRef] [Green Version]
- Youssefian, S.; Nakamura, M.; Orudgev, E.; Kondo, N. Increased cysteine biosynthesis capacity of transgenic tobacco overexpressing an O-acetylserine(thiol) lyase modifies plant responses to oxidative stress. Plant Physiol. 2001, 126, 1001–1011. [Google Scholar] [CrossRef] [Green Version]
- Baudouin, E.; Poilevey, A.; Hewage, N.I.; Cochet, F.; Puyaubert, J.; Bailly, C. The significance of hydrogen sulfide for Arabidopsis seed germination. Front. Plant Sci. 2016, 7, 930. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Ghoto, K.; Wei, M.Y.; Gao, C.H.; Liu, Y.L.; Ma, D.N.; Zheng, H.L. Unraveling hydrogen sulfide-promoted lateral root development and growth in mangrove plant Kandelia obovata: Insight into regulatory mechanism by TMT-based quantitative proteomic approaches. Tree Physiol. 2021, 41, 1749–1766. [Google Scholar] [CrossRef]
- Liu, Y.L.; Shen, Z.J.; Simon, M.; Li, H.; Ma, D.N.; Zhu, X.Y.; Zheng, H.L. Comparative proteomic analysis reveals the regulatory effects of h2s on salt tolerance of mangrove plant kandelia obovata. Int. J. Mol. Sci. 2019, 21, 118. [Google Scholar] [CrossRef] [Green Version]
- Deng, G.; Zhou, L.; Wang, Y.; Zhang, G.; Chen, X. Hydrogen sulfide acts downstream of jasmonic acid to inhibit stomatal development in Arabidopsis. Planta 2020, 251, 42. [Google Scholar] [CrossRef]
- Jin, Z.; Sun, L.; Yang, G.; Pei, Y. Hydrogen sulfide regulates energy production to delay leaf senescence induced by drought stress in Arabidopsis. Front. Plant Sci. 2018, 9, 1722. [Google Scholar] [CrossRef] [Green Version]
- Huo, J.; Huang, D.; Zhang, J.; Fang, H.; Wang, B.; Wang, C.; Liao, W. Hydrogen sulfide: A gaseous molecule in postharvest freshness. Front. Plant Sci. 2018, 9, 1172. [Google Scholar] [CrossRef]
- Yao, G.F.; Li, C.; Sun, K.K.; Tang, J.; Huang, Z.Q.; Yang, F.; Huang, G.G.; Hu, L.Y.; Jin, P.; Hu, K.D.; et al. Hydrogen sulfide maintained the good appearance and nutrition in post-harvest tomato fruits by antagonizing the effect of ethylene. Front. Plant Sci. 2020, 11, 584. [Google Scholar] [CrossRef]
- Wei, L.; Wang, C.; Liao, W. Hydrogen sulfide improves the vase life and quality of cut roses and chrysanthemums. J. Plant Growth Regul. 2021, 1–19. [Google Scholar]
- Zou, H.; Zhang, N.-N.; Lin, X.-Y.; Zhang, W.-Q.; Zhang, J.-H.; Chen, J.; Wei, G.-H. Hydrogen sulfide is a crucial element of the antioxidant defense system in Glycine max–Sinorhizobium fredii symbiotic root nodules. Plant Soil 2020, 449, 209–231. [Google Scholar] [CrossRef]
- Gotor, C.; Garcia, I.; Crespo, J.L.; Romero, L.C. Sulfide as a signaling molecule in autophagy. Autophagy 2013, 9, 609–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arif, Y.; Hayat, S.; Yusuf, M.; Bajguz, A. Hydrogen sulfide: A versatile gaseous molecule in plants. Plant Physiol. Biochem. 2021, 158, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Hancock, J.T.; Lisjak, M.; Teklic, T.; Wilson, I.D.; Whiteman, M. Hydrogen sulfide and signaling in plants. CAB Rev. 2011, 6, 1–7. [Google Scholar] [CrossRef]
- Filipovic, M.R.; Zivanovic, J.; Alvarez, B.; Banerjee, R. Chemical biology of h2s signaling through persulfidation. Chem. Rev. 2018, 118, 1253–1337. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, A.K.; Gadalla, M.M.; Sen, N.; Kim, S.; Mu, W.; Gazi, S.K.; Barrow, R.K.; Yang, G.; Wang, R.; Snyder, S.H. H2S signals through protein S-sulfhydration. Sci. Signal. 2009, 2, ra72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuevasanta, E.; Lange, M.; Bonanata, J.; Coitino, E.L.; Ferrer-Sueta, G.; Filipovic, M.R.; Alvarez, B. Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide. J. Biol. Chem. 2015, 290, 26866–26880. [Google Scholar] [CrossRef] [Green Version]
- Aroca, A.; Schneider, M.; Scheibe, R.; Gotor, C.; Romero, L.C. Hydrogen Sulfide Regulates the Cytosolic/Nuclear Partitioning of Glyceraldehyde-3-Phosphate Dehydrogenase by Enhancing its Nuclear Localization. Plant Cell Physiol. 2017, 58, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Greiner, R.; Palinkas, Z.; Basell, K.; Becher, D.; Antelmann, H.; Nagy, P.; Dick, T.P. Polysulfides link H2S to protein thiol oxidation. Antioxid. Redox Signal. 2013, 19, 1749–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iciek, M.; Bilska-Wilkosz, A.; Górny, M. Sulfane sulfur–new findings on an old topic. Acta Biochim. Pol. 2019, 66, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, N.; Fu, C.; Pappin, D.J.; Tonks, N.K. H2S-induced sulfhydration of the phosphatase PTP1B and its role in the endoplasmic reticulum stress response. Sci. Signal. 2011, 4, ra86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francoleon, N.E.; Carrington, S.J.; Fukuto, J.M. The reaction of H2S with oxidized thiols: Generation of persulfides and implications to H2S biology. Arch. Biochem. Biophys. 2011, 516, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Predmore, B.L.; Lefer, D.J.; Gojon, G. Hydrogen sulfide in biochemistry and medicine. Antioxid. Redox Signal. 2012, 17, 119–140. [Google Scholar] [CrossRef] [Green Version]
- Reyes, A.; Cuevasanta, E.; De Armas, M.I.; Mastrogiovanni, M.; Zeida, A.; Radi, R.; Alvarez, B.; Trujillo, M. Interactions of hydrogen sulfide with AhpE from Mycobacterium tuberculosis: Formation and reactions of a model peroxiredoxin persulfide. Free. Radic. Biol. Med. 2018, 128, S57. [Google Scholar] [CrossRef]
- Zhang, D.; Macinkovic, I.; Devarie-Baez, N.O.; Pan, J.; Park, C.M.; Carroll, K.S.; Filipovic, M.R.; Xian, M. Detection of protein S-sulfhydration by a tag-switch technique. Angew. Chem. Int. Ed. 2014, 53, 575–581. [Google Scholar] [CrossRef] [Green Version]
- Filipovic, M.R.; Miljkovic, J.; Nauser, T.; Royzen, M.; Klos, K.; Shubina, T.; Koppenol, W.H.; Lippard, S.J.; Ivanovic-Burmazovic, I. Chemical characterization of the smallest S-nitrosothiol, HSNO; cellular cross-talk of H2S and S-nitrosothiols. J. Am. Chem. Soc. 2012, 134, 12016–12027. [Google Scholar] [CrossRef]
- Nava, M.; Martin-Drumel, M.A.; Lopez, C.A.; Crabtree, K.N.; Womack, C.C.; Nguyen, T.L.; Thorwirth, S.; Cummins, C.C.; Stanton, J.F.; McCarthy, M.C. Spontaneous and selective formation of HSNO, a crucial intermediate linking H2S and nitroso chemistries. J. Am. Chem. Soc. 2016, 138, 11441–11444. [Google Scholar] [CrossRef]
- Aroca, A.; Benito, J.M.; Gotor, C.; Romero, L.C. Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J. Exp. Bot. 2017, 68, 4915–4927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, Y.; Chen, H.; Shen, W.; Shen, W.; Huang, L. Hydrogen peroxide is involved in hydrogen sulfide-induced lateral root formation in tomato seedlings. BMC Plant Biol. 2017, 17, 162. [Google Scholar] [CrossRef]
- Noctor, G.; Reichheld, J.P.; Foyer, C.H. ROS-related redox regulation and signaling in plants. Semin Cell Dev. Biol. 2018, 80, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Yang, G. Signal function studies of ROS, especially RBOH-dependent ROS, in plant growth, development and environmental stress. J. Plant Growth Regul. 2019, 39, 157–171. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Bedhomme, M.; Marchand, C.H.; Morisse, S.; Trost, P.; Lemaire, S.D. Redox regulation in photosynthetic organisms: Focus on glutathionylation. Antioxid. Redox Signal. 2012, 16, 567–586. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.Q.; Zhang, J.H.; Sun, L.M.; Zhu, L.F.; Abliz, B.; Hu, W.J.; Zhong, C.; Bai, Z.G.; Sajid, H.; Cao, X.C.; et al. Hydrogen sulfide alleviates aluminum toxicity via decreasing apoplast and symplast Al contents in rice. Front. Plant Sci. 2018, 9, 294. [Google Scholar] [CrossRef] [Green Version]
- Qi, Q.; Guo, Z.; Liang, Y.; Li, K.; Xu, H. Hydrogen sulfide alleviates oxidative damage under excess nitrate stress through MAPK/NO signaling in cucumber. Plant Physiol. Biochem. 2019, 135, 1–8. [Google Scholar] [CrossRef]
- Pan, D.Y.; Fu, X.; Zhang, X.W.; Liu, F.J.; Bi, H.G.; Ai, X.Z. Hydrogen sulfide is required for salicylic acid-induced chilling tolerance of cucumber seedlings. Protoplasma 2020, 257, 1543–1557. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, J.; Zhou, M.; Zhou, H.; Cui, B.; Gotor, C.; Romero, L.C.; Fu, L.; Yang, J.; Foyer, C.H.; et al. Persulfidation-based modification of cysteine desulfhydrase and the NADPH oxidase RBOHD controls guard cell abscisic acid signaling. Plant Cell 2020, 32, 1000–1017. [Google Scholar] [CrossRef]
- Laureano-Marin, A.M.; Moreno, I.; Romero, L.C.; Gotor, C. Negative regulation of autophagy by sulfide is independent of reactive oxygen species. Plant Physiol. 2016, 171, 1378–1391. [Google Scholar]
- Aroca, A.; Yruela, I.; Gotor, C.; Bassham, D.C. Persulfidation of ATG18a regulates autophagy under ER stress in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2023604118. [Google Scholar] [CrossRef]
- Šamajová, O.; Plihal, O.; Al-Yousif, M.; Hirt, H.; Samaj, J. Improvement of stress tolerance in plants by genetic manipulation of mitogen-activated protein kinases. Biotechnol. Adv. 2013, 31, 118–128. [Google Scholar] [CrossRef]
- Teige, M.; Scheikl, E.; Eulgem, T.; Doczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell. 2004, 15, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Jin, Z.; Liu, D.; Yang, G.; Pei, Y. Hydrogen sulfide alleviates the cold stress through MPK4 in Arabidopsis thaliana. Plant Physiol. Biochem. 2017, 120, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.I.; Kwak, J.M.; Allen, G.J. Guard cell abscisic acid signalling and engineering drought hardiness in plants. Nature 2001, 410, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Jacob, T.; Ritchie, S.; Assmann, S.M.; Gilroy, S. Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. USA 1999, 96, 12192–12197. [Google Scholar] [CrossRef] [Green Version]
- Franks, P.J.; Farquhar, G.D. The effect of exogenous abscisic acid on stomatal development, stomatal mechanics, and leaf gas exchange in Tradescantia virginiana. Plant Physiol. 2001, 125, 935–942. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Jia, H.; Wang, X.; Shi, C.; Wang, X.; Ma, P.; Wang, J.; Ren, M.; Li, J. Hydrogen sulfide positively regulates abscisic acid signaling through persulfidation of SnRK2.6 in guard cells. Mol. Plant 2020, 13, 732–744. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, H.; Zhou, M.; Ge, Z.; Zhang, F.; Foyer, C.H.; Yuan, X.; Xie, Y. The coordination of guard-cell autonomous ABA synthesis and DES1 function in situ regulates plant water deficit responses. J. Adv. Res. 2021, 27, 191–197. [Google Scholar] [CrossRef]
- Kim, T.H.; Bohmer, M.; Hu, H.; Nishimura, N.; Schroeder, J.I. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Ann. Rev. Plant Biol. 2010, 61, 561–591. [Google Scholar] [CrossRef] [Green Version]
- Mustilli, A.C.; Merlot, S.; Vavasseur, A.; Fenzi, F.; Giraudat, J. Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 2002, 14, 3089–3099. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, U.; Luo, X.; Zhou, W.; Shu, K. Multifaceted signaling networks mediated by abscisic acid insensitive 4. Plant Commun. 2020, 1, 100040. [Google Scholar] [CrossRef]
- Wind, J.J.; Peviani, A.; Snel, B.; Hanson, J.; Smeekens, S.C. ABI4: Versatile activator and repressor. Trends Plant Sci. 2013, 18, 125–132. [Google Scholar] [CrossRef]
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Signals from chloroplasts converge to regulate nuclear gene expression. Science 2007, 316, 715–719. [Google Scholar] [CrossRef]
- Bai, Z.; Zhang, J.; Ning, X.; Guo, H.; Xu, X.; Huang, X.; Wang, Y.; Hu, Z.; Lu, C.; Zhang, L.; et al. A kinase-phosphatase-transcription factor module regulates adventitious root emergence in Arabidopsis root-hypocotyl junctions. Mol. Plant 2020, 13, 1162–1177. [Google Scholar] [CrossRef]
- Mitula, F.; Tajdel, M.; Ciesla, A.; Kasprowicz-Maluski, A.; Kulik, A.; Babula-Skowronska, D.; Michalak, M.; Dobrowolska, G.; Sadowski, J.; Ludwikow, A. Arabidopsis aba-activated kinase mapkkk18 is regulated by protein phosphatase 2c ABI1 and the ubiquitin-proteasome pathway. Plant Cell Physiol. 2015, 56, 2351–2367. [Google Scholar] [CrossRef]
- Blume, B.; Grierson, D. Expression of ACC oxidase promoter-GUS fusions in tomato and Nicotiana plumbaginifolia regulated by developmental and environmental stimuli. Plant J. 1997, 12, 731–746. [Google Scholar] [CrossRef] [PubMed]
- Spanu, P.; Reinhardt, D.; Boller, T. Analysis and cloning of the ethylene-forming enzyme from tomato by functional expression of its mRNA in Xenopus laevis oocytes. EMBO J. 1991, 10, 2007–2013. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wu, X.; Sun, M.; Peng, F. Hydrogen sulfide alleviates waterlogging-induced damage in peach seedlings via enhancing antioxidative system and inhibiting ethylene synthesis. Front. Plant Sci. 2020, 11, 696. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Ann. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Xia, F.N.; Xiao, S. Autophagy in plants: Physiological roles and post-translational regulation. J. Integr. Plant Biol. 2020, 63, 161–179. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Bassham, D.C. New insight into the mechanism and function of autophagy in plant cells. Int. Rev. Cell Mol. Biol. 2015, 320, 1–40. [Google Scholar] [PubMed]
- Ahlberg, J.; Glaumann, H. Uptake—Microautophagy—And degradation of exogenous proteins by isolated rat liver lysosomes. Effects of pH, ATP, and inhibitors of proteolysis. Exp. Mol. Pathol. 1985, 42, 78–88. [Google Scholar] [CrossRef]
- Li, F.; Vierstra, R.D. Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012, 17, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Kuroyanagi, M.; Yamada, K.; Meshi, T.; Tsuda, S.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 2004, 305, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Chung, T.; Vierstra, R.D. Autophagy-Related11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell 2014, 26, 788–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwarten, M.; Stoldt, M.; Mohrlüder, J.; Willbold, D. Sequence-specific 1H, 13C, and 15N resonance assignment of the autophagy-related protein Atg8. Biomol. NMR Assign. 2009, 3, 137–139. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, C.; Garcia, I.; Moreno, I.; Perez-Perez, M.E.; Crespo, J.L.; Romero, L.C.; Gotor, C. Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell 2012, 24, 4621–4634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurado-Flores, A.; Romero, L.C.; Gotor, C. Label-free quantitative proteomic analysis of nitrogen starvation in arabidopsis root reveals new aspects of H2S signaling by protein persulfidation. Antioxidants 2021, 10, 508. [Google Scholar] [CrossRef]
- Laureano-Marin, A.M.; Aroca, A.; Perez-Perez, M.E.; Yruela, I.; Jurado-Flores, A.; Moreno, I.; Crespo, J.L.; Romero, L.C.; Gotor, C. Abscisic acid-triggered persulfidation of the Cys protease ATG4 mediates regulation of autophagy by sulfide. Plant Cell 2020, 32, 3902–3920. [Google Scholar] [CrossRef]
- Liu, Y.; Burgos, J.S.; Deng, Y.; Srivastava, R.; Howell, S.H.; Bassham, D.C. Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 2012, 24, 4635–4651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, I.K.; Bajeli, S.; Sahu, S.; Bhat, S.A.; Kumar, A. Hydrogen sulfide-induced GAPDH sulfhydration disrupts the CCAR2-SIRT1 interaction to initiate autophagy. Autophagy 2021, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.K.; Yun, B.W.; Kang, J.G.; Raja, M.U.; Kwon, E.; Sorhagen, K.; Chu, C.; Wang, Y.; Loake, G.J. Nitric oxide function and signalling in plant disease resistance. J. Exp. Bot. 2008, 59, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Fancy, N.N.; Bahlmann, A.K.; Loake, G.J. Nitric oxide function in plant abiotic stress. Plant Cell Environ. 2017, 40, 462–472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liao, W. Protein S-nitrosylation in plant abiotic stresses. Funct. Plant Biol. 2019, 47, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hess, D.T.; Stamler, J.S. Regulation by S-nitrosylation of protein post-translational modification. J. Biol. Chem. 2012, 287, 4411–4418. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Ye, T.; Chan, Z. Nitric oxide-activated hydrogen sulfide is essential for cadmium stress response in bermudagrass (Cynodon dactylon (L). Pers.). Plant Physiol. Biochem. 2014, 74, 99–107. [Google Scholar] [CrossRef]
- Da-Silva, C.J.; Mollica, D.C.F.; Vicente, M.H.; Peres, L.E.P.; Modolo, L.V. NO, hydrogen sulfide does not come first during tomato response to high salinity. Nitric Oxide 2018, 76, 164–173. [Google Scholar] [CrossRef]
- Iqbal, N.; Umar, S.; Khan, N.A.; Corpas, F.J. Nitric oxide and hydrogen sulfide coordinately reduce glucose sensitivity and decrease oxidative stress via ascorbate-glutathione cycle in heat-stressed wheat (Triticum aestivum L.) Plants. Antioxidants 2021, 10, 108. [Google Scholar] [CrossRef]
- Hu, J.; Huang, X.; Chen, L.; Sun, X.; Lu, C.; Zhang, L.; Wang, Y.; Zuo, J. Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2015, 167, 1731–1746. [Google Scholar] [CrossRef] [Green Version]
- Munoz-Vargas, M.A.; Gonzalez-Gordo, S.; Canas, A.; Lopez-Jaramillo, J.; Palma, J.M.; Corpas, F.J. Endogenous hydrogen sulfide (H2S) is up-regulated during sweet pepper (Capsicum annuum L.) fruit ripening. In vitro analysis shows that NADP-dependent isocitrate dehydrogenase (ICDH) activity is inhibited by H2S and NO. Nitric Oxide 2018, 81, 36–45. [Google Scholar] [CrossRef]
- Cheung, S.H.; Lau, J.Y.W. Hydrogen sulfide mediates athero-protection against oxidative stress via S-sulfhydration. PLoS ONE 2018, 13, e0194176. [Google Scholar] [CrossRef] [Green Version]
- Yun, B.W.; Feechan, A.; Yin, M.; Saidi, N.B.; Le Bihan, T.; Yu, M.; Moore, J.W.; Kang, J.G.; Kwon, E.; Spoel, S.H.; et al. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 2011, 478, 264–268. [Google Scholar] [CrossRef]
- Terron-Camero, L.C.; Rodriguez-Serrano, M.; Sandalio, L.M.; Romero-Puertas, M.C. Nitric oxide is essential for cadmium-induced peroxule formation and peroxisome proliferation. Plant Cell Environ. 2020, 43, 2492–2507. [Google Scholar] [CrossRef]
- Begara-Morales, J.C.; Sanchez-Calvo, B.; Chaki, M.; Valderrama, R.; Mata-Perez, C.; Lopez-Jaramillo, J.; Padilla, M.N.; Carreras, A.; Corpas, F.J.; Barroso, J.B. Dual regulation of cytosolic ascorbate peroxidase (APX) by tyrosine nitration and S-nitrosylation. J. Exp. Bot. 2014, 65, 527–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Du, Y.; Hou, Y.J.; Zhao, Y.; Hsu, C.C.; Yuan, F.; Zhu, X.; Tao, W.A.; Song, C.P.; Zhu, J.K. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. USA 2015, 112, 613–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wawer, I.; Bucholc, M.; Astier, J.; Anielska-Mazur, A.; Dahan, J.; Kulik, A.; Wyslouch-Cieszynska, A.; Zareba-Koziol, M.; Krzywinska, E.; Dadlez, M.; et al. Regulation of Nicotiana tabacum osmotic stress-activated protein kinase and its cellular partner GAPDH by nitric oxide in response to salinity. Biochem. J. 2010, 429, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Serrato, A.J.; Romero-Puertas, M.C.; Lazaro-Payo, A.; Sahrawy, M. Regulation by S-nitrosylation of the Calvin-Benson cycle fructose-1,6-bisphosphatase in Pisum sativum. Redox Biol. 2018, 14, 409–416. [Google Scholar] [CrossRef] [PubMed]

| Metabolic Process | Plant Species | Abiotic Stress | Protein Modified | Sites | Protein Activity | Functions | Reference |
|---|---|---|---|---|---|---|---|
| Anti-oxidant system | Solanum lycopersicum | CuO NPs | CAT1 | Cys234 | ↓ | Anti-oxidation | [7] |
| APX1 | Cys168 | ↑ | |||||
| POD5 | Cys46, Cys61 | ↑ | |||||
| Arabidopsis thaliana | Cold | MPK4 | ↑ | Enhances the signal cascade of MAPKs, thereby alleviating stress | [8] | ||
| ABA signaling | A. thaliana | Drought | DES1 | Cys44, Cys205 | ↑ | Promotes the release of H2S from DES1 | [59] |
| A. thaliana | Drought | RBOHD | Cys825, Cys890 | ↑ | Improves the ability of RBOHD to generate ROS | [59] | |
| A. thaliana | Drought | SnRK2.6 | Cys131, Cys137 | ↑ | Strengthens the interplay between SnRK2.6 and ABF2, therefore promotes ABA signaling | [59] | |
| A. thaliana | Drought | ABI4 | Cys250 | ↑ | Enhances the transcription of MAPKKK18, therefore promote MAPK cascade | [9] | |
| ETH signaling | S. lycopersicum | Osmotic | LeACO1 | Cys60 | ↓ | Inhibits the ETH synthesis | [10] |
| S. lycopersicum | Osmotic | LeACO2 | ↓ | Inhibits the ETH synthesis | [10] | ||
| Autophagy pathway | A. thaliana | Nitrogen starvation | ATG4a | Cys170 | ↓ | Weakens the catalytic effect of ATG8 and PE conjunction, therefore inhibits autophagy | [60] |
| A. thaliana | ER-phagy | ATG18a | Cys103 | ↑ | Enhanced ATG18a affinity and co-localization time with phagophore membranes, thus delaying the release and maturation of autophagosomes | [61] |
| Plant Species | Protein Modified | Persulfidation/ S-nitrosation Sites | Persulfidated/ S-nitrosated Activity | Reference |
|---|---|---|---|---|
| Capsicum annuum L. | NADP-ICDH | --/ Cys75 | ↓/↓ | [101] |
| Arabidopsis thaliana | SnRK2.6 | Cys131, Cys137/ Cys137 | ↑/↓ | [68,100] |
| A. thaliana | RBOHD | Cys825, Cys890/ Cys890 | ↑/↓ | [59,103] |
| Solanum lycopersicum * A. thaliana # | CAT1 | Cys234/-- | ↓/↓ | [7,104] |
| S. lycopersicum * Pisum sativum L. # | APX1 | Cys168/ Cys32 | ↑/↑ | [7,105] |
| A. thaliana | GapC1 | Cys156/ Cys160 | ↑/↓ | [41] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Fang, H.; Gao, R.; Liao, W. Protein Persulfidation in Plants: Function and Mechanism. Antioxidants 2021, 10, 1631. https://doi.org/10.3390/antiox10101631
Wang P, Fang H, Gao R, Liao W. Protein Persulfidation in Plants: Function and Mechanism. Antioxidants. 2021; 10(10):1631. https://doi.org/10.3390/antiox10101631
Chicago/Turabian StyleWang, Peng, Hua Fang, Rong Gao, and Weibiao Liao. 2021. "Protein Persulfidation in Plants: Function and Mechanism" Antioxidants 10, no. 10: 1631. https://doi.org/10.3390/antiox10101631
APA StyleWang, P., Fang, H., Gao, R., & Liao, W. (2021). Protein Persulfidation in Plants: Function and Mechanism. Antioxidants, 10(10), 1631. https://doi.org/10.3390/antiox10101631







